Abstract
Background
This case-control study concerns a molecular biological method based on the data gathered from a group of Korean subjects to examine the distribution of Malassezia yeasts in seborrheic dermatitis (SD) patients. Cultures for Malassezia yeasts were taken from the foreheads, cheeks and chests of 60 patients with SD and in 60 healthy controls of equivalent age.
Objective
The purpose of this study is to identify the relationship between certain species of Malassezia and SD. This was done by analyzing the differences in the distribution of Malassezia species in terms of age and body parts of the host with healthy controls.
Methods
26S rDNA PCR-RFLP, a fast and accurate molecular biological method, was used to overcome the limits of morphological and biochemical methods.
Results
The positive Malassezia culture rate was 51.7% in patients with SD, which was lower than that of healthy adults (63.9%). M. restricta was dominant in patients with SD (19.5%). Likewise, M. restricta was identified as a common species (20.5%) in healthy controls. In the ages 31~40, M. restricta was found to be the most common species (31.6%) among SD patients.
Conclusion
According to the results of the study, the most frequently isolated species was M. restricta (19.5%) in patients with SD. There was no statistically significant difference in the distribution of Malassezia species between the SD patients and healthy control groups.
Keywords: 26S rDNA PCR-RFLP, Malassezia yeasts, Seborrheic dermatitis
INTRODUCTION
Seborrheic dermatitis (SD) is a sub-acute or chronic superficial eczematous dermatitis, characterized by erythematous plaques with a dry or oily scale1. It develops in the areas of increased activity of sebaceous glands, usually occuring on the scalp, face, ears, chest, and axillary areas. The causes of SD have not yet been completely elucidated, however hypersecretion of sebaceous glands is seen as the major cause, as it primarily occurs where sebaceous glands are abundant. There are also other hypotheses, including a relationship with the lipophilic Malassezia yeasts as resident flora of the skin, and abnormalities of neurotransmitters and epidermal hyperplasia. Furthermore, there are several studies stating that clinical symptoms showed improvement along with decreased amounts of yeasts when treating SD with azole antimycotics2,3. In an animal experiment, when Malassezia yeasts are applied to the skin, a lesion similar to SD appeared, indicating a need for further study into the relationship between SD and Malassezia yeasts4.
In this research of subjects with SD, 26S rDNA PCR-RFLP was applied to identify and classify Malassezia yeasts. In light of the group of healthy controls, this study reveals how the frequency and distribution of the Malassezia yeasts changes depending on both age and which body parts are affected, thus clarifying the relationship between the Malassezia yeasts and SD.
MATERIALS AND METHODS
Study participants
There were 60 healthy adults and 60 patients with SD (30 males, 30 females) who participated in this study. The patients were chosen among those who had visited the dermatology outpatient clinic at Konkuk University Medical Center, from July 2005 to June 2008. The healthy adults were grouped depending on their age (20 patients in each age group: 31~40 years, 41~50 and 51~60). Excluded patients included those who had been administered adrenocortical hormones, those who had undergone light therapy or antimycotic treatment within 2 months of the examination date, as well as those who had been treated with topical antifungals within 1 month and/or with topical corticosteroids within 1 week. All participants were told not to wash their face or use moisturizer (not to apply any products) on the examination day. According to the Declaration of Helsinki, the participants were informed of the physical and mental risks that could happen during the study, and written consents were obtained before their participation.
Specimen sampling
Specimens were gathered from forehead, cheek and chest, and were sampled by scrub-wash technique, based on the method that Williamson and Kligman suggested5,6. A stainless tube, which has an interior area of 4.909 cm2, was set on the selected skin: forehead, cheek and chest, and then 1 ml of detergent7 (0.01% NaH2PO4H2O2, 1.01% Na2HPO4, 0.1% Triton X-100 [pH 7.9]) was put into the tube. After rubbing the skin by a glass rod for 1 minute, the sample was taken out by a pipette and stored in a different container. Then, 1 ml of the detergent was put into the stainless tube, and the specimen was repetitively sampled and added to the first sample. 100 µl of the sampled specimen was then mixed with 900 µl of the detergent with 50% concentration, and 100 µl was taken from the mixture, evenly applied on the Leeming-Notman medium, and cultured at 34℃ for 14 days8.
