Abstract
Lamin A is formed from prelamin A by four post-translational processing steps—farnesylation, release of the last three amino acids of the protein, methylation of the farnesylcysteine and the endoproteolytic release of the C-terminal 15 amino acids (including the farnesylcysteine methyl ester). When the final processing step does not occur, a farnesylated and methylated prelamin A accumulates in cells, causing a severe progeroid disease, restrictive dermopathy (RD). Whether RD is caused by the retention of farnesyl lipid on prelamin A, or by the retention of the last 15 amino acids of the protein, is unknown. To address this issue, we created knock-in mice harboring a mutant Lmna allele (LmnanPLAO) that yields exclusively non-farnesylated prelamin A (and no lamin C). These mice had no evidence of progeria but succumbed to cardiomyopathy. We suspected that the non-farnesylated prelamin A in the tissues of these mice would be strikingly mislocalized to the nucleoplasm, but this was not the case; most was at the nuclear rim (indistinguishable from the lamin A in wild-type mice). The cardiomyopathy could not be ascribed to an absence of lamin C because mice expressing an otherwise identical knock-in allele yielding only wild-type prelamin A appeared normal. We conclude that lamin C synthesis is dispensable in mice and that the failure to convert prelamin A to mature lamin A causes cardiomyopathy (at least in the absence of lamin C). The latter finding is potentially relevant to the long-term use of protein farnesyltransferase inhibitors, which lead to an accumulation of non-farnesylated prelamin A.
INTRODUCTION
Lamin A, a key component of the nuclear lamina, is formed from a precursor protein, prelamin A, by four enzymatic post-translational processing steps—farnesylation of a cysteine located four amino acids from the C terminus of the protein, endoproteolytic cleavage of the last three amino acids of the protein, carboxyl methylation of the newly exposed farnesylcysteine and endoproteolytic release of the last 15 amino acids of the protein (including the farnesylcysteine methyl ester) (1–5). The last endoproteolytic cleavage step, which releases mature lamin A, is carried out by ZMPSTE24, a membrane zinc metalloproteinase of the endoplasmic reticulum (6–9).
Defects in prelamin A processing and lamin A biogenesis have been linked to progeroid disorders in humans. The classic pediatric progeroid disease, the Hutchinson–Gilford progeria syndrome (HGPS), is caused by a LMNA point mutation that alters mRNA splicing, resulting in a 50 amino acid internal deletion in prelamin A (10). This deletion does not affect the C-terminal farnesylation or methylation, but it does remove the site for the last ZMPSTE24 cleavage event. The elimination of the final processing step causes an accumulation of a truncated prelamin A, commonly called progerin, that retains a C-terminal farnesylcysteine methyl ester (11,12). Aside from the farnesylcysteine methyl ester, progerin contains four additional C-terminal residues that are not found in mature lamin A.
Progerin adversely affects the integrity and function of the nuclear lamina, leading to misshapen nuclei in cultured cells and a host of aging-like disease phenotypes (10,13). The frequency of misshapen nuclei in HGPS fibroblasts is reduced with a protein farnesyltransferase inhibitor (FTI) (14–18). Recently, Yang et al. (19) generated knock-in mice expressing farnesylated progerin (LmnaHG/+) and tested whether their progeria-like disease phenotypes might be ameliorated by an FTI. In three independent studies, FTI treatment lessened the severity of disease phenotypes in LmnaHG/+ mice (19–21). These mouse experiments prompted a clinical trial of FTI therapy in children with HGPS (22).
The favorable results with FTI therapy in LmnaHG/+ mice have raised hopes for a positive outcome of the FTI trial in humans. However, recent genetic studies in mice have suggested that the FTI treatment strategy may be destined to fall short of a complete cure. Yang et al. (12) generated knock-in mice expressing a ‘non-farnesylated’ version of progerin (LmnanHG/+) and showed that those animals developed severe progeria (although somewhat less severe than in mice producing farnesylated progerin). The finding that non-farnesylated progerin elicits disease suggested that an altered primary structure (i.e. the 50 amino acid deletion)—and not merely the farnesyl lipid—contributes to the pathogenesis of HGPS.
A more severe progeroid syndrome, restrictive dermopathy (RD), is caused by a deficiency in ZMPSTE24. ZMPSTE24 deficiency abolishes the final processing step that releases mature lamin A, resulting in an accumulation of a farnesylated and methylated prelamin A. Unlike HGPS, the prelamin A in RD does not have an internal deletion. It does, however, retain the extra 15 amino acids at its C terminus—the segment that is normally absent in mature lamin A.
Currently, no one understands why the accumulation of prelamin A in RD patients (and ZMPSTE24-deficient mice) elicits a progeroid syndrome. One possibility is that disease is largely due to the retention of the farnesyl lipid at the C terminus of prelamin A. However, there are other possibilities. For example, one could reasonably speculate that the extra 15 amino acids at prelamin A's C terminus—and not the lipid anchor—elicit disease. The proposition that an altered primary structure of prelamin A might elicit progeria is not farfetched, given that non-farnesylated progerin causes progeria in mice (12) and given that certain missense mutations in lamins A/C cause progeria in humans (10,23).
In the current study, we tested the hypothesis that the retention of the last 15 amino acids of prelamin A is sufficient to elicit progeria in mice. To approach this issue, we created and analyzed Lmna knock-in mice that produce exclusively non-farnesylated prelamin A.
RESULTS
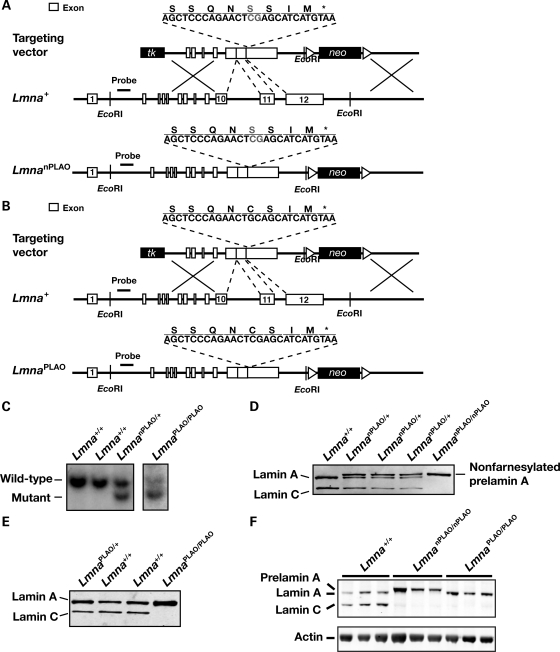
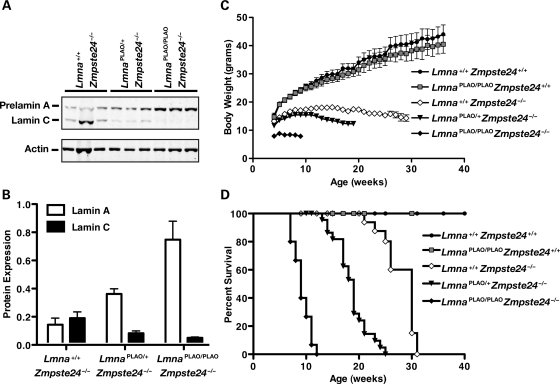
We created Lmna knock-in mice expressing exclusively non-farnesylated prelamin A or exclusively wild-type prelamin A. To create a mutant allele yielding only non-farnesylated prelamin A (‘non-farnesylated prelamin A-only’ allele, LmnanPLAO), we used gene targeting to remove introns 10 and 11 (thereby abolishing lamin C synthesis) and to change the cysteine in the CaaX motif to a serine (abolishing protein prenylation; Fig. 1A). To create an allele yielding only wild-type prelamin A (‘prelamin A-only’ allele, LmnaPLAO), we used the same gene-targeting vector except that the cysteine within the CaaX motif was not altered (Fig. 1B). Targeted embryonic stem (ES) cell clones (Fig. 1C) were used to create chimeric mice, which were bred to produce heterozygous mice (LmnanPLAO/+ and LmnaPLAO/+). Heterozygotes were then intercrossed to produce homozygotes. LmnanPLAO/+ embryonic fibroblasts produced lamin A, lamin C and prelamin A, whereas LmnanPLAO/nPLAO fibroblasts produced exclusively prelamin A (Fig. 1D). Extracts of LmnaPLAO/PLAO embryonic fibroblasts contained only mature lamin A (produced from the processing of prelamin A; Fig. 1E). Western blots of extracts from the hearts of LmnanPLAO/nPLAO and LmnaPLAO/PLAO mice yielded the expected lamin proteins in the expected amounts (Fig. 1F).
