Abstract
To identify type 2 diabetes (T2D) susceptibility loci, we conducted genome-wide association (GWA) scans in nested case–control samples from two prospective cohort studies, including 2591 patients and 3052 controls of European ancestry. Validation was performed in 11 independent GWA studies of 10 870 cases and 73 735 controls. We identified significantly associated variants near RBMS1 and ITGB6 genes at 2q24, best-represented by SNP rs7593730 (combined OR = 0.90, 95% CI = 0.86–0.93; P = 3.7 × 10−8). The frequency of the risk-lowering allele T is 0.23. Variants in this region were nominally related to lower fasting glucose and HOMA-IR in the MAGIC consortium (P < 0.05). These data suggest that the 2q24 locus may influence the T2D risk by affecting glucose metabolism and insulin resistance.
INTRODUCTION
Type 2 diabetes (T2D) is one of the fastest growing public health problems worldwide (1). Although environmental factors have clearly contributed to the recent rise in prevalence of the disease, there is growing evidence that genetic factors also have an important impact (2,3). Unlike the monogenic forms of diabetes that result from a few rare and highly penetrant variants, the genetic basis of the common forms of T2D is more complex (4). Multiple susceptibility loci have been identified for T2D from genome-wide association studies (GWASs) in populations of European and Asian origin (5–8), with common variants being modestly associated with disease risk. However, the reported genetic variants account for only a small proportion of the heritability of T2D. It is possible that a large number of common genetic variants for diabetes remain to be identified.
To identify additional T2D susceptibility loci, we carried out a GWAS of T2D in two well-established prospective cohorts: the Nurses' Health Study (NHS) and the Health Professional Follow-Up Study (HPFS). We followed up on a potentially novel locus by in silico analysis of 11 independent GWA scans. Furthermore, we assessed the relationship between SNPs in the new locus and biomarkers of glucose metabolism and insulin resistance in collaboration with MAGIC [the Meta-Analysis of Glucose and Insulin-related traits Consortium (9)] and examined the gene expression profiles.
RESULTS
In the discovery stage, 3529 women (NHS) and 2668 men (HPFS) were independently genotyped for 909 622 SNPs, using the Affymetrix Genome-Wide Human 6.0 array (Santa Clara, CA, USA). After applying quality control filters, 3221 (1467 cases, 1754 controls) women and 2422 (1124 cases, 1298 controls) men of genetically inferred European ancestry were included in the final analysis (Table 1). SNPs with minor allele frequency (MAF) of <2%, missing call rate of >2%, significant deviation from Hardy–Weinberg equilibrium (HWE, P < 1 × 10−4), significant plate associations and high discordance rates were excluded, leaving 704 409 SNPs in NHS and 706 040 SNPs in HPFS for analysis.
Table 1.
Studies included in the genome-wide association analysis of type 2 diabetes
| No. of subjects |
Location | Study design | ||
|---|---|---|---|---|
| Cases | Controls | |||
| Discovery | ||||
| NHS | 1467 | 1754 | USA | Nested case–control |
| HPFS | 1124 | 1298 | USA | Nested case–control |
| In total | 2591 | 3052 | – | – |
| Replication | ||||
| DIAGRAM | 8130 | 38987 | UK, Iceland, Germany, Finland, Sweden, France, Netherlands, Croatia, Italy | Case–control (meta-analysis) |
| WGHS | 1370 | 20684 | USA | Cohort |
| FHS | 674 | 7644 | USA | Cohort |
| ARIC | 696 | 6420 | USA | Cohort |
| In total | 10870 | 73735 | – | – |
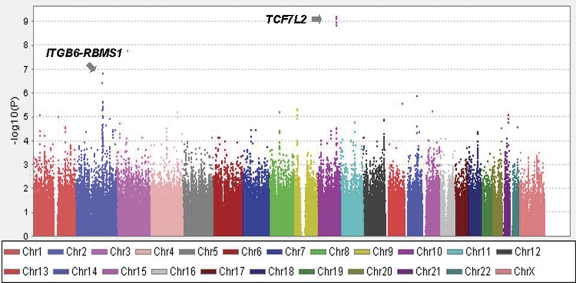
For each cohort, the Cochran–Armitage test for trend (one degree of freedom) was used to examine the association between each SNP and T2D. We observed no evidence of systematic bias in the distribution of P-values for association tests (genomic inflation factors were 1.02 in NHS and 1.01 in HPFS; Supplementary Material, Fig. S1). To improve power, the results for SNPs common to both cohorts (n = 683 509) were combined for a meta-analysis. A graphical summary of the meta-analysis results is displayed in Figure 1. Consistent with previous reports (10), several SNPs in/near TCF7L2 (chromosome 10) were significantly associated with T2D (the best P-value = 2.13E − 09). Using the same study samples, we have reported in our earlier analysis the significant associations for some other loci such as HHEX, CDKAL1, IGF2BP2, SLC30A8, CDKN2A/B, PPARG and KCNJ11 (11). A second cluster of SNPs on 2q24 showed strong association with T2D with P-values reaching 1.47 × 10−7 (SNP rs1020731) and defined a candidate region that encompassed the RBMS1 and ITGB6 genes (Fig. 2). The interactions between the SNPs and gender were not significant (adjusted P for interaction = 0.3), although the associations appeared stronger in women. We noted that a signal on chr 3 (rs2371765) also showed strong association with T2D (Fig. 1). In our first effort of replication, we were more interested in 2q24 because this locus encompasses a cluster of SNPs with strong associations with diabetes, whereas there is only one SNP in the chr 3 locus that we suspected to be a false positive.