DNA extraction & PCR
For DNA extraction and PCR analysis of skin isolates, this study adopted colony PCR analysis which was developed to extract DNA directly from a colony of a PCR tube and to amplify 26S rDNA at the same time instead of direct genomic DNA extraction methods. A colony of Malassezia yeast was taken and transferred to a PCR tube and warmed up 3 times a day for 1 minute, in a double boiler by using a microwave, and then the tube was moved into ice water. The PCR reaction mixture (0.25 mM deoxynucleoside triphosphate, 10X PCR buffer, 5X Q buffer, 0.5 µM primers, 1.25 U Hot StarTaq polymerase, 20 mM MgSO4) was added and vortex mixing was done. Then, PCR, using Mastercycler 5333 (Eppendorf, Hamburg, Germany), was performed immediately. To amplify 26S rDNA, a primer that can amplify all 11 standard strains at once was chosen. The sequence was: forward, 5'-TAACAAGGATTCCCCTAGTA-3' and reverse, 5'-ATTACGCCAGCATCCTAAG-3'9. The conditions in the early stage of the reaction were at 95℃ for 14 minutes for predenaturation, at 94℃ for 45 seconds for denaturation, at 55℃ for 45 seconds for annealing, at 72℃ for 1 minute for extension of 40 cycles, and at 72℃ for 7 minutes for the last extension. Amplified DNA was visualized by electrophoresis on a 1.5% (w/v) agarose gel with Ethidium bromide (0.5 µg/ml) by using 1 X TAE migrating buffer (pH 8.0, 40 mM Tri-acetate 1 mM EDTA).
Restriction fragment length polymorphism (RFLP) analysis
After checking the amplified 26S rDNA, the PCR product was purified using the Accu-Prep PCR purification kit (Bioneer, Daejeon, Korea). For the 26S rDNA RFLP analysis of Malassezia yeasts, 2 restriction enzymes were used: Hha I (Takara Biomedicals, Otsu, Japan) and BtsC I (SibEnzyme, Novosibirsk, Russia). The restriction enzyme reaction was conditioned with 10X PCR buffer, 10 U restriction enzyme, and reaction mixture including 7.5 µl of the PCR product to become 20 µl. After 3 hours of reaction at 37℃, electrophoresis was performed in the TAE buffer with 3.5% (w/v) NuSieve GTG agarose gel (FMC, Rockland, ME, USA) by 100 volts. Then, it was stained with ethidium bromide, and the size and number of DNA fragment were checked by a UV transilluminator to analyze RFLP patterns10.
Statistical analysis
Chi-squared tests (SPSS) were used to find the difference in the distribution of the Malassezia species between patients with SD and healthy adults from equivalent age groups. The results are considered to be statistically significant when the p-value is less than 0.05.
RESULTS
Comparison of identification rates of Malassezia yeasts between patients with SM and healthy adults
Specimens were collected from 3 body sites: forehead, cheek, and chest, in 60 SD patients and 60 healthy adults. Malassezia yeasts of the patient group were cultured in 93 out of 180 specimens, and thus the identification rate was 51.7%. Five Malassezia species (M. sympodialis, M. restricta, M. furfur, M. globosa, M. dermatis) were identified in the group of SD patients (Fig. 1, 2). The healthy control group, on the other hand, showed 7 Malassezia species (M. globosa, M. restricta, M. sympodialis, M. furfur, M. dermatis, M. slooffiae, M. obtusa), and the identification rate was 63.9% (115/180) (Table 1, 2). Within age groups, a high identification rate was shown in of the 30s (31~40 years) in both patient and healthy adult groups: 66.7% (40/60) in the patient group, 71.7% (43/60) in the healthy adult group. Within body parts, in both patient and healthy adult groups, a high identification rate was shown in the forehead: 56.7% (34/60) in the patient group and 66.7% (40/60) in the healthy adult group.
