Abstract
The primary circulating form of vitamin D, 25-hydroxy-vitamin D [25(OH)D], is associated with multiple medical outcomes, including rickets, osteoporosis, multiple sclerosis and cancer. In a genome-wide association study (GWAS) of 4501 persons of European ancestry drawn from five cohorts, we identified single-nucleotide polymorphisms (SNPs) in the gene encoding group-specific component (vitamin D binding) protein, GC, on chromosome 4q12-13 that were associated with 25(OH)D concentrations: rs2282679 (P = 2.0 × 10−30), in linkage disequilibrium (LD) with rs7041, a non-synonymous SNP (D432E; P = 4.1 × 10−22) and rs1155563 (P = 3.8 × 10−25). Suggestive signals for association with 25(OH)D were also observed for SNPs in or near three other genes involved in vitamin D synthesis or activation: rs3829251 on chromosome 11q13.4 in NADSYN1 [encoding nicotinamide adenine dinucleotide (NAD) synthetase; P = 8.8 × 10−7], which was in high LD with rs1790349, located in DHCR7, the gene encoding 7-dehydrocholesterol reductase that synthesizes cholesterol from 7-dehydrocholesterol; rs6599638 in the region harboring the open-reading frame 88 (C10orf88) on chromosome 10q26.13 in the vicinity of ACADSB (acyl-Coenzyme A dehydrogenase), involved in cholesterol and vitamin D synthesis (P = 3.3 × 10−7); and rs2060793 on chromosome 11p15.2 in CYP2R1 (cytochrome P450, family 2, subfamily R, polypeptide 1, encoding a key C-25 hydroxylase that converts vitamin D3 to an active vitamin D receptor ligand; P = 1.4 × 10−5). We genotyped SNPs in these four regions in 2221 additional samples and confirmed strong genome-wide significant associations with 25(OH)D through meta-analysis with the GWAS data for GC (P = 1.8 × 10−49), NADSYN1/DHCR7 (P = 3.4 × 10−9) and CYP2R1 (P = 2.9 × 10−17), but not C10orf88 (P = 2.4 × 10−5).
INTRODUCTION
The primary form of circulating vitamin D, 25-hydroxy-vitamin D [25(OH)D], is a modifiable quantitative trait associated with multiple medical outcomes, including osteoporosis, multiple sclerosis, selected malignancies, and especially colorectal cancer, with rickets being the most common expression of severe clinical vitamin D deficiency in children (1). The concentration of 25(OH)D in blood, which reflects endogenous generation through ultraviolet B (UVB) exposure as well as exogenous dietary and supplemental vitamin D intake, is considered the best indicator of vitamin D status. Following metabolic activation to 1,25-dihydroxy-vitamin D [1,25(OH)2D] through multiple hydroxylation steps (2), vitamin D has pleiotropic effects in addition to its traditional role in calcium homeostasis; for example, vitamin D receptor response elements directly or indirectly influence cell cycling and proliferation, differentiation and apoptosis (3).
Common genetic variants that influence circulating 25(OH)D levels could be important for identifying persons at risk for vitamin D deficiency and enhancing our understanding of the observed associations between vitamin D status and several diseases. Previously, twin studies suggested a heritable component to circulating vitamin D levels (4–6), and investigations of common genetic variation in candidate genes in relation to 25(OH)D concentrations have been based on small study samples and were inconclusive. Here, we report an analysis of 4501 individuals in a meta-analysis of five genome-wide association studies (GWASs) within five cohorts (Table 1) with prospectively collected 25(OH)D levels: a case–control study of lung cancer in the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC) (7); a case–control study of prostate cancer [Cancer Genetic Markers of Susceptibility (CGEMS)] in the Prostate, Lung, Colorectal, Ovarian Cancer Screening Trial (PLCO) (8); three case–control studies of pancreatic cancer nested within ATBC, Cancer Prevention Study-II (CPS-II) (9) and Give Us a Clue to Cancer and Heart Disease Study (CLUE II) (10); and case–control studies of breast cancer (CGEMS) and type 2 diabetes (T2D) nested within the Nurses' Health Study (NHS) (11). Markers near genes known to be involved in vitamin D synthesis or activation that showed strong evidence for association with 25(OH)D levels in the GWAS were genotyped in an additional 2221 individuals with serologic vitamin D levels from the case–control studies of colon polyps and colorectal cancer nested in the NHS and a prostate cancer case–control study nested within the Health Professionals Follow-up Study (HPFS) (12–14).
Table 1.