Figure 1.
Plot of the −log10P-values for the combined analysis for NHS and HPFS GWA trend tests. Each point represents an SNP from the 683 509 SNPs remaining after quality control filters and overlapped between the two study samples. Different bands are used to differentiate SNPs on consecutive autosomal and X chromosomes.
Figure 2.
Association signals at 2q24 locus: (A) association signals from the combined analysis for NHS and HPFS GWA trend tests, across a 500 kb region centering on rs1020731. The vertical axis representing the −log10P-values from the Armitage trend test. SNP rs1020731 is shown in blue and rs7953730 is shown in red, labeled with discovery stage P-values. Estimated recombination rates from HapMap are plotted to reflect the local LD structure. Genes were extracted from the HapMap Genome Browser; (B) LD in the CEU HapMap population. Disequilibrium coefficient values for HapMap Phase II data (v. 22) were generated with Haploview.
To validate and further refine the evidence for association at the 2q24 locus, all SNPs that were within 50 kb of SNP rs1020731 and strongly associated with T2D in NHS and HPFS samples (combined P < 1 × 10−4) were subject to in silico replications. Nine additional SNPs at 2q24 were identified (Supplementary Material, Table S1) and were within weak (r2 = 0.43) to strong (r2 = 0.92) LD with the target SNP rs1020731 (Supplementary Material, Table S2).
The replication included GWA scans from a total of 10 870 cases and 73 735 controls from DIAGRAM, which is comprised of eight independent GWA scans (WTCCC, DGI, FUSION, DECODE, DCDG, ERGO EUROSPAN and KORA), atherosclerosis risk in communities (ARIC), FHS and WGHS (Table 1). SNP rs7593730 (r2 = 0.58 with rs1020731) showed a strongest association in the replication sets (combined OR = 0.92, 95% CI 0.88–0.96; P = 9.09 × 10−5). SNP rs1020731 was also nominally associated with T2D in the replication samples (OR = 0.94, 95% CI 0.90–0.97; P = 8.97 × 10−4). The directions of effect in the replication samples were consistent with the discovery analyses. Combining results from the discovery and replication stages strengthened the association (Table 2). Each T allele of SNP rs7593730 was associated with a 10% (95% CI: 7–14%, P = 3.7 × 10−8) lower odds of T2D. SNPs rs1020731, rs2925757, rs4077463 and rs4589705 also showed near genome-wide significant associations with T2D risk, with P-values of 9.44 × 10−8, 7.94 × 10−8, 6.41 × 10−8 and 7.24 × 10−8, respectively (Supplementary Material, Table S3).
Table 2.
Summary of associations between SNP rs7593730 at 2q24 and risk of type 2 diabetes in discovery and replication samples
| MAF (%) |
Odds ratios (95% CI) | P-values | ||
|---|---|---|---|---|
| Cases | Controls | |||
| Discovery | ||||
| NHS | 0.18 | 0.23 | 0.74 (0.66–0.84) | 1.15E − 06 |
| HPFS | 0.22 | 0.24 | 0.90 (0.79–1.03) | 1.36E − 01 |
| Pooled, NHS and HPFS | – | – | 0.81 (0.74–0.89) | 4.51E − 06 |
| Replication | ||||
| DIAGRAM | – | – | 0.93 (0.89–0.98) | 6.00E − 03 |
| WGHS | 0.21 | 0.22 | 0.90 (0.79–1.04) | 1.60E − 01 |
| FHS | 0.2 | 0.23 | 0.82 (0.71–0.94) | 5.49E − 03 |
| ARIC | 0.21 | 0.22 | 0.92 (0.80–1.07) | 2.82E − 01 |
| Pooled, replication | – | – | 0.92 (0.88–0.96) | 9.09E − 05 |
| Pooled, all studies | – | – | 0.90 (0.86–0.93) | 3.70E − 08 |
The bold values are odds ratios and P-values of the combined associations in discovery and replication stages, and in all studies together.
The MAF is for T allele.