Figure 1.
Production of knock-in mice that express a non-farnesylated version of prelamin A or wild-type prelamin A. (A) Gene-targeting strategy to create a mutant Lmna allele, LmnanPLAO, that yields exclusively a non-farnesylated version of prelamin A. This vector removes introns 10 and 11 (thereby abolishing synthesis of lamin C) and introduces a point mutation that changes the cysteine in the CaaX motif to a serine (thereby abolishing protein prenylation). (B) Gene-targeting strategy to create a mutant Lmna allele, LmnaPLAO, that yields exclusively wild-type prelamin A. This vector removes introns 10 and 11 but leaves the CaaX motif intact. (C) Southern blot identification of targeting events in ES cell clones. The genomic DNA was cleaved with EcoRI, and the blot was hybridized with the indicated 5′ flanking probe. (D) Western blot of Lmna+/+, LmnanPLAO/+ and LmnanPLAO/nPLAO fibroblasts with a lamin A/C-specific antibody. (E) Western blot of Lmna+/+, LmnaPLAO/+ and LmnaPLAO/PLAO fibroblasts with a lamin A/C-specific antibody. (F) A western blot, with an antibody against lamin A/C, showing prelamin A, lamin A and lamin C in the hearts of Lmna+/+, LmnanPLAO/nPLAO and LmnaPLAO/PLAO mice (n = 3 mice/genotype). Actin levels were measured as a loading control.
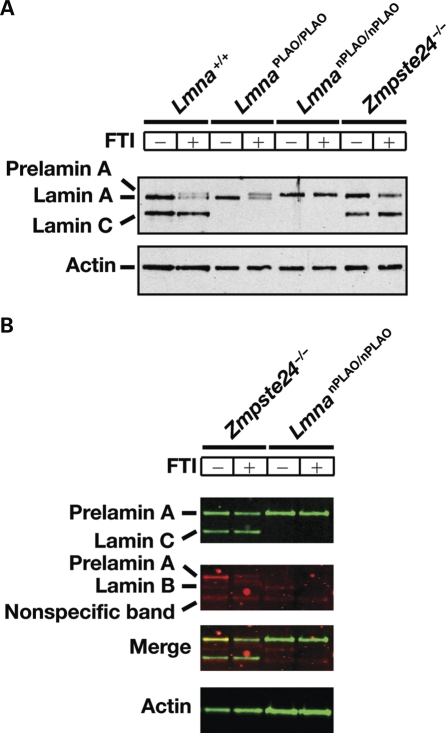
Treatment of fibroblasts with an FTI prevents farnesylation of prelamin A and thus prevents the multistep pathway that converts prelamin A to mature lamin A (12,18). When Lmna+/+ and LmnaPLAO/PLAO fibroblasts were incubated with an FTI, non-farnesylated prelamin A accumulated and was easily detectable by western blotting (i.e. the lamin protein that migrates above mature lamin A; Fig. 2A). The FTI did not affect the migration of the non-farnesylated prelamin A in LmnanPLAO/nPLAO fibroblasts (Fig. 2A). To verify that the prelamin A in LmnanPLAO/nPLAO fibroblasts was not farnesylated, we incubated LmnanPLAO/nPLAO and Zmpste24−/− fibroblasts with a farnesol analog, 8-anilinogeraniol (AG) (24). After entering cells, AG is incorporated into anilinogeranyl diphosphate (AGPP), which is used as a substrate by protein farnesyltransferase (FTase) for modifying CaaX proteins, including prelamin A. The prelamin A in Zmpste24−/− fibroblasts was labeled with AG, and this labeling was largely eliminated with an FTI (Fig. 2B). As expected, there was no AG labeling of the prelamin A in LmnanPLAO/nPLAO fibroblasts, with or without an FTI (Fig. 2B).
Figure 2.
Prelamin A processing in LmnaPLAO/PLAO and LmnanPLAO/nPLAO fibroblasts. (A) Western blots, with an antibody against lamin A/C, showing lamin A, prelamin A and lamin C in Lmna+/+, LmnaPLAO/PLAO, LmnanPLAO/nPLAO and Zmpste24−/− fibroblasts in the presence and the absence of an FTI (ABT-100; 5 µm). Actin was used as a loading control. (B) Metabolic labeling experiment with a farnesol analog, AG to assess the farnesylation of prelamin A. Fibroblasts were incubated with AG (50 µm) in the presence (+) or the absence (−) of an FTI (ABT-100, 5 µm). Western blots were performed on cell extracts with an antibody specific for lamin A/C (green) and an antibody specific for AG (red). As expected, the FTI blocked the incorporation of AG into the prelamin A in Zmpste24−/− fibroblasts. No farnesylation of prelamin A in LmnanPLAO/nPLAO cells was detected. In the AG western blot, lamin B1 was detected (electrophoretic mobility intermediate between lamin A and lamin C); this band disappeared with an FTI. A non-specific band was also observed in the AG western blot (electrophoretic band immediately beneath lamin C).
In wild-type mouse fibroblasts, lamin A was present at the nuclear rim, but significant amounts were also found in the nucleoplasm (Fig. 3A). Similarly, in LmnaPLAO/PLAO fibroblasts, most of lamin A was located at the nuclear rim (Fig. 3A). In LmnanPLAO/nPLAO fibroblasts, some of the prelamin A was at the rim, but substantial amounts were located in the nucleoplasm (Fig. 3A). Also, a higher percentage of the nuclei of LmnanPLAO/nPLAO fibroblasts was misshapen, with blebs and folds (P < 0.0001; Fig. 3B and C). Although the nuclei of LmnanPLAO/nPLAO fibroblasts were misshapen, the stiffness of LmnanPLAO/nPLAO nuclei was not significantly different than nuclei of wild-type fibroblasts (Fig. 3D and E).
Figure 3.