Descriptive characteristics of the participating GWAS cohorts
| Cohort | n (cases/controls) | Location | % Female | Years of blood draw | Type of specimen | Median (inter-quartile range) of 25(OH)D (nmol/l) | Vitamin D assay (CV) | % of 25(OH)D deficiency (<25 nmol/l) | % of vitamin D supplement use |
|---|---|---|---|---|---|---|---|---|---|
| Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC) (7) | 1372 (865/507) | Finland | 0 | 1985–1988 | Serum | 24.2 (37.3–52.9) | CLIA (CV = 9.5–12.7%) | 26 | 7 |
| Cancer Prevention Study (CPS-II) (8) | 121 (0/121) | USA | 42 | 1998–2001 | Serum | 45.2 (57.7–70.0) | CLIA (CV = 3.8–14.7%) | 6 | 61 |
| Give Us a Clue to Cancer and Heart Disease Study (CLUE II) (9) | 95 (0/95) | Washington County, MD, USA | 54 | 1989 | Serum | 44.0 (59.2–76.1) | CLIA (CV = 6.0–16.4%) | 5 | 23 |
| Prostate, Lung, Colorectal, Ovarian Cancer Screening Trial (PLCO) (10) | 1319 (730/589) | Multi-center, USA | 3 | 1993–2001 | Serum | 45.0 (56.8–70.0) | CLIA (CV = 8.1%) | 3 | 39 |
| Nurses' Health Study (NHS) (11) | 1594 (801/793) | USA | 100 | 1989–1990 | Plasma | 66.6 (49.0–90.0) | RIA (CV = 8.7–17.6%) | 2 | 38 |
RESULTS
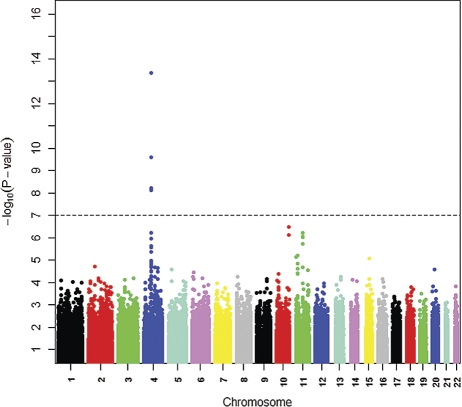
A Manhattan plot of genetic signal for 25(OH)D levels shows a genome-wide significance for single-nucleotide polymorphisms (SNPs) on chromosome 4 (Fig. 1). In the region harboring the group-specific component gene (GC, encoding vitamin D-binding protein, DBP) on chromosome 4q12-q13, a set of SNPs were associated with circulating 25(OH)D levels at the genome-wide significance level. The strongest association was for rs2282679 (Table 2).
Figure 1.
Genome-wide associations of circulating 25-hydroxy vitamin D graphed by chromosome position and −log10 P-value based on the five GWAS cohorts (n = 4501). Only P values of >10−16 are plotted, with the most significant variants in the GC gene on chromosome 4 not being shown (rs2282679, rs1155563 and rs7041; P-values of 1.96 × 10−30, 3.56 × 10−25 and 4.07 × 10−22, respectively).
Table 2.
Association of circulating 25(OH)D concentrations with GC and other vitamin D-related genes in the GWAS and replication study cohorts
| SNPa | Risk allele (freq) | Chr | Locationb | Gene neighborhood | Population | P-valuec | Mean 25(OH)D levels (nmol/l) by genotype (copies of risk allele) |
β-valued | SEd | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | |||||||||
| GWASs | |||||||||||
| rs2282679 | C (0.26) | 4 | 72827247 | GC | Total | 4.1 × 10−22 | −0.36 | 0.05 | |||
| ATBC | 1.5 × 10−8 | 42.1 | 37.2 | 35.2 | −0.38 | 0.07 | |||||
| CPS-II | 0.09 | 60.1 | 57.4 | 46.0 | −0.31 | 0.18 | |||||
| CLUE II | 2.2 × 10−3 | 67.5 | 58.4 | 44.3 | −0.69 | 0.22 | |||||
| PLCO | 5.8 × 10−13 | 61.5 | 55.3 | 53.4 | −0.37 | 0.05 | |||||
| NHS-CGEMS | 1.2 × 10−7 | 86.8 | 77.3 | 72.5 | −0.46 | 0.09 | |||||
| NHS-T2D | 9.5 × 10−4 | 58.8 | 55.8 | 48.8 | −0.24 | 0.07 | |||||
| Replication studies | Total | 5.4 × 10−21 | −0.41 | 0.04 | |||||||
| NHS-Polyps | 3.4 × 10−5 | 70.5 | 65.5 | 57.5 | −0.35 | 0.09 | |||||
| NHS-CRC | 2.2 × 10−6 | 71.5 | 67.3 | 56.3 | −0.42 | 0.09 | |||||
| HPFS | 3.1 × 10−13 | 65.5 | 56.8 | 53.5 | −0.44 | 0.06 | |||||
| Combined meta-analysis | 1.8 × 10−49 | −0.38 | 0.03 | ||||||||
| GWASs | |||||||||||
| rs3829251 | A (0.19) | 11 | 70872207 | NADSYN1 | Total | 8.8 × 10−7 | −0.17 | 0.04 | |||
| ATBC | 2.0 × 10−2 | 41.4 | 38.8 | 39.4 | −0.14 | 0.06 | |||||
| CPS-II | 0.75 | 61.1 | 64.9 | 61.0 | 0.07 | 0.21 | |||||
| CLUE II | 0.60 | 56.2 | 61.7 | 46.8 | 0.16 | 0.30 | |||||
| PLCO | 3.5 × 10−4 | 59.3 | 56.2 | 53.0 | −0.22 | 0.06 | |||||
| NHS-CGEMS | 9.2 × 10−3 | 83.3 | 77.8 | 81.3 | −0.28 | 0.11 | |||||
| NHS-T2D | 0.39 | 57.3 | 55.3 | 55.8 | −0.08 | 0.09 | |||||
| Replication studies | |||||||||||
| rs11234027e | A (0.16) | 70911755 | Total | 1.0 × 10−3 | −0.19 | 0.06 | |||||
| NHS-Polyps | 0.30 | 68.3 | 66.5 | 61.8 | −0.11 | 0.10 | |||||
| NHS-CRC | 0.16 | 69.0 | 67.3 | 51.8 | −0.17 | 0.12 | |||||
| HPFS | 1.7 × 10−3 | 62.0 | 57.3 | 57.5 | −0.25 | 0.08 | |||||
| Combined meta-analysis | 3.4 × 10−9 | −0.18 | 0.03 | ||||||||