To gain insight into potential mechanisms by which the polymorphisms at 2q24 locus impact T2D risk, we examined the associations with T2D-related quantitative traits in MAGIC (9), a consortium of GWA scans (n = 7082–46 186 for various traits) on fasting and 2 h stimulated glucose and insulin, as well as insulin resistance measured by homeostasis model assessment (HOMA-IR). We examined the best-associated SNP rs7592730 and another SNP rs2925757, which showed nearly genome-wide significant association with T2D but was in relatively low LD (r2 < 0.8) with rs7593730. Each T allele for SNP rs7593730 was associated with a 0.009 mmol/l (P = 4.35 × 10−2) lower fasting glucose. Each G allele for SNP rs2925757 was associated with a 0.0107 mmol/l (P = 2.27 × 10−2) lower fasting glucose and a 0.0104 unit lower HOMA-IR (P = 4.34 × 10−2).
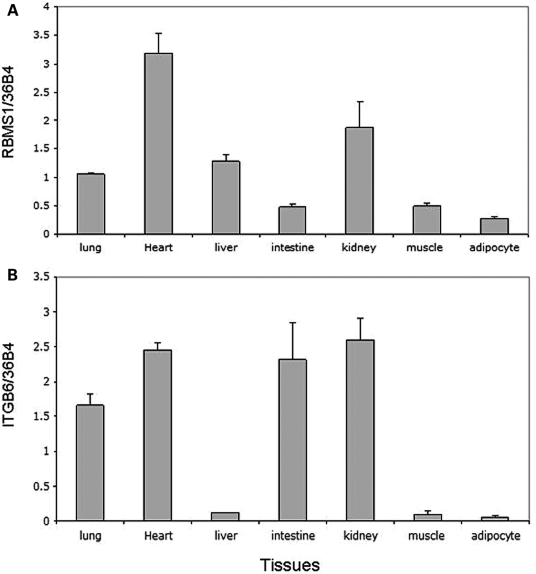
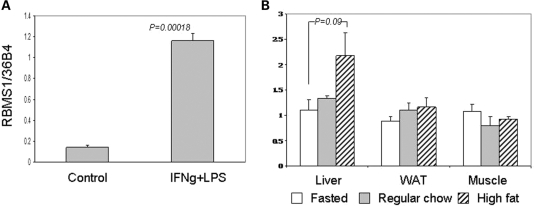
We examined the expression profile of genes RBMS1 and ITGB6 in human tissues. RBMS1 and ITGB6 are expressed in many tissues relevant to T2D (i.e. heart, muscle, liver and adipose; Fig. 3). We also examined the expression of the genes in response to inflammation and high fat-diet. When mouse bone marrow macrophages were treated in vitro with the inflammatory agents interferon (IFN)-γ and lipopolysaccharide (LPS), the expression of RBMS1 was induced (P = 0.00018; Fig. 4), whereas ITGB6 expression was unaffected (data not shown). The liver of mice fed a high fat-diet for 2 months showed a trend toward higher expression of RBMS1 relative to their fasting and regular chow-fed counterparts (P = 0.09; Fig. 4).
Figure 3.
The relative expression levels of (A) RBMS1 and (B) ITGB6 genes in human tissues. The expression levels were determined by RT–PCR analysis.
Figure 4.
(A) RBMS1 is induced by pro-inflammatory stimulants in the macrophages. Bone marrow-derived wild-type mouse macrophages were incubated with or without interferon-γ (IFN-γ) (10 ng/ml) overnight followed by lipopolysaccharide (LPS) treatment (10 ng/ml) for 24 h. The experiments were done in triplicate. *P < 0.05; (B) RBMS1 expression profiles in the liver, white adipose tissue (WAT) and muscle of fasted, regular chow-fed and high fat-diet-fed (HFD, 2 months) mice.
DISCUSSION
Considerable progress has been made in identifying the genetic factors contributing to the development of T2D. This progress is largely attributable to the advent of GWAS, which have uncovered multiple susceptibility loci in populations of European and Asian descent (5–8). Nevertheless, a large proportion of the heritability of T2D remains unexplained, suggesting other loci have yet to be discovered.
In the present study, we carried out GWA scans for T2D in 2591 patients and 3052 controls of European ancestry from two prospective cohorts, and performed validation analyses in 11 independent GWA scans of a total of 10 870 cases and 73 735 controls of European origin. We identified a novel locus at 2q24 with genome-wide significant association at the 5 × 10−8 threshold.
Locus 2q24 encompasses two genes: RBMS1 and ITGB6. The best-associated SNP (rs7593730) is located in intron 3 of RBMS1. This gene encodes RNA-binding motif, single-stranded interacting protein 1, a member of a small family of proteins that bind single-stranded DNA/RNA. While these proteins have been implicated in DNA replication, gene transcription, cell cycle progression and apoptosis (12), a role of these proteins in diabetogenic pathways has not been reported. ITGB6 encodes integrin, beta 6. Integrins are transmembrane glycoprotein receptors that mediate dynamic linkages between the actin cytoskeleton and the extracellular matrix and also transduce signals to and from the cell interior (13). Expression of the beta 6 isoform may affect secretion of matrix metalloproteinases and tumor progression (14,15). On the basis of the current literature, there is little support for an obvious role of proteins encoded by the 2q24 locus in T2D etiology.