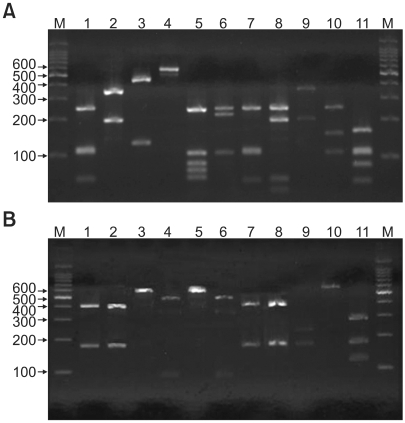
Fig. 1.
PCR-RFLP patterns of 26S rDNA PCR digested with Hha I (A), BtsC I (B) of 11 Malassezia standard strains. Lanes: M: molecular marker, 1: M. furfur (KCTC 7743), 2: M. sympodialis (KCTC 7985), 3: M. globosa (CBS 7966), 4: M. restricta (KCTC 7848), 5: M. slooffiae (KCTC 17431), 6: M. pachydermatis (KCTC 17008), 7: M. japonica (CBS 9432), 8: M. nana (JCM 12085), 9: M. dermatis (JCM 11348), 10: M. obtusa (KCTC 7847), 11: M. yamatoensis (CBS 9725).
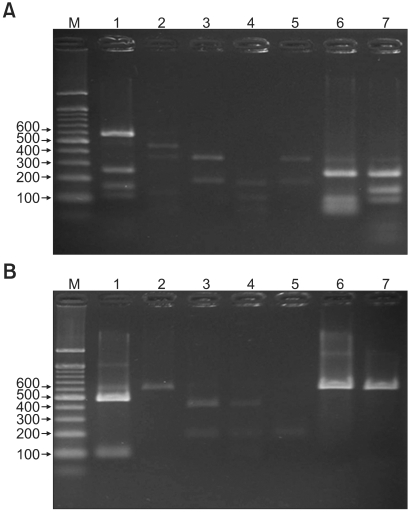
Fig. 2.
PCR-RFLP patterns of 26S rDNA PCR digested with Hha I (A), BtsC I (B) of Malassezia yeasts from seborrheic dermatitis patients and healthy controls. Lanes: M: molecular marker, 1: M. restricta, 2: M. globosa, 3: M. sympodialis, 4: M. furfur, 5: M. dermatis, 6: M. slooffiae (from healthy control), 7: M. obtusa (from healthy control).
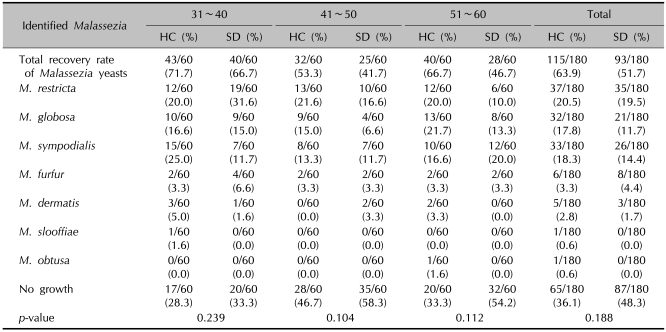
Table 1.
Identified Malassezia species by ages - 26S rDNA PCR-RFLP analysis
HC: healthy controls, SD: seborrheic dermatitis. p<0.05 is considered as significant.
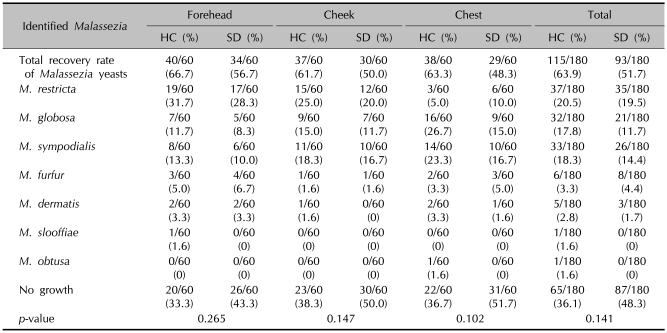
Table 2.
Identified Malassezia species by body sites - 26S rDNA PCR-RFLP analysis
HC: healthy controls, SD: seborrheic dermatitis. p<0.05 is considered as significant.