| GWASs | |||||||||||
| rs6599638 | G (0.49) | 10 | 124694139 | C10orf88 | Total | 3.3 × 10−7 | −0.14 | 0.03 | |||
| ATBC | 5.0 × 10−4 | 41.8 | 39.6 | 39.5 | −0.19 | 0.06 | |||||
| CPS-II | 0.95 | 57.3 | 58.4 | 56.8 | −0.01 | 0.16 | |||||
| CLUE II | 0.11 | 65.6 | 61.8 | 58.0 | −0.33 | 0.20 | |||||
| PLCO | 1.3 × 10−2 | 60.3 | 57.8 | 57.6 | −0.11 | 0.05 | |||||
| NHS-CGEMS | 2.2 × 10−3 | 83.3 | 83.0 | 77.5 | −0.24 | 0.08 | |||||
| NHS-T2D | 0.12 | 58.5 | 56.0 | 55.5 | −0.10 | 0.07 | |||||
| Replication Studies | Total | 9.3 × 10−1 | 0.00 | 0.04 | |||||||
| NHS-Polyps | 0.70 | 68.3 | 67.3 | 66.5 | −0.03 | 0.07 | |||||
| NHS-CRC | 0.66 | 66.5 | 70.0 | 66.8 | −0.04 | 0.09 | |||||
| HPFS | 0.55 | 60.5 | 60.3 | 61.3 | 0.03 | 0.06 | |||||
| Combined meta-analysis | 2.4 × 10−5 | −0.09 | 0.04 | ||||||||
| GWASs | |||||||||||
| rs2060793 | A (0.41) | 11 | 14866810 | CYP2R1 | Total | 1.4 × 10−5 | 0.12 | 0.03 | |||
| ATBC | 0.28 | 39.5 | 40.7 | 40.1 | 0.06 | 0.06 | |||||
| CPS-II | 0.78 | 57.9 | 55.8 | 63.6 | 0.05 | 0.19 | |||||
| CLUE II | 0.92 | 62.1 | 57.7 | 71.1 | −0.03 | 0.25 | |||||
| PLCO | 0.15 | 57.1 | 59.1 | 58.6 | 0.07 | 0.05 | |||||
| NHS-CGEMS | 3.5 × 10−4 | 77.0 | 83.3 | 80.5 | 0.28 | 0.08 | |||||
| NHS-T2D | 8.9 × 10−4 | 53.8 | 58.0 | 59.3 | 0.22 | 0.07 | |||||
| Replication studies | |||||||||||
| rs1993116e | A (0.39) | 14866810 | Total | 1.6 × 10−17 | 0.34 | 0.04 | |||||
| NHS-Polyps | 6.2 × 10−9 | 62.5 | 68.8 | 75.0 | 0.39 | 0.07 | |||||
| NHS-CRC | 5.2 × 10−4 | 65.5 | 68.8 | 73.8 | 0.29 | 0.08 | |||||
| HPFS | 1.6 × 10−8 | 56.0 | 62.3 | 67.0 | 0.33 | 0.06 | |||||
| Combined meta-analysis | 2.9 × 10−17 | 0.25 | 0.05 | ||||||||
P-values for heterogeneity were >0.05, except for rs2060793 (P = 0.01).
aSNP identifier based on NCBI dbSNP.
bChromosomal location based on NCBI Human Genome Build 35 coordinates.
cThe result of 1 df test based on linear regression, using an additive model, after square-root transformation of 25(OH)D levels—adjusted for age, vitamin D assay batch, study, case–control status, sex, body mass index, season of blood collection, vitamin D supplement intake, dietary vitamin D intake, region/latitude and chosen eigenvectors to control population stratification. The pooled P-value is based on the combined Wald statistics weighted by the square-root of the corresponding sample size.
dSE: standard error; the regression β and standard error were based on square-root scale. The pooled β and standard error summary calculated based on the random-effect model.
er2 = 1 in the HapMap CEU panel with the SNP from the initial GWAS.
Two additional SNPs in GC (rs7041 and rs1155563), in moderate linkage disequilibrium (LD; r2 = 0.4; Supplementary Material, Fig. S1), reached the genome-wide significance in the GWAS data meta-analysis (P = 4.1 × 10−22 and 3.5 × 10−25, respectively). In the four-study pooled analysis (ATBC, CPS-II, CLUE II and PLCO), we fit the regression model including all three genome-wide significant SNPs in GC along with the covariates used in the GWAS analysis and observed the strongest association signal for rs2282679, with much weaker signals from the other two SNPs (P = 3.8 × 10−4, 0.075 and 0.75 for rs2282679, rs7041 and rs1155563, respectively). We also conducted a stratified analysis by conditioning on genotype of rs2282679, and the two SNPs again showed markedly weaker evidence for association after stratification (P = 0.002 and 0.027 for rs7041 and rs1155563, respectively). Together, these observations suggest that the three common variants confer an association signal derived from a common source, with rs2282679 exhibiting the strongest association with 25(OH)D levels. This association was confirmed in the pooled analysis of the replication studies (P = 5.4 × 10−21) and resulted in a final combined meta-analysis significance of P = 1.8 × 10−49. The difference in mean 25(OH)D levels between carriers of two copies of the minor allele versus those of none across the GWAS and replication studies ranged from −6.4 to −34.4% (median −18.3%; Table 2).