To gain insight into the potential mechanism by which these novel loci contribute to T2D, we conducted a series of quantitative trait and gene expression analyses. The less common alleles of variants at 2q24, which were associated with reduced T2D risk, were also related to lower levels of fasting glucose and/or HOMA-IR. These data suggest that the 2q24 locus may encompass variants affecting glucose metabolism and insulin resistance, but further investigation is warranted.
Our gene expression experiments demonstrate that RBMS1 and ITGB6 are expressed in tissues of biological relevance to T2D such as the heart, muscle, liver and adipose tissue. RBMS1 also appeared responsive to inflammation, as demonstrated in vitro when cells were directly exposed to inflammatory agents and in vivo. Since inflammation has been implicated in T2D (16), it is possible that the association between variants encompassing RBMS1 with T2D is mediated through a role in inflammatory pathways. In addition, a high fat-diet tended to induce the expression of RBMS1. We caution the exploratory nature of these experiments and the need for more extensive follow-up before proposing such a mechanism.
The present study emphasizes that follow-up of tentative associations identified by GWA scans in large replication studies can reliably identify additional susceptibility loci with modest effects on T2D. Because previous GWASs followed and reported only the most significant signals from their discovery scans, it is not surprising that 2q24 locus was not identified in these studies. Distinct exposure to environment factors including age, BMI, and lifestyle may differentially affect the expression of the disease phenotype in these studies. The robustness of the 2q24 result is strengthened by its documentation in samples ascertained with different study designs, including cohort and case–control studies. However, power calculations indicate that our discovery sets had only 58% power to detect variants with a relative risk of 1.1 and an MAF of 20% at P < 0.05, suggesting that other T2D loci should be detectable by further combined analyses at the discovery stage.
In conclusion, we identified a new T2D susceptibility locus on chromosome 2q24. Genetic variants in this locus were nominally associated with fasting glucose and insulin resistance. Detailed fine-mapping and functional analyses will be needed to establish the causal variants at the 2q24 locus. Mechanistic studies will also be necessary to characterize the role of this locus in T2D development. Although many bona fide T2D loci have been identified recently, the current study supports the notion that many T2D loci remain to be discovered and further demonstrate the need for large sample sizes in both the discovery and follow-up stages of GWASs.
MATERIALS AND METHODS
Discovery study populations
Details of the NHS and HPFS cohorts have been described previously (17,18). Briefly, the NHS was established in 1976 when 121 700 female registered nurses aged 30–55 years and residing in 11 US states completed a mailed questionnaire on their medical history and lifestyle. The lifestyle factors, including smoking, menopausal status and postmenopausal hormone therapy and body weight, have been updated by validated questionnaires every 2 years. A total of 32 826 women provided blood samples between 1989 and 1990. The HPFS is a prospective cohort study of 51 529 US male health professionals aged 40–75 years at study initiation in 1986. Information about health and disease is assessed biennially by a self-administered questionnaire. Between 1993 and 1999, 18 159 men provided blood samples. The present study was approved by the institutional review board of the Brigham and Women's Hospital and the Human Subjects Committee Review Board of Harvard School of Public Health.
NHS and HPFS participants for the current study were selected among those with a blood sample using a ‘nested’ case–control study design (11,19). Diabetes cases were defined as self-reported diabetes confirmed by a validated supplementary questionnaire. For cases before 1998, diagnosis was made using criteria consistent with those proposed by the National Diabetes Data Group [NDDG (20)]. We used the American Diabetes Association diagnostic criteria for diagnosis of diabetes cases during the 1998 and 2000 cycles (21). A 98% of self-reported cases were confirmed by medical records review in both cohorts (22). Controls were defined as those free of diabetes at the time of diagnosis of the case and remained unaffected through follow-up (2006). Although controls were originally matched per case (by gender, year of birth, month of blood collection and fasting status), matched pairs were broken because not all subjects gave informed consent for submission of their GWAS data to dbGaP.
Genotyping and quality control
The NHS and HPFS T2D GWA scans are a component of the Gene Environment-Association Studies (GENEVA) under the NIH Genes, Environment and Health Initiative (GEI). Genotyping for 2745 patients with T2D and 3148 healthy controls from NHS and HPFS was done at the Broad Center for Genotyping and Analysis using the Affymetrix Genome-Wide Human 6.0 array (Santa Clara, CA, USA) and the Birdseed calling algorithm (23). Genotypic data first passed Broad's initial QC which included SNP fingerprints for sample tracking and early detection of sample misidentification, missing call rates of ≥5%, the use of a HapMap control to check genotype quality independent of study samples and tracking of reagent and instrumental performance. Genotype data were subsequently released for further QC to the GENEVA Coordinating Center at the University of Washington.