Phenotypes of fibroblasts from LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice. (A) Immunostaining of fibroblasts from Lmna+/+, LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice with lamin A-specific (red) and prelamin A-specific (green) antibodies (clone 3C8). Nuclei were counterstained with DAPI (blue). Images were taken with a confocal microscope. Scale bars represent 15 µm. Bar graphs showing the percentage of nuclei with blebs in LmnaPLAO/PLAO (B) and LmnanPLAO/nPLAO (C) fibroblasts compared with Lmna+/+ control cells. (D and E) Analysis of nuclear stiffness in Lmna+/+ and LmnanPLAO/nPLAO fibroblasts. We examined fibroblasts that had been immortalized by repeated passaging (D) and also low-passage number primary embryonic fibroblasts (E). In both cases, we observed no significant difference in nuclear deformation under uniaxial strain application, compared with Lmna+/+ cells. The extent of nuclear deformations was measured and is expressed as a ratio of nuclear strain to applied membrane strain (normalized nuclear strain).
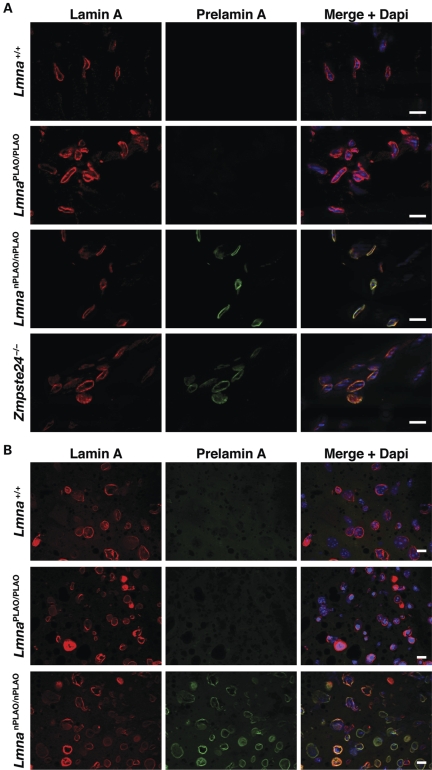
Immunofluorescence microscopy of tissues from wild-type, LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice did not uncover differences in the patterns of lamin A/prelamin A localization. In wild-type and LmnaPLAO/PLAO tissues, virtually all the lamin A was at the nuclear rim (Fig. 4). Similarly, virtually all the prelamin A in LmnanPLAO/nPLAO tissues was located at the nuclear rim (Fig. 4).
Figure 4.
Nuclear rim localization of lamin A and prelamin A in tissues of LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice. (A) Immunostaining of heart from Lmna+/+, LmnaPLAO/PLAO, LmnanPLAO/nPLAO and Zmpste24−/− mice with lamin A-specific (red) and prelamin A-specific (clone 3C8; green) antibodies. Nuclei were counterstained with DAPI (blue). Scale bars represent 20 µm. (B) Immunostaining of liver from Lmna+/+, LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice with lamin A-specific (red) and prelamin A-specific (clone 7G11; green) antibodies. Nuclei were counterstained with DAPI (blue). Scale bars represent 20 µm.
Body weight curves in male Lmna+/+, LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice were not significantly different (Fig. 5A), but female LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice appeared to have slightly lower body weights than Lmna+/+ littermates (Fig. 5B). The survival of LmnaPLAO/PLAO mice was not different than that of wild-type mice. Male and female LmnanPLAO/nPLAO mice appeared to be entirely normal for the first 20 weeks of life. After that, however, we observed premature death in males and females; all male LmnanPLAO/nPLAO mice succumbed by 44 weeks of age (median survival, 38.5 weeks) and all female mice succumbed by 80 weeks (median survival, 49.5 weeks). Kaplan–Meier survival analysis revealed that the survival of LmnanPLAO/nPLAO mice was significantly reduced, compared with Lmna+/+ or LmnaPLAO/PLAO mice (P < 0.0001 for both males and females; Fig. 5C and D).
Figure 5.
Body weight and survival curves for LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice. (A) Body weight curves for male Lmna+/+ (n = 27), LmnaPLAO/PLAO (n = 12) and LmnanPLAO/nPLAO mice (n = 20). (B) Body weight curves for female Lmna+/+ (n = 28), LmnaPLAO/PLAO (n = 12) and LmnanPLAO/nPLAO mice (n = 22). Body weights of female LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice were slightly lower than those of Lmna+/+ mice (P < 0.001). (C) Kaplan–Meier survival plots for male Lmna+/+ (n = 21), LmnaPLAO/PLAO (n = 12) and LmnanPLAO/nPLAO mice (n = 12). Survival time for male LmnanPLAO/nPLAO mice was shorter than for Lmna+/+ or LmnaPLAO/PLAO mice (P < 0.0001). (D) Kaplan–Meier survival plots for female Lmna+/+ (n = 33), LmnaPLAO/PLAO (n = 16) and LmnanPLAO/nPLAO mice (n = 38). Survival time for female LmnanPLAO/nPLAO mice was shorter than that for Lmna+/+ or LmnaPLAO/PLAO mice (P < 0.0001).
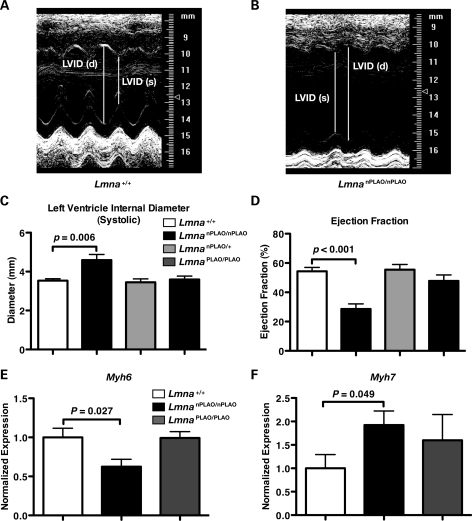
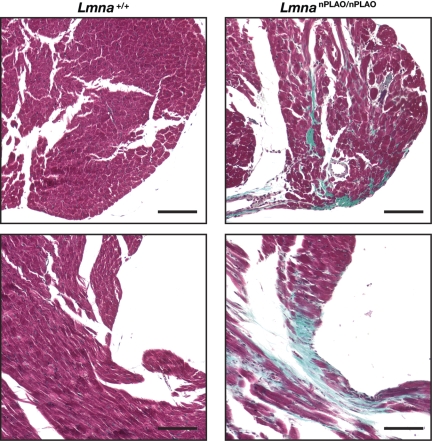
The premature death in LmnanPLAO/nPLAO mice suggested that non-farnesylated prelamin A might elicit cardiomyopathy. Indeed, echocardiography revealed that LmnanPLAO/nPLAO mice had a dilated cardiomyopathy (Fig. 6A and B), with an increased left ventricular (LV) chamber size during systole (Fig. 6C) and a reduced LV ejection fraction (Fig. 6D). In keeping with these findings, the expression of myosin heavy chain alpha (Myh6), which typically falls in the setting of cardiomyopathy (25), was reduced in LmnanPLAO/nPLAO mice (Fig. 6E). The expression of myosin heavy chain beta (Myh7), a marker of cardiomyopathy (25), was increased in LmnanPLAO/nPLAO mice (Fig. 6F). This cardiomyopathy phenotype, with a greater severity in males, is similar to the phenotype of Lmna knock-in mice carrying an H222P substitution (26). LmnaH222P/H222P mice were reported to have a ‘marked increase in fibrosis’ in the left and right ventricles. We examined cardiac pathology in an LmnanPLAO/nPLAO mouse that had severe cardiomyopathy by echocardiography and found significant fibrosis in the papillary muscle of the left ventricle, as judged by a Gomori trichrome stain (Fig. 7). No such fibrosis was observed in the papillary muscle of an age-matched Lmna+/+ control mouse (Fig. 7). We would characterize the fibrosis in the LmnanPLAO/nPLAO mouse as being mild–moderate, and in some mice, fibrosis was minimal.