GC protein (DBP) is a serum glycoprotein that belongs to the albumin family. It binds to 25(OH)D and other blood vitamin D sterol metabolites and transports them in circulation to target organs. It is plausible that rs2282679, in the intron 12, located near the actin subdomain III (15), may differentially affect GC binding of 25(OH)D (16). In previous candidate gene studies, we (17) and others (18,19) reported that the three GC SNPs were associated with circulating 25(OH)D concentrations, with strongest P-values for rs2282679 (P = 10−3–10−5). Nonetheless, it is not clear whether the 25(OH)D concentration differentials by the number of GC risk alleles also reflect differences in binding and bioavailability of 25(OH)D, activated 1,25(OH)2D and other vitamin D metabolites. On the basis of the pooled analysis of ATBC, PLCO, CPS-II and CLUE II, compared with having the wild type (AA), a heterozygote for the variant (C) has a nearly 2-fold higher risk (OR = 1.83) of having clinically deficient 25(OH)D levels (i.e. <25 nmol/l). Under a linear-additive genetic model for the square-root-transformed 25(OH)D level, rs2282679 explains an additional 1.0% of the variance after adjusting for other variables based on the pooled analysis of GWAS and replication studies.
Although not reaching the genome-wide significance, three other SNPs in or near genes involved in vitamin D synthesis or activation had P-values <10−5 in the initial GWASs (Table 2). In the 15th intron of NADSYN1 (chromosome 11q13.4; Supplementary Material, Fig. S2), which encode nicotinamide adenine dinucleotide (NAD) synthetase-1, rs3829251 was associated with 25(OH)D levels (P = 8.8 × 10−7) in the GWAS. NAD synthetase-1 catalyzes the final step in the biosynthesis of NAD, which involves a coenzyme in common metabolic redox reactions and a substrate for protein post-translational modifications (20). [A SNP in high LD (r2 = 0.84–0.87) with rs3829251, rs1790349, located in DHCR7, the gene encoding 7-dehydrocholesterol reductase, an enzyme catalyzing the production in skin of cholesterol from 7-dehydrocholesterol (21) using NADPH in 25(OH)D de novo synthesis, was also associated with 25(OH)D concentrations (P = 1.8 × 10−6). Because of the biological relevance of DHCR7 to vitamin D metabolism, we subsequently refer to the signal from this region as the ‘NADSYN1/DHCR7 locus’.] The finding for the NADSYN1/DHCR7 locus was confirmed in our replication set (for rs11234027, r2 = 1.0 with rs3829251 in the HapMap CEU samples: P = 1.0 × 10−3) and yielded a combined genome-wide significance of P = 3.4 × 10−9. The difference in mean 25(OH)D levels between homozygote minor and homozygote major allele genotypes across the GWASs ranged from −16.7 to 0% (median −2.4%) and across replication studies from −7.3 to −24.9% (median −9.5%), and 1.2% of the variance in 25(OH)D concentrations was accounted for by variation in this SNP across studies.
In the region harboring the open-reading frame 88 (C10orf88) on chromosome 10q26.13, two highly correlated SNPs (r2 = 1) were associated with circulating 25(OH)D levels in the GWAS [rs6599638 and rs1079458, with P = 3.3 × 10−7 and 1.8 × 10−6, respectively; (Table 2)]. The role of this gene is not well-characterized, but these two SNPs are located in the vicinity of ACADSB (acyl-coenzyme A dehydrogenase), which is involved in cholesterol and vitamin D synthesis. There were no notable associations between SNPs in ACADSB and vitamin D concentrations, however, and the association for rs6599638 was not confirmed in the pooled analysis of the replication studies (P = 9.3 × 10−1; meta-analysis with GWAS, P = 2.4 × 10−5).
The variant rs2060793, located in the 5′ untranslated region of the CYP2R1 gene encoding microsomal vitamin D 25-hydroxylase, was also associated with circulating 25(OH)D levels in the GWASs (P = 1.4 × 10−5; Table 2). This SNP, located on chromosome 11p15.2, is in complete LD (r2 = 1 in the HapMap CEU samples) with rs1993116 (Supplementary Material, Fig. S3), which has been inconsistently associated with 25(OH)D levels in the literature (22,23). We confirmed a significant association between the latter SNP and vitamin D levels (P = 1.6 × 10−17) in our replication sample and found a genome-wide significance for the combined studies from meta-analysis (P = 2.9 × 10−17). The difference in mean 25(OH)D levels between homozygote minor and homozygote major allele genotypes across the GWASs ranged from 1.5 to 14.4% (median 7.2%) and across the replication studies from 12.7 to 20.0% (median 19.6%), but only 0.6% of the variance in 25(OH)D concentrations was accounted for by variation in this SNP.