Relatedness was evaluated using pairwise identity-by descent estimation using 80k SNPs in a method of moments approach implemented in PLINK software (24). In NHS, five pairs of duplicate samples were identified and removed. One pair of full siblings and eight sets (six pairs and two triplets) of possible first cousins were also identified. Gender was confirmed by examining the mean of the intensities of SNP probes on the X and Y chromosomes. One male sample was mis-identified as a female sample and was excluded. Twenty-seven subjects with highly variable intensity data, determined by analyzing relative intensity (‘LogRRatio’) and allelic imbalance (‘BAlleleFreq’, BAF) (25), and 22 samples having a missing call rate of ≥2% were also removed. In HPFS, four pairs of duplicate samples were excluded. Three pairs of full sibs and one pair of possible cousins were identified. Six samples with evidence of contamination and 20 with highly variable intensity data were excluded; as were 13 with missing call rates ≥2%.
Population structure was investigated by principal component analysis (26). We used a set of 12 021 SNPs selected to have very low levels of linkage disequilibrium and to have minor allele frequencies >0.05 in Caucasians (27). Unrelated genetically inferred European ancestral women and men passing QC were included in the current study. An additional 65 cases in NHS and 65 cases in HPFS suspected of having type 1 diabetes were excluded, leaving 3221 NHS samples (1467 cases and 1754 controls) and 2422 HPFS samples (1124 cases and 1298 controls) for the final analysis.
Of the 909 622 SNP probes on the array, 879 071 passed the Broad's technical QC standards for NHS samples, and 874 517 SNP probes passed this QC stage for HPFS samples. We applied the same QC parameters to both scans: excluding SNPs which were monomorphic, had a missing call rate of ≥2%, more than one discordance, significant deviations from HWE (P < 1 × 10−4) and an MAF of <0.02. Duplicate SNPs (assayed with different probes) were also removed. A total of 704 409 SNPs for NHS samples and 706 040 SNPs for HPFS samples passed QC and were included for analysis. Cluster plots of all significant SNPs considered for replication were also manually inspected for quality assurance.
Statistical analysis
We performed logistic regression to analyze the association between each SNP (coded as counts of minor alleles) and T2D risk using PLINK software (24). Both NHS and HPFS were examined separately. The genomic inflation factor λ was estimated from the median χ2 statistic. To control for potential confounding by population stratification, we performed further analyses by including the top principal components of genetic variation chosen for each study in the models. Adjusting for the top three and four eigenvectors for NHS and HPFS, respectively, made no material difference to the GWA results.
In silico replication study populations
The in silico replications were restricted to samples of European ancestry. All individuals provided informed consent and all studies were approved by local ethics committees.
Diabetes genetics replication and meta-analysis (diagram) consortium
In silico replication data from the DIAGRAM consortium (Supplementary Material) was derived from a meta-analysis of eight T2D GWAS comprising 8130 T2D cases and 38 987 controls of European descent. T2D case–control GWA data from the WTCCC, DGI and FUSION scans [the subjects of a previous joint analysis (5)] were combined with those from the previously published scans performed by DeCODE (28) and DCDG (10), as well as unpublished T2D case–control analyses from the KORA, Rotterdam and EUROSPAN groups. Case and control status were defined using study-specific criteria. The meta-analysis included data from a total of 2 255 857 imputed and genotyped SNPs combined within a fixed effect and additive genetic model. Data from each sample were genome-control-adjusted prior to inclusion in the meta-analysis.
Atherosclerosis risk in communities
The ARIC study is a bi-racial (European American and African American) cohort study of 15 792 persons aged 45–64 years at baseline (1987–1989), randomly chosen from four US communities. Details of the ARIC study samples and GWAS have been described previously (29). Briefly, the replication analysis included 696 T2D cases and 6420 non-cases ascertained at the baseline examination. Diabetes was defined as self-reported physician diagnosis of diabetes, self-reported use of diabetes medications in the last 2 weeks, fasting glucose ≥126 mg/dl or casual glucose ≥200 mg/dl. Non-cases had fasting glucose <110 mg/dl.
ARIC samples were genotyped using the Affymetrix Genome-Wide Human 6.0 array. Prior to SNP imputation, samples with call rate <95% and SNPs with an MAF <0.01 and significant deviations from HWE (P < 1 × 10−4) were excluded. A total of 708 116 SNPs passed the QC criteria. In addition, participants were excluded from the analysis for gender misidentification, high discordance with previously genotyped markers, first-degree relatedness to another included individual and extreme values (i.e. genetic outlier) based on allele sharing and principal components analyses. Only participants of European ancestry were included in the present analysis.