Figure 6.
Dilated cardiomyopathy in LmnanPLAO/nPLAO mice. Representative M-mode echocardiograms of Lmna+/+ (A) and LmnanPLAO/nPLAO (B) mice. Lines depict the systolic (s) and diastolic (d) LV internal diameters (LVIDs). Quantification of systolic LVID (C) and LV ejection fraction (D) in Lmna+/+, LmnanPLAO/nPLAO, LmnanPLAO/+ and LmnaPLAO/PLAO mice (n = 6/genotype). Bar graphs show mean ± SEM. Myh6 (E) and Myh7 (F) expression levels in the hearts of Lmna+/+, LmnanPLAO/nPLAO and LmnaPLAO/PLAO mice (n = 7/genotype). Expression levels were measured by quantitative RT–PCR and normalized to Gapdh (mean ± SEM).
Figure 7.
Fibrosis within an LV papillary muscle from a male LmnanPLAO/nPLAO mouse, as judged by a Gomori trichrome stain. This LmnanPLAO/nPLAO mouse had severe LV dysfunction, as judged by echocardiography. Fibrotic tissue is stained green. No papillary muscle fibrosis was observed in an age- and sex-matched littermate Lmna+/+ mouse. Scale bars represent 100 µm.
In humans, lamin-related progeroid syndromes are associated with osteolytic lesions in bones, a severe growth defect, and reduced survival (10,27,28). Zmpste24−/− mice share these phenotypes. Indeed, osteolytic lesions and spontaneous rib fractures are prominent in Zmpste24−/− mice, which accumulate the farnesylated form of prelamin A (Fig. 8). Interestingly, LmnanPLAO/nPLAO mice did not have osteolytic lesions in any of their bones, and rib fractures were never observed (Fig. 8). Bone lesions were also absent in LmnaPLAO/PLAO mice (Fig. 8). Zmpste24−/− and LmnanPLAO/nPLAO mice also differed in their body weight curves. Zmpste24−/− mice were far smaller than wild-type mice, and they invariably die from severe, progressive inanition (6,8,9; also see Fig. 9). At 5–6 months of age, Zmpste24−/− mice typically weigh only 11–17 g (6,8,9; also see Fig. 9), whereas 5–6-month-old LmnanPLAO/nPLAO typically weigh 30–40 g (Fig. 5). Thus, the non-farnesylated prelamin A in LmnanPLAO/nPLAO mice elicits none of the progeria-like disease phenotypes found in Zmpste24−/− mice, but it does cause cardiomyopathy.
Figure 8.
Surface renderings of µCT scans showing the lack of bone abnormalities in LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice. (A) Scans showing characteristic rib fractures in 7-month-old Zmpste24−/− mice. Arrowheads indicate rib fractures and surrounding callus. No rib fractures were observed in Lmna+/+ (B), LmnaPLAO/PLAO (C) or LmnanPLAO/nPLAO (D) mice. Scale bars represent 2.5 mm.
Figure 9.
The LmnaPLAO allele worsens the progeria-like disease phenotypes in Zmpste24−/− mice in a dose-dependent manner. (A) Western blots (with an antibody against lamin A/C) showing prelamin A and lamin C in the kidneys of Lmna+/+Zmpste24−/−, LmnaPLAO/+Zmpste24−/− and LmnaPLAO/PLAOZmpste24−/− mice (n = 3/genotype). (B) Quantitative analysis of prelamin A and lamin C expression in the kidney in Lmna+/+Zmpste24−/−, LmnaPLAO/+Zmpste24−/− and LmnaPLAO/PLAOZmpste24−/− mice (bar graphs depict prelamin A:actin and lamin C:actin ratios). (C) Body weight curves for Lmna+/+Zmpste24+/+ (n = 25), LmnaPLAO/PLAOZmpste24+/+ (n = 17), Lmna+/+Zmpste24−/− (n = 24), LmnaPLAO/+Zmpste24−/− (n = 24) and LmnaPLAO/PLAOZmpste24−/− (n = 15) mice. Body weight curves for Lmna+/+Zmpste24+/+ and LmnaPLAO/PLAOZmpste24+/+ mice were not significantly different. Body weight curves for all other genotypes were significantly different from wild-type mice and from each other (P < 0.01 between Lmna+/+Zmpste24−/− and LmnaPLAO/+Zmpste24−/− mice; P < 0.001 for all other comparisons). (D) Kaplan–Meier survival plots for Lmna+/+Zmpste24+/+ (n = 26), LmnaPLAO/PLAOZmpste24+/+ (n = 16), Lmna+/+Zmpste24−/− (n = 23), LmnaPLAO/+Zmpste24−/− (n = 24) and LmnaPLAO/PLAOZmpste24−/− (n = 15) mice. Survival times for Lmna+/+Zmpste24+/+ and LmnaPLAO/PLAOZmpste24+/+ mice were not statistically different. Survival times for all other genotypes were significantly different from the wild-type mice and from each other (P < 0.001 for all comparisons).
The LmnaPLAO allele yields exclusively prelamin A, whereas a wild-type Lmna allele yields both lamin C and prelamin A. Because all of the output of the LmnaPLAO allele is channeled into prelamin A production, we predicted that the LmnaPLAO allele would increase prelamin A levels in Zmpste24−/− mice and worsen disease phenotypes. Indeed, this was the case. Prelamin A levels increased with each additional LmnaPLAO allele (Fig. 9A and B), and both body weight and survival worsened (Fig. 9C and D).
DISCUSSION
Finding progeria-like disease phenotypes in ‘non-farnesylated progerin mice’ (12) was intriguing because it suggested that the primary structure of progerin, and not merely its farnesyl lipid anchor, could be central to the pathogenesis of HGPS. Those findings immediately raised parallel questions about the pathogenesis of RD (ZMPSTE24 deficiency). When ZMPSTE24 is absent, are the disease phenotypes elicited by the retention of the hydrophobic lipid anchor on prelamin A or are they due to the extra 15 amino acids at the protein's C terminus? To address this issue, we created ‘knock-in mice’ (LmnanPLAO/nPLAO) that synthesize exclusively non-farnesylated prelamin A. Given the unequivocal findings with ‘non-farnesylated progerin mice’, we hypothesized that the ‘non-farnesylated prelamin A mice’ would manifest all the same progeria-like disease phenotypes found in Zmpste24−/− mice. To our surprise, this hypothesis was proven wrong. Even though LmnanPLAO/nPLAO mice expressed very high levels of non-farnesylated prelamin A, they bore no resemblance to Zmpste24−/− mice. Unlike Zmpste24−/− mice, LmnanPLAO/nPLAO mice maintained a normal or near-normal body weight, and they had no osteolytic lesions or bone fractures. In contrast to the Zmpste24−/− mice, LmnanPLAO/nPLAO mice did develop a dilated cardiomyopathy. The most parsimonious interpretation of our findings is that the lipid anchor on prelamin A is central to disease phenotypes in Zmpste24−/− mice, whereas the extra 15 amino acid residues at the end of prelamin A are not. In addition, we conclude that non-farnesylated prelamin A leads to a distinct disease phenotype, dilated cardiomyopathy, which is not found in Zmpste24−/− mice. Clearly, non-farnesylated prelamin A is not functionally equivalent to mature lamin A.