Comparison of the Malassezia species by different ages
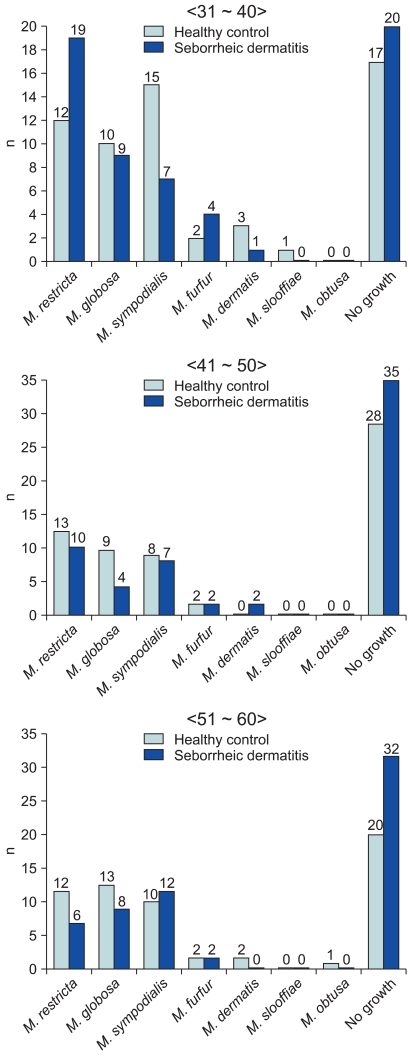
Identifying and classifying the Malassezia yeasts cultured on each body part according to ages, in the patients in their 30s and 40s, M. restricta (30s: 31.6%, 40s: 16.6%) was identified as the major species, and in their 50s, M. sympodialis (20.0%) was identified as the major species. In the healthy adult groups, M. sympodialis (25.0%), M. restricta (21.6%) and M. globosa (21.7%) were identified to be predominant in the 30s, 40s, and 50s, respectively (Table 1). Comparing the difference in the distribution of the Malassezia species between the patient and control groups by equivalent ages, there was no statistically significant difference (Fig. 3).
Fig. 3.
Identified Malassezia species from seborrheic dermatitis group compared with healthy control group, by ages.
Comparison of the Malassezia species by different body sites
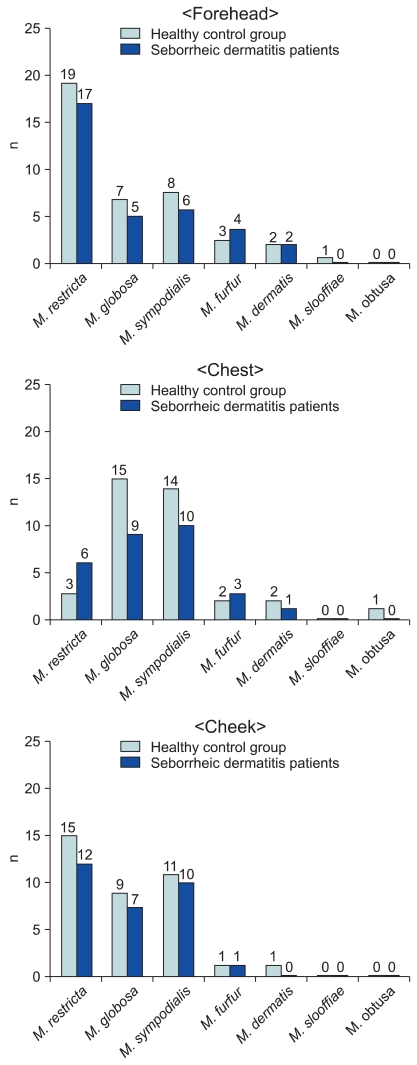
Identifying the Malassezia yeasts cultured from patients' body parts, in whole parts M. restricta was identified more than other species (19.5%, 35 cases). Both in the experiment and control groups by different body parts, M. restricta was identified frequently in the forehead and cheek. In the chest, on the other hand, M. sympodialis (16.7%) and M. globosa (26.7%) were identified as common species in both the patient and healthy adult groups, respectively (Table 2). The results showed no significant difference in the distribution of the Malassezia species in different body parts in either the patient or healthy control groups (Fig. 4).
Fig. 4.