The microsomal enzyme CYP2R1 catalyzes C-25 hydroxylation of vitamin D3 to an active vitamin D receptor ligand in the liver and other organs (24). An inherited mutation in this gene causes the substitution of a proline for an evolutionarily conserved leucine at amino acid 99 in the CYP2R1 protein and the proline to valine shift has been related to rickets (25), mediated through this defect in vitamin D C-25 hydroxylation (24).
In combination, rs2282679 in GC, rs3829251 (rs11234027) in the NADSYN1/DHCR7 locus, rs6599638 in C10orf88 and rs2060793 (rs1993116) in CYP2R1 accounted for 2.8% of the variance in circulating vitamin D levels, based on the estimate in data pooled from ATBC, CPS-II, CLUE II and PLCO. A sensitivity analysis of 1312 non-cancer case controls in ATBC, CPS-II, CLUE II and PLCO gave similar β estimates (Supplementary Material, Table S1). We also found a significant signal with clinical vitamin D deficiency [i.e. 25(OH)D <25 nmol/l] for GC (one risk allele OR = 1.83; P = 2.5 × 10−8), and weak or null associations for C10orf88 (OR 1.17; P = 8.1 × 10−2), NADSYN1/DHCR7 (OR 1.18; P = 0.11) and CYP2R1 (OR 0.89; P = 0.21), based on 2907 subjects in ATBC, PLCO, CPS-II and CLUE II (Supplementary Material, Table S2). It should be noted, however, that the prevalence of vitamin D deficiency in three of these four cohorts was very low (i.e. 3–6%), and examination of the dichotomized 25(OH)D outcome reduced statistical power for this comparison. The odds ratio for clinical deficiency for having one risk allele for each of the four gene loci was 2.97 (95% CI: 2.00–4.39), and the joint effect from the four loci is significant (P = 3.2 × 10−8).
DISCUSSION
In this GWAS analysis, we found and replicated conclusive evidence that variants in GC, the NADSYN1/DHCR7 locus and CYP2R1 that encode the vitamin D carrier protein and enzymes in the vitamin D metabolic pathway are associated with circulating 25(OH)D levels, with the latter two genes representing novel findings. An initial signal for C10orf88 (in the vicinity of ACADSB) was not replicated. The three SNPs we found associated with vitamin D levels and with clinical deficiency to some extent, along with nearby variants in LD with them, could be included in studies of Mendelian randomization of vitamin D status and disease outcomes, although the modest differences by number of minor alleles suggest that very large sample sizes would be needed to confirm the expected small associations. Strengths of the study include its large sample size, with findings confirmed in multiple independent populations, and having the vitamin D phenotype measured with good precision. Heterogeneity was, however, observed for the 25(OH)D association with CYP2R1. Potential limitations of our study include there being only one blood sample assayed for 25(OH)D for each person—measurements from multiple time points would likely provide more valid estimates of usual circulating cholecalciferol levels. Also, two laboratory methods were used to determine 25(OH)D concentrations, and for the RIA method, this was done for three separate parent studies, possibly contributing to some of the cohort differences in the study findings. Follow-up studies, as well as future meta-analyses in other populations with GWAS that have measured 25(OH)D concentrations and possibly DBP, are likely to uncover more common alleles associated with circulating plasma 25(OH)D levels and vitamin D status.
Insights gained from the study of circulating vitamin D are likely to have implications for the examination of complex diseases such as osteoporosis, cardiovascular disease and cancer. Further investigation of the biological mechanisms underlying the associations observed here, and replication of the findings in other populations, including those of African and Asian descent, is required.
MATERIALS AND METHODS
We conducted a meta-analysis of five GWASs within five cohorts (Table 1) with prospectively collected 25(OH)D levels and replicated the findings in three prospective case–control studies. Analyses were restricted to subjects of European ancestry. The five GWASs were: a case–control study of lung cancer in the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC) (7); a case–control study of prostate cancer [Cancer Genetic Markers of Susceptibility (CGEMS)] in the Prostate, Lung, Colorectal, Ovarian Cancer Screening Trial (PLCO) (8); three case–control studies of pancreatic cancer nested within ATBC, Cancer Prevention Study-II (CPS-II) (9) and Give Us a Clue to Cancer and Heart Disease Study (CLUE II) (10); and case–control studies of breast cancer (CGEMS) and type 2 diabetes (T2D) nested within the Nurses' Health Study (NHS) (11). GWAS genotyping used the Illumina 550K (or higher version) platform with the exception of the T2D study, which was genotyped using the Affymetrix 6.0 platform. Genotypes for markers that were on the Illumina 550K platform but not the Affymetrix 6.0 were imputed in the T2D study using the hidden-Markov model algorithm implemented in the MACH and the HapMap CEU reference panel (Rel 22). Quality-control assessment of genotypes, including sample completion and SNP call rates, concordance rates, deviation from fitness for the Hardy–Weinberg proportions in control DNA and final sample selection for association analyses, are described elsewhere (26–29). For the majority (92%) of the PLCO, ATBC, CPS-II and CLUE II samples, serum 25(OH)D concentrations were measured by competitive chemiluminescence immunoassay (CLIA) in a single laboratory (Heartland Assays, Ames, IA, USA) (30), with coefficients of variation (CV) for 25(OH)D in the blinded duplicate quality-control samples of 9.3% (intrabatch) and 12.7% (interbatch). Previously available 25(OH)D measurements for 492 ATBC subjects from other serologic substudies showed similar CVs [9.5% (intrabatch) and 13.6% (interbatch)]. For the NHS-CGEMS samples, plasma 25(OH)D levels were measured by radioimmunoassay (RIA) (30) in three batches (CVs 8.7–17.6%): two in the laboratory of Dr Michael Holick at Boston University School of Medicine (31) and the third in the laboratory of Dr Bruce Hollis at the Medical University of South Carolina in Charleston, SC (31). Plasma levels of 25(OH)D in the NHS-T2D samples were measured in the Nutrition Evaluation Laboratory in the Human Nutrition Research Center on Aging at Tufts University (with CV = 8.7%) by rapid extraction followed by an equilibrium I-125 RIA procedure (DiaSorin Inc., Stillwater, MN, USA) as specified by the manufacturer's procedural documentation and analyzed on a gamma counter (Cobra II, Packard).