The involvement of Lmna in cardiomyopathy is not without precedent. In humans, many LMNA missense mutations cause muscular dystrophy and/or dilated cardiomyopathy with conduction system disease (29–31). Several of the missense mutations observed in humans have been introduced into mouse Lmna, and homozygous mice for those mutations manifest cardiomyopathy and premature death (26,32). The available data suggest that lamin A/C-related cardiomyopathy could be caused by increased nuclear fragility and changes in gene expression, in particular the MAPK pathway (33). Interestingly, no one has yet found a case of cardiomyopathy due to a missense mutation involving the last 15 amino acids of prelamin A, nor has anyone identified a CaaX motif mutation leading to the production of non-farnesylated prelamin A.
The conclusion that non-farnesylated prelamin A leads to cardiomyopathy, but not progeria, is subject to several caveats. First, we would point out that LmnanPLAO/nPLAO mice are incapable of producing lamin C. Thus, one could conceivably postulate that the cardiomyopathy is elicited by the inability to make that lamin isoform. However, this explanation seems unlikely, given the complete absence of cardiomyopathy or decreased survival in LmnaPLAO/PLAO mice (which also cannot make lamin C). It remains a formal possibility that cardiomyopathy is caused by non-farnesylated prelamin A, but only in the absence of lamin C. Second, it is important to note that prelamin A farnesylation was eliminated in LmnanPLAO/nPLAO mice by introducing a cysteine-to-serine substitution in prelamin A's CaaX motif. That amino acid substitution is remarkably subtle, amounting to replacing a single sulfur atom with an oxygen atom. Nevertheless, one could propose that this amino acid substitution is solely responsible for the cardiomyopathy. Against that idea, however, is the absence of cardiomyopathy in ‘non-farnesylated progerin mice’ (12), which share the identical cysteine-to-serine substitution in the CaaX motif.
The ideal scenario would be to create ‘non-farnesylated prelamin A mice’ without introducing any mutation into prelamin A's CaaX motif. Unfortunately, we are not aware of any strategy that would make this possible. In theory, one could achieve this goal by eliminating the expression of FTase, a genetic intervention that would lead to the accumulation of non-farnesylated prelamin A (34). For such an approach to be practical, however, FTase deficiency could not elicit any of its own untoward consequences. Unfortunately, this is not the case. FTase has >60 protein substrates (35,36), and a deficiency in this enzyme elicits pathology in multiple cell types. A complete deficiency of FTase causes early embryonic lethality (37). Although initially reported that a knockout of FTase in adult mice leads to little if any pathology (37), it is now clear that this is not the case. FTase deficiency in skin keratinocytes yields severe alopecia (34), and an absence of FTase in liver causes severe hepatocellular disease (Shao H. Yang, unpublished observations). Thus, an attempt to produce ‘non-farnesylated prelamin A mice’ by eliminating FTase expression would be fraught with significant complications.
The absence of disease phenotypes in LmnaPLAO/PLAO mice implies that lamin C synthesis is dispensable in the laboratory mouse. In earlier studies, Fong et al. (38) found no disease phenotypes in mice synthesizing exclusively lamin C (LmnaLCO/LCO), leading them to conclude that lamin A synthesis is dispensable. An absence of ‘both’ lamin A and lamin C causes muscular dystrophy and cardiomyopathy (39). Thus, a key lesson from the LmnaPLAO/PLAO and LmnaLCO/LCO models is that lamin A alone, or lamin C alone, is capable of preventing disease. Also, when we state that each isoform is ‘dispensable’, we simply mean only that an absence of one isoform does not lead to obvious disease. In the future, it is conceivable that newer and more sensitive assays could uncover unique and important physiologic roles for each of the two protein isoforms.
Dogma has held that prelamin A's hydrophobic lipid anchor is important for the targeting of the protein to the inner nuclear membrane (which is adjacent to the nuclear lamina). In part, this conclusion rested on the observation that prelamin A appears in the nucleoplasm of cultured fibroblasts, rather than at the nuclear rim, when protein farnesylation is inhibited (40). Also, two groups reported that when a ‘non-farnesylated prelamin A construct’ was transfected into cells, the prelamin A did not reach the nuclear rim (5,41). A third study found that some non-farnesylated prelamin A reached the nuclear rim, but with delayed kinetics (42). Recently, however, the conclusion that farnesylation is important for the nuclear envelope targeting of prelamin A has been challenged. Lamin C, which is never farnesylated, reaches the nuclear rim, even in the absence of lamin A, suggesting that farnesylation is not a requirement for nuclear rim localization of that nuclear lamin (38). Lee et al. (34) generated keratinocyte-specific FTase knockout mice and showed that FTase-deficient keratinocytes accumulate substantial amounts of non-farnesylated prelamin A. Taking advantage of a new monoclonal antibody against mouse prelamin A, Lee et al. (34) showed that the non-farnesylated prelamin A in FTase-deficient keratinocytes is located largely or exclusively at the nuclear rim, indistinguishable from the localization of mature lamin A in wild-type keratinocytes. Little if any non-farnesylated prelamin A was found in the nucleoplasm. In the current study, we observed consistent findings. As judged by immunofluorescence microscopy, virtually all of the non-farnesylated prelamin A in the heart and liver of LmnanPLAO/nPLAO mice was located at the nuclear rim. Indeed, the distribution of non-farnesylated prelamin A in those tissues was identical to the distribution of mature lamin A in LmnaPLAO/PLAO tissues. Thus, prelamin A's farnesyl lipid anchor is not required for targeting to the nuclear rim. In LmnanPLAO/nPLAO fibroblasts, non-farnesylated prelamin A was also found at the nuclear rim, although the amount was somewhat less than the amount of mature lamin A at the nuclear rim in wild-type fibroblasts.
Several years ago, Fong et al. (8,38) showed that the disease phenotypes in Zmpste24−/− mice could be eliminated by reducing farnesyl-prelamin A production, either by introducing an Lmna knockout allele or an LmnaLCO allele (an allele yielding exclusively lamin C). Those findings supported the concept that farnesyl-prelamin A is toxic and provided the first suggestion that strategies to reduce prelamin A production could be therapeutically useful in progeria. The current studies with the LmnaPLAO allele further underscore the toxicity of farnesyl-prelamin A. Because the LmnaPLAO allele yields exclusively prelamin A, Zmpste24−/− mice harboring that allele synthesize increased amounts of farnesyl-prelamin A. Larger amounts of farnesyl-prelamin A clearly lead to more toxicity: the disease phenotypes in Zmpste24−/−LmnaPLAO/PLAO mice were far more severe than those in Zmpste24−/−LmnaPLAO/+ mice, which in turn were more severe than those in Zmpste24−/−Lmna+/+ mice.
From the standpoint of therapy, the current study would suggest that an FTI has the potential to be a double-edged sword. On the one hand, our findings suggest that reducing protein farnesylation with an FTI might be useful, in that non-farnesylated prelamin A appears to be devoid of the capacity to elicit progeria-like disease phenotypes. That conclusion is consistent with an earlier study showing that an FTI ameliorates disease in Zmpste24−/− mice (43). On the other hand, our studies offer a darker possibility—that non-farnesylated prelamin A carries its own distinctive toxicity, dilated cardiomyopathy. Thus, it is theoretically possible that any accumulation of non-farnesylated prelamin A, such as would occur with a long-term FTI therapy, could adversely affect the heart. Fortunately, we are not aware of any reports of cardiomyopathy with FTI therapy in humans, nor have we observed any evidence of cardiomyopathy when treating mice with an FTI, but any physician contemplating long-term FTI treatment for progeroid disorders should be attuned to this possibility.