Framingham heart study
The Framingham heart study (FHS) is a community-based study with a family component, including the original (n = 5209, recruited in 1948), offspring (n = 5214, recruited in 1971) and third generation cohorts (n = 4995, recruited in 2002). The replication analysis included 674 cases and 7644 controls from all three generations of Framingham participants. Diabetes was defined as: (i) cohort (Generation 1): casual glucose ≥200 mg/dl at any examination 1–22 or taking diabetes medication (oral or insulin) at any examination; (ii) offspring (Generation 2): fasting plasma glucose ≥126 mg/dl at any examination 1–7 or diabetes treatment at any examination (>99% of diabetes in Framingham is T2D) and (iii) Generation 3: fasting ≥8 h and fasting plasma glucose ≥126 mg/dl at examination 1 or diabetes treatment at examination 1.
FHS samples were genotyped using the Affymetrix 500K and MIPS 50K SNPs. Samples were excluded if the call rate was <97%, heterozygosity >5 standard deviations from the mean or they had an extreme number of Mendelian errors (>1000). SNPs were excluded if call rate <95%, MAF < 0.01 in the combined sample or HWE P < 1 × 10−6. A total of 378 163 genotyped SNPs passed QC criteria, and the genomic inflation factor was estimated as 1.04.
Women's genome health study
The Women's genome health study (WGHS) is an ongoing prospective cohort GWAS that derives from the Women's Health Study and includes more than 25 000 initially healthy North American health care professionals of 45 years of age or older at enrollment (30). The WGHS participants have been followed for over 13 years for development of common disorders including diabetes. The current data were derived from 22 054 WGHS participants with confirmed, self-reported European ancestry for whom genotype information was available at the time of analysis. Cases of diabetes were initially identified by self-report on yearly follow-up questionnaires and were subsequently verified through telephone interview (31). Cases were confirmed if 1 or more of the following conditions were met: (i) the presence of >1 classic symptom of hyperglycemia and either a fasting plasma glucose of ≥126 mg/dl or random plasma glucose ≥200 mg/dl; (ii) in the absence of symptoms, two or more elevated plasma glucose concentrations (fasting ≥126 mg/dl, random ≥200 mg/dl or 2 h OGT ≥200 mg/dl) or (iii) use of insulin or an oral hypoglycemic agent (21).
WGHS samples were genotyped using the HumanHap300 Duo ‘+’ chips or the combination of the HumanHuman300 Duo and iSelect chips (Illumina, San Diego, CA, USA) with the Infinium II protocol. Samples were excluded if the call rate was <98%. SNPs were excluded if the call rate was <90%, MAF was ≤0.01 or calls deviated from HWE (P < 1 × 10−6). A total of 339 913-genotyped SNPs passed QC criteria.
Imputation and analysis in replication studies
DIAGRAM, ARIC and FHS used MACH (v1.0.15/16) to impute up to ∼2.5 million SNPs with NCBI build 36 of Phase II HapMap CEU data (release 22) as the reference panel. MACH dosage files were used for analysis of imputed data. Each SNP was tested for an association with T2D by logistic regression, assuming an additive genetic model and adjusting for age (ARIC), gender (ARIC, FHS), field center, BMI, and physical activity (ARIC), cohort (FHS) and gender × cohort (FHS). FHS further accounted for relatedness in analysis. Genomic inflation factors for imputed results were 1.07, 1.02 and 1.02 for DIAGRAM, ARIC and FHS, respectively.
Meta-analysis
We used a meta-analysis to combine the T2D association results for the discovery stage, including the NHS and HPFS samples, and for combining the discovery and replication samples. For the discovery studies, we combined study-specific β-estimates from GWAs using the inverse of the variance of the study-specific β-estimates to weight the contribution of each study, with the fixed effect model using SAS 9.1. The same method was used to combine the discovery and replication associations. Individual studies were corrected for residual inflation of the test statistic using genomic control methods (32) and the heterogeneity in effect size across studies was tested using the Cochran's Q-test (33). The overall results of the meta-analysis were visualized using HAPLOVIEW.
Quantitative trait analyses
Quantitative trait analyses were carried out in MAGIC (9). Fasting glucose and HOMA-IR results were based on a meta-analysis of 21 studies: CHS, FHS, TwinsUK, DGI, BLSA, FUSION, SardiNIA, CoLaus, GEMS, InCHIANTI, deCODE, NFBC1966, NTR/NESDA, Rotterdam Study, KORA F4, PROCARDIS, Sorbs, ERF, CROAS (Vis Study), ORCADES (Orkney), and MICROS (Tyrol). The results for 2 h glucose were based on nine GWA data sets: ARIC, BLSA, CHS-stage 1&2, CoLaus, DGI, Fenland, FHS, FUSION and Sorbs. Effective sample sizes varied by traits and ranged from 7082 to 46 186. A linear regression model was used to analyze the association of each SNP and the continuous measures of the quantitative traits. The details of genotyping and metabolic measurements for MAGIC studies have been described elsewhere (9).