Four markers selected for replication were genotyped by TaqMan in three case–control samples: NHS colon polyp study (13) (n = 403/407 cases/controls), colorectal cancer study (12) (173/371 cases/controls) and a study of prostate cancer in the Health Professionals Follow-up Study (14) (431/436 cases/controls). There was no overlap of participants in the NHS-CGEMS, T2D, colon polyp or colorectal cancer studies. Plasma 25(OH)D concentrations in these studies were measured by RIA in the laboratory of Dr Bruce Hollis [CVs 7.5, 11.8 and 5.4–5.6% (two-batch), respectively].
Concentrations of 25(OH)D were similar across CPS-II, CLUE II and PLCO (Tables 1 and 2). In ATBC, the average population concentration was lower, likely due to reduced UVB solar radiation exposure at that northern latitude and no blood collection during July and some of June and August. The NHS had overall higher values, likely the result of having had blood samples analyzed in a different laboratory using a different assay. Nonetheless, there was a substantial overlap with the observed range of 25(OH)D values across all studies.
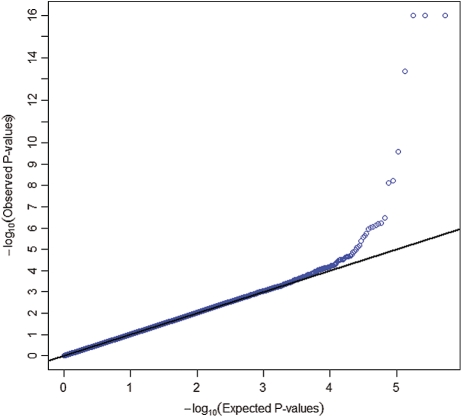
We conducted a pooled analysis (1) of four GWASs (ATBC, CPS-II, CLUE II and PLCO) and tested the association between 593 253 SNP markers that passed quality-control filters and the square-root-transformed value of circulating 25(OH)D using linear regression under an additive genetic model. We adjusted for age, vitamin D assay batch, study, case–control status, sex, body mass index (<20, 20–25, 25–30, 30+ kg/m2), season of blood collection (December–February; March–May; June–August; September–November), vitamin D supplement intake (missing, 0, 0–400 and 400+ IU/day), dietary vitamin D intake (missing, <100, 100–200, 200–300, 300–400 and 400+ IU/day), region/latitude and three eigenvectors to control for population stratification. Usual dietary intake and other covariate information were collected through self-administered food frequency questionnaires and baseline risk factor questionnaires, respectively. The square-root transformation was very close to the most optimal transformation identified by the Box-Cox procedure and was used to ensure normality of the residuals. The Wald test was used for testing the association between each SNP and the outcome. A similar approach was used for analysis of each of the following two GWASs, NHS-CGEMS (2) and NHS-T2D (3). Imputed markers in the NHS-T2D study were analyzed using genotype dosages (expected allele counts). We conducted the meta-analysis of the GWASs, and of the GWAS and replication studies combined, by averaging the signed Wald statistics weighted by the square root of the corresponding sample sizes; this analysis is robust to differences in scale across different techniques for measuring circulating 25(OH)D (32). We also used the random-effect model to estimate the common effect size and to assess heterogeneity among results from different studies. The quantile–quantile plot of P-values from the GWAS meta-analysis showed no evidence of systematic type-I error inflation (λGC = 1.0007; Fig. 2).
Figure 2.
Quantile–quantile plot of 593 253 SNPs from the meta-analysis of the five GWAS scans (n = 4501). Each blue dot represents an observed statistic [defined as the −log10(P)] versus the corresponding expected statistic. The blue dots correspond to the distribution of test statistics. The black line corresponds to the null distribution. Observed P-values <10−16 are plotted at the 10−16 significance level.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
The Alpha-Tocopherol, Beta-Carotene Cancer Prevention (ATBC) Study is supported by the Intramural Research Program of the National Cancer Institute, NIH, and by US Public Health Service contracts N01-CN-45165, N01-RC-45035, N01-RC-37004 and HHSN261201000006C from the National Cancer Institute, NIH, DHHS. The Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO) is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and by contracts from the Division of Cancer Prevention, National Cancer Institute (NCI), US National Institutes of Health (NIH), Department of Health and Human Services (DHHS). The Nurses’ Health Study (NHS) is supported by NIH grants P01CA087969 and 5U01HG004399-2, and the Health Professionals Follow-up Study (HPFS) is supported by NIH grant P01CA055075. The American Cancer Society (ACS) study is supported by U01 CA098710. K.C.S. is supported by NIH Kirschstein-NRSA T32 ES016645-01. The genome-wide scans have been supported by the NCI, NIH, under contract N01-CO-12400. Funding to pay the Open Access publication charges for this article was provided by the Division of Cancer Epidemiology and Genetics, NCI, NIH.