MATERIALS AND METHODS
Generation of knock-in mice
To create a mutant Lmna allele yielding only non-farnesylated prelamin A (LmnanPLAO), we used a gene-targeting vector similar to one used to create a mutant allele yielding exclusively progerin (LmnaHG) (18). Introns 10 and 11 of Lmna were deleted by site-directed mutagenesis (QuikChange; Stratagene, La Jolla, CA, USA). The oligonucleotide 5′-GATGGAGAAGAGCTCCTCCATCACCACCGTGGTTCCCACTGCAGCGGCTCGGGGGACCCC-3′ and its reverse complement were used to delete intron 10; oligonucleotide 5′-CTCCTGGGCAACTCCAGTCCCCGGAGCCAGAGCTCCCAGAACTGCAGCATCATGTAATCT-3′ and its reverse complement were used to delete intron 11. Also, a point mutation was introduced into exon 12 that changes the cysteine of prelamin A's CaaX motif to a serine (i.e. −CSIM to −SSIM). The mutation was introduced into the 5′ arm of the vector by site-directed mutagenesis with the QuikChange kit (Stratagene) and oligonucleotide 5′-CAGAGCTCCCAGAACTCGAGCATCATGTAATCT-3′ (and the complementary primer). To create a mutant Lmna allele yielding only wild-type prelamin A (LmnaPLAO), the same gene-targeting vector was used, except that the CaaX motif was not changed. The integrity of both gene-targeting vectors was verified by restriction endonuclease digestion and DNA sequencing.
Gene-targeting vectors were linearized with NotI and electroporated into 129/OlaHsd mouse ES cells. Targeted clones were identified by Southern blotting with EcoRI-cleaved genomic DNA with a 348 bp 5′ flanking probe (12). Targeted ES cells were microinjected into C57BL/6 blastocysts, and the resulting chimeras were bred with C57BL/6 females to generate LmnaPLAO/+ and LmnanPLAO/+ mice. Heterozygous mice were intercrossed to generate homozygous mice (LmnaPLAO/PLAO and LmnanPLAO/nPLAO). LmnaPLAO/+ mice were also bred with Zmpste24+/− mice (44) and their offspring intercrossed to obtain Lmna+/+Zmpste24+/+, LmnaPLAO/PLAOZmpste24+/+, Lmna+/+Zmpste24−/−, LmnaPLAO/+Zmpste24−/− and LmnaPLAO/PLAOZmpste24−/− mice. Genotyping of mice was performed by PCR using DNA from tail biopsies (18). All mice were fed a chow diet and housed in a virus-free barrier facility with a 12 h light–dark cycle. All mouse experiments were approved by UCLA's Animal Research Committee.
Western blots and metabolic labeling
Primary mouse embryonic fibroblasts were prepared from embryonic day 13.5 embryos (6). To block protein farnesylation, we added a highly selective FTI, ABT-100 (5 µm), to the cell culture medium.
Urea-soluble extracts were prepared as described previously (45). Extracts were size-fractionated on 4–12% gradient polyacrylamide Bis–Tris gels (Invitrogen), and the separated proteins transferred to nitrocellulose membranes for western blotting. Antibody dilutions were 1:200 for anti-lamin A/C goat IgG (sc-6215, Santa Cruz Biotechnology), 1:1000 for anti-actin goat IgG (sc-1616, Santa Cruz Biotechnology) and 1:5000 for IRDye 800 anti-goat IgG (Rockland Immunochemicals). The IR-coupled antibodies were detected with an Odyssey infrared imaging scanner and quantified according to the manufacturer's instructions.
To assess protein farnesylation, fibroblasts were incubated for 48 h with an analog of farnesol, AG (50 µm in DMSO) (24). AG is incorporated into AGPP and used as a substrate by protein farnesyltransferase. The incorporation of AG into CaaX proteins such as prelamin A can be detected by western blotting with an AG-specific mouse monoclonal antibody (1:5000) (24) and an IRDye 680 anti-mouse IgG (1:2500) (Li-Cor).
Analysis of nuclear shape in fibroblasts
To assess nuclear shape abnormalities, early-passage mouse embryonic fibroblasts were grown on coverslips and then fixed and permeabilized (8). Cells were incubated with antibodies against lamin A (1:50, sc-20680, Santa Cruz Biotechnology) and prelamin A (1:50, clone 3C8) for 2 h. After washing, cells were incubated with Alexa Fluor 568-conjugated anti-rabbit antibody (1:500, Jackson ImmunoResearch Laboratories), Alexa Fluor 488-conjugated anti-rat antibody (1:500, Invitrogen) and DAPI to visualize DNA. Images were obtained on an Axiovert 200 MOT microscope (Zeiss) or on a Leica SP2 1P-FCS confocal microscope and nuclear shape was assessed (∼1000 cells/genotype).
Analysis of nuclear stiffness in fibroblasts
Nuclear strain studies were performed as described previously (46), with the following modification. Cells were subjected to ∼20% uniaxial strain rather than biaxial strain. Membrane and nuclear deformations were computed based on bright-field and fluorescent images acquired before, during and after strain application. Normalized nuclear strain was defined as the ratio of nuclear strain to membrane strain to compensate for small variations in applied membrane strain. Cells that were damaged during or after strain application were excluded from the analysis.
Immunohistochemistry
To assess the location of lamin A and prelamin A in tissues, 6–10-µm-thick frozen sections were prepared and placed on glass slides. The sections were fixed with cold (−20°C) methanol for 10 min, rinsed with acetone, permeabilized with 0.1% Tween in PBS, and then incubated in a PBS blocking buffer containing 0.1% Tween and 5% donkey serum for 1 h at room temperature. The sections were incubated with primary antibodies in blocking buffer overnight at a dilution of 1:50 for a rabbit anti-lamin A antibody (Santa Cruz Biotechnology), 1:50 for a rat anti-prelamin A monoclonal antibody (3C8) and 1:50 for a rat anti-prelamin A antibody (7G11). Monoclonal antibodies 7G11 and 3C8 were derived from rats immunized with a C-terminal peptide of mouse prelamin A (34). After washing the sections, they were incubated for 1 h at room temperature with fluorescently labeled secondary antibodies (1:200 Alexa488-labeled anti-rat IgG; 1:200 Alexa568-labeled anti-rabbit IgG). The sections were then washed, post-fixed with 4% paraformaldehyde in PBS and stained with DAPI to visualize DNA. Images were obtained with an Axiovert 200 MOT microscope equipped with an Apotome and processed with AxoVision 4.2 software (Zeiss, Germany).
Analysis of mouse phenotypes
Body weights were measured weekly for 20 weeks and then every other week; numbers of surviving mice were recorded weekly. To assess osteolytic lesions and rib fractures, the thoracic cages of Zmpste24−/−, Lmna+/+, LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice were imaged by X-ray microtomography (µCT) with the Skyscan 1172 system (Skyscan).
Echocardiograms
Echocardiograms were obtained on Lmna+/+, LmnaPLAO/PLAO and LmnanPLAO/nPLAO mice anesthetized with 2% isoflurane with the Vevo 770 Imaging system and 35 mHz scan probe (Visualsonics). Data were analyzed using the Vevo analysis program. LV chamber diameters during systole (LV Vol:s) and diastole (LV Vol:d) were measured from M-mode recordings. LV ejection fraction (%) was calculated as [(LV Vol:d − LV Vol:s)/LV Vol:d] × 100.