Real time-PCR and analyses of experimental data
Fasted (24 h), regular chow-fed and high fat-diet-fed mice were age matched (6 months old). Animals were kept on the high fat-diet for 2 months (Bio-Ser, diet # F3282, 5.286 kcal/g, 59.36% fat, 24.48% carbohydrate and 16.16% protein). Primary mouse macrophages were differentiated from bone marrow and maintained in Dulbecco's modification of Eagle's medium and 10% fetal bovine serum. Macrophage activation was achieved by treating cells with interferon-γ (IFN-γ, 10 ng/ml) and LPS (10 ng/ml) for 24 h. RNA was isolated using Trizol (Invitrogen) and reverse transcribed. The human tissue cDNA panel was purchased from Clontech. Real-time quantitative PCR was conducted using SYBR green in triplicate (Applied Biosystems). The expression of the 36B4 housekeeping gene was used for normalization to obtain relative expression levels. Expression values are presented as means ± SE and group means were compared using Student's t-test (two-tailed). Statistical significance was established at P < 0.05. Animal studies were approved by the Harvard Medical Area Standing Committee on Animals.
SUPPLEMENTARY MATERIAL
FUNDING
The NHS/HPFS T2D GWAS (U01HG004399) is a component of a collaborative project that includes 13 other GWAS funded as part of the Gene Environment-Association Studies (GENEVA) under the NIH Genes, Environment and Health Initiative (GEI) (U01HG004738, U01HG004422, U01HG004402, U01HG004729, U01HG004726, U01HG004735, U01HG004415, U01HG004436, U01HG004423, U01HG004728, RFAHG006033) with additional support from individual NIH (NIDCR: U01DE018993, U01DE018903; NIAAA: U10AA008401, NIDA: P01CA089392, R01DA013423; NCI: CA63464, CA54281, CA136792, Z01CP010200). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Genotyping was performed at the Broad Institute of MIT and Harvard, with funding support from the NIH GEI (U01HG04424), and Johns Hopkins University Center for Inherited Disease Research, with support from the NIH GEI (U01HG004438) and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (HHSN268200782096C). Additional funding for the current research was provided by the National Cancer Institute (NCI, P01CA087969, P01CA055075), and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, R01DK058845). The WGHS is supported by HL043851 and HL69757 from the National Heart, Lung, and Blood Institute (NHLBI) and CA047988 from NCI, the Donald W. Reynolds Foundation and the Fondation Leducq, with collaborative scientific support and funding for genotyping provided by Amgen. The ARIC Study is carried out as a collaborative study supported by NHLBI contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367 and R01HL086694 and NIH contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. W.H.L.K. was supported by K01-DK067207. The FHS was partially supported by the NHLBI's Framingham Heart Study (contract no. N01-HC-25195) and its contract with Affymetrix, Inc. for genotyping services (contract no. N02-HL-6-4278), and the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. FHS was also supported by the NIDDK: R01DK078616 to J.B.M., J.D. and J.C.F.; NIDDK K24 DK080140 to J.B.M. and NIDDK Research Career Award K23 DK65978, a Massachusetts General Hospital Physician Scientist Development Award and a Doris Duke Charitable Foundation Clinical Scientist Development Award to J.C.F. L.Q. is supported by National Institutes of Health grants RO1 HL71981, American Heart Association Scientist Development Award and the Boston Obesity Nutrition Research Center (DK46200). M.C.C. is a recipient of a Canadian Institutes of Health Research Fellowship. C.H.L. is supported by the American Heart Association and NIH (R01DK075046). K.J.S. is supported by NIH Roadmap training grant R90DK071507.
Supplementary Material
ACKNOWLEDGEMENT
We are indebted to the staff and participants of the NHS, the HPFS, the ARIC Study, the WGHS Study, the FHS Study, MAGIC and the DIAGRAM Consortium for their dedication and contributions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Stumvoll M., Goldstein B.J., van Haeften T.W. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. doi:10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 2.Qi L., Hu F.B., Hu G. Genes, environment, and interactions in prevention of type 2 diabetes: a focus on physical activity and lifestyle changes. Curr. Mol. Med. 2008;8:519–532. doi: 10.2174/156652408785747915. doi:10.2174/156652408785747915. [DOI] [PubMed] [Google Scholar]
- 3.McCarthy M.I., Zeggini E. Genome-wide association studies in type 2 diabetes. Curr. Diab. Rep. 2009;9:164–171. doi: 10.1007/s11892-009-0027-4. doi:10.1007/s11892-009-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarthy M.I. Progress in defining the molecular basis of type 2 diabetes mellitus through susceptibility-gene identification. Hum. Mol. Genet. 2004;13(Spec. 1):R33–R41. doi: 10.1093/hmg/ddh057. [DOI] [PubMed] [Google Scholar]