Supplementary Material
REFERENCES
- 1.Food and Nutrition Board, Institute of Medicine. Dietary Reference Intakes for Calcium, Phosphorus, Magnesium, Vitamin D, and Fluoride. Washington, DC: National Academies Press; 1999. [PubMed] [Google Scholar]
- 2.DeLuca H.F., Zierold C. Mechanisms and functions of vitamin D. Nutr. Rev. 1998;56:S4–S10. doi: 10.1111/j.1753-4887.1998.tb01686.x. [DOI] [PubMed] [Google Scholar]
- 3.Lin R., White J.H. The pleiotropic actions of vitamin D. Bioessays. 2004;26:21–28. doi: 10.1002/bies.10368. doi:10.1002/bies.10368. [DOI] [PubMed] [Google Scholar]
- 4.Orton S.M., Morris A.P., Herrera B.M., Ramagopalan S.V., Lincoln M.R., Chao M.J., Vieth R., Sadovnick A.D., Ebers G.C. Evidence for genetic regulation of vitamin D status in twins with multiple sclerosis. Am. J. Clin. Nutr. 2008;88:441–447. doi: 10.1093/ajcn/88.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter D., De Lange M., Snieder H., MacGregor A.J., Swaminathan R., Thakker R.V., Spector T.D. Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J. Bone Miner. Res. 2001;16:371–378. doi: 10.1359/jbmr.2001.16.2.371. doi:10.1359/jbmr.2001.16.2.371. [DOI] [PubMed] [Google Scholar]
- 6.Shea M.K., Benjamin E.J., Dupuis J., Massaro J.M., Jacques P.F., D'Agostino R.B., Sr, Ordovas J.M., O'Donnell C.J., Dawson-Hughes B., Vasan R.S., et al. Genetic and non-genetic correlates of vitamins K and D. Eur. J. Clin. Nutr. 2009;63:458–464. doi: 10.1038/sj.ejcn.1602959. doi:10.1038/sj.ejcn.1602959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The ATBC Cancer Prevention Study Group. The alpha-tocopherol, beta-carotene lung cancer prevention study: design, methods, participant characteristics, and compliance. Ann. Epidemiol. 1994;4:1–10. doi: 10.1016/1047-2797(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 8.Hayes R.B., Sigurdson A., Moore L., Peters U., Huang W.Y., Pinsky P., Reding D., Gelmann E.P., Rothman N., Pfeiffer R.M., et al. Methods for etiologic and early marker investigations in the PLCO trial. Mutat. Res. 2005;592:147–154. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Calle E.E., Rodriguez C., Jacobs E.J., Almon M.L., Chao A., McCullough M.L., Feigelson H.S., Thun M.J. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94:2490–2501. doi: 10.1002/cncr.101970. doi:10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 10.Gallicchio L., Chang H., Christo D.K., Thuita L., Huang H.Y., Strickland P., Ruczinski I., Hoffman S.C., Helzlsouer K.J. Single nucleotide polymorphisms in inflammation-related genes and mortality in a community-based cohort in Washington County, Maryland. Am. J. Epidemiol. 2008;167:807–813. doi: 10.1093/aje/kwm378. doi:10.1093/aje/kwm378. [DOI] [PubMed] [Google Scholar]
- 11.Willett W.C., Stampfer M.J., Colditz G.A., Rosner B.A., Hennekens C.H., Speizer F.E. Moderate alcohol consumption and the risk of breast cancer. N. Engl. J. Med. 1987;316:1174–1180. doi: 10.1056/NEJM198705073161902. [DOI] [PubMed] [Google Scholar]
- 12.Feskanich D., Ma J., Fuchs C.S., Kirkner G.J., Hankinson S.E., Hollis B.W., Giovannucci E.L. Plasma vitamin D metabolites and risk of colorectal cancer in women. Cancer Epidemiol. Biomarkers Prev. 2004;13:1502–1508. [PubMed] [Google Scholar]
- 13.Platz E.A., Hankinson S.E., Hollis B.W., Colditz G.A., Hunter D.J., Speizer F.E., Giovannucci E. Plasma 1,25-dihydroxy- and 25-hydroxyvitamin D and adenomatous polyps of the distal colorectum. Cancer Epidemiol. Biomarkers Prev. 2000;9:1059–1065. [PubMed] [Google Scholar]
- 14.Platz E.A., Leitzmann M.F., Hollis B.W., Willett W.C., Giovannucci E. Plasma 1,25-dihydroxy- and 25-hydroxyvitamin D and subsequent risk of prostate cancer. Cancer Causes Control. 2004;15:255–265. doi: 10.1023/B:CACO.0000024245.24880.8a. doi:10.1023/B:CACO.0000024245.24880.8a. [DOI] [PubMed] [Google Scholar]
- 15.Otterbein L.R., Cosio C., Graceffa P., Dominguez R. Crystal structures of the vitamin D-binding protein and its complex with actin: structural basis of the actin-scavenger system. Proc. Natl Acad. Sci. USA. 2002;99:8003–8008. doi: 10.1073/pnas.122126299. doi:10.1073/pnas.122126299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnaud J., Constans J. Affinity differences for vitamin D metabolites associated with the genetic isoforms of the human serum carrier protein (DBP) Hum. Genet. 1993;92:183–188. doi: 10.1007/BF00219689. [DOI] [PubMed] [Google Scholar]