Histology
To assess fibrosis, Lmna+/+ and LmnanPLAO/nPLAO mice were perfused with 4% paraformaldehyde, and the hearts were removed and post-fixed in 3% formalin. Fixed tissues were embedded in paraffin, sectioned and stained with Gomori trichrome and hematoxylin.
Quantitative reverse transcribed–PCR
Total RNA was prepared from mouse hearts with the RNeasy kit (Qiagen). RNA was treated with DNase I (Ambion) and reverse transcribed (RT) with a mixture of random primers, oligo(dT) and Superscript III (Invitrogen). Primers 5′-CGCATCAAGGAGCTCACC-3′ and 5′-CCTGCAGCCGCATTAAGT-3′ were used to amplify the mouse Myh6 cDNA, and primers 5′-GGAAGACAGGAAGAACCTACTGC-3′ and 5′-AACTTGGACAGGTTGGTGTTG-3′ were used to amplify mouse Myh7 cDNA. Expression was normalized to GAPDH (amplified with the primers 5′- GCACAGTCAAGGCCGAGAAT-3′ and 5′-GCCTTCTCCATGGTGGTGAA-3′). PCRs were performed in triplicate on a 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). Gene-expression levels were calculated with the comparative cycle threshold (CT) method.
Statistical analyses
Differences in misshapen nuclei were analyzed using the χ2 test. Variations in nuclear strain assays were analyzed by one-way ANOVA with Tukey's multiple comparison post-test. The number of surviving male and female mice was recorded weekly. Differences were assessed by the log-rank (Mantel–Cox) and the Gehan–Breslow–Wilcoxon tests. Body weight curves were compared with repeated-measures ANOVA. Echocardiographic data and gene-expression data were analyzed with the Student's t-test.
FUNDING
This work was supported by the National Institutes of Health (HL76839, HL86683, HL089781, GM66152, AG035626, HL082792, NS059348); a March of Dimes Grant (6-FY2007-1012); the Ellison Medical Foundation Senior Scholar Program; a post-doctoral fellowship award from the American Heart Association, Western States Affiliate; and a Scientist Development Grant from the American Heart Association (0635359N).
ACKNOWLEDGEMENT
We thank Dr David Frost from Abbott Pharmaceuticals for ABT-100.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Casey P.J. Biochemistry of protein prenylation. J. Lipid Res. 1992;33:1731–1740. [PubMed] [Google Scholar]
- 2.Casey P.J., Seabra M.C. Protein prenyltransferases. J. Biol. Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 3.Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu. Rev. Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. doi:10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- 4.Davies B.S., Fong L.G., Yang S.H., Coffinier C., Young S.G. The posttranslational processing of prelamin A and disease. Annu. Rev. Genomics Hum. Genet. 2009;10:153–174. doi: 10.1146/annurev-genom-082908-150150. doi:10.1146/annurev-genom-082908-150150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinensky M., Fantle K., Trujillo M., McLain T., Kupfer A., Dalton M. The processing pathway of prelamin A. J. Cell Sci. 1994;107:61–67. doi: 10.1242/jcs.107.1.61. [DOI] [PubMed] [Google Scholar]
- 6.Bergo M.O., Gavino B., Ross J., Schmidt W.K., Hong C., Kendall L.V., Mohr A., Meta M., Genant H., Jiang Y., et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl Acad. Sci. USA. 2002;99:13049–13054. doi: 10.1073/pnas.192460799. doi:10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corrigan D.P., Kuszczak D., Rusinol A.E., Thewke D.P., Hrycyna C.A., Michaelis S., Sinensky M.S. Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem. J. 2005;387:129–138. doi: 10.1042/BJ20041359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fong L.G., Ng J.K., Meta M., CotÇ N., Yang S.H., Stewart C.L., Sullivan T., Burghardt A., Majumdar S., Reue K., et al. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc. Natl Acad. Sci. USA. 2004;101:18111–18116. doi: 10.1073/pnas.0408558102. doi:10.1073/pnas.0408558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pendas A.M., Zhou Z., Cadinanos J., Freije J.M.P., Wang J., Hultenby K., Astudillo A., Wernerson A., Rodriguez F., Tryggvason K., et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. doi:10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dechat T., Shimi T., Adam S.A., Rusinol A.E., Andres D.A., Spielmann H.P., Sinensky M.S., Goldman R.D. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc. Natl Acad. Sci. USA. 2007;104:4955–4960. doi: 10.1073/pnas.0700854104. doi:10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang S.H., Andres D.A., Spielmann H.P., Young S.G., Fong L.G. Progerin elicits disease phenotypes of progeria in mice whether or not it is farnesylated. J. Clin. Invest. 2008;118:3291–3300. doi: 10.1172/JCI35876. doi:10.1172/JCI35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C.L., Munnich A., Le Merrer M., et al. Lamin A truncation in Hutchinson–Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. doi:10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 14.Capell B.C., Erdos M.R., Madigan J.P., Fiordalisi J.J., Varga R., Conneely K.N., Gordon L.B., Der C.J., Cox A.D., Collins F.S. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. USA. 2005;102:12879–12884. doi: 10.1073/pnas.0506001102. doi:10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glynn M.W., Glover T.W. Incomplete processing of mutant lamin A in Hutchinson–Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum. Mol. Genet. 2005;14:2959–2969. doi: 10.1093/hmg/ddi326. doi:10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- 16.Mallampalli M.P., Huyer G., Bendale P., Gelb M.H., Michaelis S. Inhibiting farnesylation reverses the nuclear morphology defect in a HeLa cell model for Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. USA. 2005;102:14416–14421. doi: 10.1073/pnas.0503712102. doi:10.1073/pnas.0503712102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toth J.I., Yang S.H., Qiao X., Beigneux A.P., Gelb M.H., Moulson C.L., Miner J.H., Young S.G., Fong L.G. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl Acad. Sci. USA. 2005;102:12873–12878. doi: 10.1073/pnas.0505767102. doi:10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S.H., Bergo M.O., Toth J.I., Qiao X., Hu Y., Sandoval S., Meta M., Bendale P., Gelb M.H., Young S.G., et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson–Gilford progeria syndrome mutation. Proc. Natl Acad. Sci. USA. 2005;102:10291–10296. doi: 10.1073/pnas.0504641102. doi:10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S.H., Meta M., Qiao X., Frost D., Bauch J., Coffinier C., Majumdar S., Bergo M.O., Young S.G., Fong L.G. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson–Gilford progeria syndrome mutation. J. Clin. Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. doi:10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S.H., Chang S.Y., Andres D.A., Spielmann H.P., Young S.G., Fong L.G. Assessing the efficacy of protein farnesyltransferase inhibitors in mouse models of progeria. J. Lipid Res. 2010;51:400–405. doi: 10.1194/jlr.M002808. doi:10.1194/jlr.M002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S.H., Qiao X., Fong L.G., Young S.G. Treatment with a farnesyltransferase inhibitor improves survival in mice with a Hutchinson–Gilford progeria syndrome mutation. Biochim. Biophys. Acta. 2008;1781:36–39. doi: 10.1016/j.bbalip.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kieran M.W., Gordon L., Kleinman M. New approaches to progeria. Pediatrics. 2007;120:834–841. doi: 10.1542/peds.2007-1356. doi:10.1542/peds.2007-1356. [DOI] [PubMed] [Google Scholar]