- 5.Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. doi:10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 7.Scott L.J., Mohlke K.L., Bonnycastle L.L., Willer C.J., Li Y., Duren W.L., Erdos M.R., Stringham H.M., Chines P.S., Jackson A.U., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unoki H., Takahashi A., Kawaguchi T., Hara K., Horikoshi M., Andersen G., Ng D.P., Holmkvist J., Borch-Johnsen K., Jorgensen T., et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat. Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. doi:10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 9.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. doi:10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. doi:10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis M.C., Qi L., Zhang C., Kraft P., Manson J., Cai T., Hunter D.J., Hu F.B. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann. Intern. Med. 2009;150:541–550. doi: 10.7326/0003-4819-150-8-200904210-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takai T., Nishita Y., Iguchi-Ariga S.M., Ariga H. Molecular cloning of MSSP-2, a c-myc gene single-strand binding protein: characterization of binding specificity and DNA replication activity. Nucleic. Acids Res. 1994;22:5576–5581. doi: 10.1093/nar/22.25.5576. doi:10.1093/nar/22.25.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hynes R.O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. doi:10.1016/0092-8674(92)90115-S. [DOI] [PubMed] [Google Scholar]
- 14.Niu J., Dorahy D.J., Gu X., Scott R.J., Draganic B., Ahmed N., Agrez M.V. Integrin expression in colon cancer cells is regulated by the cytoplasmic domain of the beta6 integrin subunit. Int. J. Cancer. 2002;99:529–537. doi: 10.1002/ijc.10397. doi:10.1002/ijc.10397. [DOI] [PubMed] [Google Scholar]
- 15.Li X., Yang Y., Hu Y., Dang D., Regezi J., Schmidt B.L., Atakilit A., Chen B., Ellis D., Ramos D.M. Alphavbeta6-Fyn signaling promotes oral cancer progression. J. Biol. Chem. 2003;278:41646–41653. doi: 10.1074/jbc.M306274200. doi:10.1074/jbc.M306274200. [DOI] [PubMed] [Google Scholar]
- 16.Shoelson S.E., Lee J., Goldfine A.B. Inflammation and insulin resistance. J. Clin. Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. doi:10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colditz G.A., Hankinson S.E. The Nurses' Health Study: lifestyle and health among women. Nat. Rev. Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. doi:10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 18.Rimm E.B., Giovannucci E.L., Willett W.C., Colditz G.A., Ascherio A., Rosner B., Stampfer M.J. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338:464–468. doi: 10.1016/0140-6736(91)90542-w. doi:10.1016/0140-6736(91)90542-W. [DOI] [PubMed] [Google Scholar]
- 19.Qi L., Kang K., Zhang C., van Dam R.M., Kraft P., Hunter D., Lee C.H., Hu F.B. Fat mass-and obesity-associated (FTO) gene variant is associated with obesity: longitudinal analyses in two cohort studies and functional test. Diabetes. 2008;57:3145–3151. doi: 10.2337/db08-0006. doi:10.2337/db08-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance. National Diabetes Data Group. Diabetes. 1979;28:1039–1057. doi: 10.2337/diab.28.12.1039. [DOI] [PubMed] [Google Scholar]
- 21.American Diabetes Association. Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 22.Hu F.B., Leitzmann M.F., Stampfer M.J., Colditz G.A., Willett W.C., Rimm E.B. Physical activity and television watching in relation to risk for type 2 diabetes mellitus in men. Arch. Intern. Med. 2001;161:1542–1548. doi: 10.1001/archinte.161.12.1542. doi:10.1001/archinte.161.12.1542. [DOI] [PubMed] [Google Scholar]
- 23.Korn J.M., Kuruvilla F.G., McCarroll S.A., Wysoker A., Nemesh J., Cawley S., Hubbell E., Veitch J., Collins P.J., Darvishi K., et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. doi:10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peiffer D.A., Le J.M., Steemers F.J., Chang W., Jenniges T., Garcia F., Haden K., Li J., Shaw C.A., Belmont J., et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. doi:10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. doi:10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu K., Wang Z., Li Q., Wacholder S., Hunter D.J., Hoover R.N., Chanock S., Thomas G. Population substructure and control selection in genome-wide association studies. PLoS One. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. doi:10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., Walters G.B., Styrkarsdottir U., Gretarsdottir S., Emilsson V., Ghosh S., et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007;39:770–775. doi: 10.1038/ng2043. doi:10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 29.Psaty B.M., O'Donnell C.J., Gudnason V., Lunetta K.L., Folsom A.R., Rotter J.I., Uitterlinden A.G., Harris T.B., Witteman J.C.M., Boerwinkle E., et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet. 2009;Vol. 2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. doi:10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridker P.M., Chasman D.I., Zee R.Y., Parker A., Rose L., Cook N.R., Buring J.E. Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin. Chem. 2008;54:249–255. doi: 10.1373/clinchem.2007.099366. doi:10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 31.Pradhan A.D., Manson J.E., Rifai N., Buring J.E., Ridker P.M. C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. Jama. 2001;286:327–334. doi: 10.1001/jama.286.3.327. doi:10.1001/jama.286.3.327. [DOI] [PubMed] [Google Scholar]
- 32.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. doi:10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 33.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. doi:10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.