- 17.Ahn J., Albanes D., Berndt S.I., Peters U., Chatterjee N., Freedman N.D., Abnet C.C., Huang W., Kibal A., Crawford E.D., et al. Vitamin D-related genes, serum vitamin D concentrations, and prostate cancer risk. Carcinogenesis. 2008;30:8. doi: 10.1093/carcin/bgp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman C.D., Fingerlin T.E., Langefeld C.D., Hicks P.J., Rich S.S., Wagenknecht L.E., Bowden D.W., Norris J.M. Genetic and environmental determinants of 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D levels in Hispanic and African Americans. J. Clin. Endocrinol. Metab. 2008;93:3381–3388. doi: 10.1210/jc.2007-2702. doi:10.1210/jc.2007-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinotte M., Diorio C., Berube S., Pollak M., Brisson J. Genetic polymorphisms of the vitamin D binding protein and plasma concentrations of 25-hydroxyvitamin D in premenopausal women. Am. J. Clin. Nutr. 2009;89:634–640. doi: 10.3945/ajcn.2008.26445. doi:10.3945/ajcn.2008.26445. [DOI] [PubMed] [Google Scholar]
- 20.Hara N., Yamada K., Terashima M., Osago H., Shimoyama M., Tsuchiya M. Molecular identification of human glutamine- and ammonia-dependent NAD synthetases. Carbon-nitrogen hydrolase domain confers glutamine dependency. J. Biol. Chem. 2003;278:10914–10921. doi: 10.1074/jbc.M209203200. doi:10.1074/jbc.M209203200. [DOI] [PubMed] [Google Scholar]
- 21.Waterham H.R., Wanders R.J. Biochemical and genetic aspects of 7-dehydrocholesterol reductase and Smith-Lemli-Opitz syndrome. Biochim. Biophys. Acta. 2000;1529:340–356. doi: 10.1016/s1388-1981(00)00159-1. [DOI] [PubMed] [Google Scholar]
- 22.Ramos-Lopez E., Bruck P., Jansen T., Herwig J., Badenhoop K. CYP2R1 (vitamin D 25-hydroxylase) gene is associated with susceptibility to type 1 diabetes and vitamin D levels in Germans. Diabetes Metab. Res. Rev. 2007;23:631–636. doi: 10.1002/dmrr.719. doi:10.1002/dmrr.719. [DOI] [PubMed] [Google Scholar]
- 23.Ramos-Lopez E., Kahles H., Weber S., Kukic A., Penna-Martinez M., Badenhoop K., Louwen F. Gestational diabetes mellitus and vitamin D deficiency: genetic contribution of CYP27B1 and CYP2R1 polymorphisms. Diabetes Obes. Metab. 2008;10:683–685. doi: 10.1111/j.1463-1326.2008.00879.x. [DOI] [PubMed] [Google Scholar]
- 24.Cheng J.B., Levine M.A., Bell N.H., Mangelsdorf D.J., Russell D.W. Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc. Natl Acad. Sci. USA. 2004;101:7711–7715. doi: 10.1073/pnas.0402490101. doi:10.1073/pnas.0402490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strushkevich N., Usanov S.A., Plotnikov A.N., Jones G., Park H.W. Structural analysis of CYP2R1 in complex with vitamin D3. J. Mol. Biol. 2008;380:95–106. doi: 10.1016/j.jmb.2008.03.065. doi:10.1016/j.jmb.2008.03.065. [DOI] [PubMed] [Google Scholar]
- 26.Thomas G., Jacobs K.B., Yeager M., Kraft P., Wacholder S., Orr N., Yu K., Chatterjee N., Welch R., Hutchinson A., et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. doi:10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 27.Amundadottir L., Kraft P., Stolzenberg-Solomon R.Z., Fuchs C.S., Petersen G.M., Arslan A.A., Bueno-de-Mesquita H.B., Gross M., Helzlsouer K., Jacobs E.J., et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landi M.T., Chatterjee N., Yu K., Goldin L.R., Goldstein A.M., Rotunno M., Mirabello L. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Gen. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollis B.W. Quantitation of 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D by radioimmunoassay using radioiodinated tracers. Methods Enzymol. 1997;282:174–186. doi: 10.1016/s0076-6879(97)82106-4. doi:10.1016/S0076-6879(97)82106-4. [DOI] [PubMed] [Google Scholar]
- 31.Bertone-Johnson E.R., Chen W.Y., Holick M.F., Hollis B.W., Colditz G.A., Willett W.C., Hankinson S.E. Plasma 25-hydroxyvitamin D and 1,25-dihydroxyvitamin D and risk of breast cancer. Cancer Epidemiol. Biomarkers Prev. 2005;14:1991–1997. doi: 10.1158/1055-9965.EPI-04-0722. doi:10.1158/1055-9965.EPI-04-0722. [DOI] [PubMed] [Google Scholar]
- 32.Whitlock M.C. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. doi:10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.