- 23.Verstraeten V.L.R.M., Broers J.L.V., van Steensel M.A.M., Zinn-Justin S., Ramaekers F.C.S., Steijlen P.M., Kamps M., Kuijpers H.J.H., Merckx D., Smeets H.J.M., et al. Compound heterozygosity for mutations in LMNA causes a progeria syndrome without prelamin A accumulation. Hum. Mol. Genet. 2006;15:2509–2522. doi: 10.1093/hmg/ddl172. doi:10.1093/hmg/ddl172. [DOI] [PubMed] [Google Scholar]
- 24.Troutman J.M., Roberts M.J., Andres D.A., Spielmann H.P. Tools to analyze protein farnesylation in cells. Bioconjug. Chem. 2005;16:1209–1217. doi: 10.1021/bc050068+. doi:10.1021/bc050068+ [DOI] [PubMed] [Google Scholar]
- 25.Lowes B.D., Minobe W., Abraham W.T., Rizeq M.N., Bohlmeyer T.J., Quaife R.A., Roden R.L., Dutcher D.L., Robertson A.D., Voelkel N.F., et al. Changes in gene expression in the intact human heart. Downregulation of alpha-myosin heavy chain in hypertrophied, failing ventricular myocardium. J. Clin. Invest. 1997;100:2315–2324. doi: 10.1172/JCI119770. doi:10.1172/JCI119770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arimura T., Helbling-Leclerc A., Massart C., Varnous S., Niel F., Lacene E., Fromes Y., Toussaint M., Mura A.M., Keller D.I., et al. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet. 2005;14:155–169. doi: 10.1093/hmg/ddi017. doi:10.1093/hmg/ddi017. [DOI] [PubMed] [Google Scholar]
- 27.DeBusk F.L. The Hutchinson–Gilford progeria syndrome. Report of 4 cases and review of the literature. J. Pediatr. 1972;80:697–724. doi: 10.1016/s0022-3476(72)80229-4. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Palazzi F., McLaren A.T., Slowie D.F. Report on a case of Hutchinson–Gilford progeria, with special reference to orthopedic problems. Eur. J. Pediatr. Surg. 1992;2:378–382. doi: 10.1055/s-2008-1063486. doi:10.1055/s-2008-1063486. [DOI] [PubMed] [Google Scholar]
- 29.Bonne G., Barletta M.R.D., Varnous S., Becane H.-M., Hammouda E.-H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.-A., et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. doi:10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 30.Muchir A., Bonne G., van der Kooi A.J., van Meegen M., Baas F., Bolhuis P.A., de Visser M., Schwartz K. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B) Hum. Mol. Genet. 2000;9:1453–1459. doi: 10.1093/hmg/9.9.1453. doi:10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- 31.Vytopil M., Benedetti S., Ricci E., Galluzzi G., Dello Russo A., Merlini L., Boriani G., Gallina M., Morandi L., Politano L., et al. Mutation analysis of the lamin A/C gene (LMNA) among patients with different cardiomuscular phenotypes. J. Med. Genet. 2003;40:e132–e132. doi: 10.1136/jmg.40.12.e132. doi:10.1136/jmg.40.12.e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mounkes L.C., Kozlov S.V., Rottman J.N., Stewart C.L. Expression of an LMNA-N195K variant of A-type lamins results in cardiac conduction defects and death in mice. Hum. Mol. Genet. 2005;14:2167–2180. doi: 10.1093/hmg/ddi221. doi:10.1093/hmg/ddi221. [DOI] [PubMed] [Google Scholar]
- 33.Muchir A., Pavlidis P., Decostre V., Herron A.J., Arimura T., Bonne G., Worman H.J. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J. Clin. Invest. 2007;117:1282–1293. doi: 10.1172/JCI29042. doi:10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee R., Chang S.Y., Trinh H., Tu Y., White A.C., Davies B.S., Bergo M.O., Fong L.G., Lowry W.E., Young S.G. Genetic studies on the functional relevance of the protein prenyltransferases in skin keratinocytes. Hum. Mol. Genet. 2010;19:1603–1617. doi: 10.1093/hmg/ddq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurer-Stroh S., Koranda M., Benetka W., Schneider G., Sirota F.L., Eisenhaber F. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput. Biol. 2007;3:e66. doi: 10.1371/journal.pcbi.0030066. doi:10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reid T.S., Terry K.L., Casey P.J., Beese L.S. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J. Mol. Biol. 2004;343:417–433. doi: 10.1016/j.jmb.2004.08.056. [DOI] [PubMed] [Google Scholar]
- 37.Mijimolle N., Velasco J., Dubus P., Guerra C., Weinbaum C.A., Casey P.J., Campuzano V., Barbacid M. Protein farnesyltransferase in embryogenesis, adult homeostasis, and tumor development. Cancer Cell. 2005;7:313–324. doi: 10.1016/j.ccr.2005.03.004. doi:10.1016/j.ccr.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Fong L.G., Ng J.K., Lammerding J., Vickers T.A., Meta M., CotÇ N., Gavino B., Qiao X., Chang S.Y., Young S.R., et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J. Clin. Invest. 2006;116:632–634. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. doi:10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beck L.A., Hosick T.J., Sinensky M. Incorporation of a product of mevalonic acid metabolism into proteins of Chinese hamster ovary cell nuclei. J. Cell Biol. 1988;107:1307–1316. doi: 10.1083/jcb.107.4.1307. doi:10.1083/jcb.107.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutz R.J., Trujillo M.A., Denham K.S., Wenger L., Sinensky M. Nucleoplasmic localization of prelamin A: implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc. Natl Acad. Sci. USA. 1992;89:3000–3004. doi: 10.1073/pnas.89.7.3000. doi:10.1073/pnas.89.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hennekes H., Nigg E.A. The role of isoprenylation in membrane attachment of nuclear lamins. A single point mutation prevents proteolytic cleavage of the lamin A precursor and confers membrane binding properties. J. Cell Sci. 1994;107:1019–1029. doi: 10.1242/jcs.107.4.1019. [DOI] [PubMed] [Google Scholar]
- 43.Fong L.G., Frost D., Meta M., Qiao X., Yang S.H., Coffinier C., Young S.G. A protein farnesyltransferase inhibitor ameliorates disease in a mouse model of progeria. Science. 2006;311:1621–1623. doi: 10.1126/science.1124875. doi:10.1126/science.1124875. [DOI] [PubMed] [Google Scholar]
- 44.Leung G.K., Schmidt W.K., Bergo M.O., Gavino B., Wong D.H., Tam A., Ashby M.N., Michaelis S., Young S.G. Biochemical studies of Zmpste24-deficient mice. J. Biol. Chem. 2001;276:29051–29058. doi: 10.1074/jbc.M102908200. doi:10.1074/jbc.M102908200. [DOI] [PubMed] [Google Scholar]
- 45.Dalton M., Sinensky M. Expression systems for nuclear lamin proteins: farnesylation in assembly of nuclear lamina. In: Patrick J.C., Janice E.B., editors. Methods in Enzymology. Vol. 250. San Diego, CA, USA: Academic Press; 1995. pp. 134–148. [DOI] [PubMed] [Google Scholar]
- 46.Lammerding J., Fong L.G., Ji J.Y., Reue K., Stewart C.L., Young S.G., Lee R.T. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. doi:10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]