Identified Malassezia species from seborrheic dermatitis group compared with healthy control group, by body sites.
DISCUSSION
Malassezia yeasts, found in 75~80% of healthy adults, are lipophilic fungi regarded as normal flora of the skin11-13. The genera Pityrosporum and Malassezia yeasts had long been considered as morphologically heterogeneous and thus are classified and distinguished from one another14.
Though, since the early days, morphologic diversity has been pointed out in previous studies of Malassezia yeasts, besides M. furfur, only the lipid-independent M. pachydermatis was classified as Malassezia for a long time15. As morphological, immunological, physiological, and molecular biological research on Malassezia yeasts continued, a need was developed for the re-classification of the genus Malassezia. Thus, Gueho et al. isolated M. furfur and re-classified into 7 species, considering the morphology, microstructure analysis, physiology, and molecular biology of each: M. furfur, M. pachydermatis, M. sympodialis, M. globosa, M. obtusa, M. restricta, M. slooffiae15-20. Moreover, 4 more Malassezia species in Japan; M. dermatis, M. japonica, M. nana, and M. yamatoensis, and 2 more species in Europe; M. caprae, M. equina have been recently identified, thus Malassezia yeasts are classified into 13 species at present9,19,21-25.
It has been reported that Malassezia yeasts are associated with dermatoses such as pityriasis versicolor, SD, and Malassezia folliculitis. Recently reported is the implication of Malassezia yeasts in atopic dermatitis and psoriasis12,13,21,26,27. Furthermore, confluent and reticulated papillomatosis28,29 and onychomycosis have been identified as being related to Malassezia yeasts30,31. The pathogenicity of Malassezia yeasts is therefore becoming significantly understood to be an underlying factor in certain generalized infections, with the yeasts occurring in premature infants who have received fluids containing fatty acids through vein catheter, as well as in adults with immune deficiency32,33.
SD occurs on sites with increased activity of sebaceous glands, and is characterized by erythematous plaques accompanied with dry or oily scales. Thus, it is indicated that SD may result from either: abnormalities of the skin's immune response to normal lipophilic Malassezia yeasts, from distribution changes, or from abnormal proliferation of certain Malassezia yeasts (the underlying cause unknown at this time). These are etiological theories and have not yet been proven.
In this research, 26S rDNA PCR-RFLP, a relatively fast and accurate molecular biological method, was used to identify Malassezia yeasts and also to examine the differences in the distribution of Malassezia yeasts between patients with SD and healthy controls, according to age and body parts. Compared with healthy adults, the group of patients with SD had 5 Malassezia yeasts (M. sympodialis, M. restricta, M. furfur, M. globosa, M. dermatis). Seven Malassezia yeasts (M. globosa, M. restricta, M. sympodialis, M. furfur, M. dermatis, M. slooffiae, M. obtusa) were identified in the healthy controls. In both groups, M. restricta was predominant. And in the healthy controls and SD patients, the sum of the identification rates of 3 Malassezia species, M. restricta, M. sympodialis, and M. globosa, was 88.6% (102/115) and 88.1% (82/93) respectively, constituting a majority of the identified Malassezia species. Moreover, according to findings regarding the frequency of Malassezia by different ages and body parts, no statistically significant difference was found in either SD patients or healthy controls.
In clarifying how normal lipophilic Malassezia yeasts could lead to SD, the results indicate that the difference of the Malassezia species is not the cause but rather the increased colonies of certain Malassezia yeasts, due to the destruction of skin barriers and to abnormalities of the immune system. Previous studies have examined lesions of patients with SD by using real-time PCR assay, which enables a quantitative analysis on Malassezia yeasts34. The finding is that lesions of patients with SD had more M. restricta than did the skin of the normal patients. This implies that a hyperproliferation of specific Malassezia yeasts may induce inflammation and result in SD.
In clarifying the relation between SD and Malassezia yeasts, further studies are needed. This is necessary not only to compare the qualitative difference of the Malassezia yeasts by body sites but also to etiologically analyze the quantitative difference. Studies may have to cover immunological features as well as abnormalities of skin barrier functions which result in proliferation of specific Malassezia yeasts.
Footnotes
This paper was supported by Konkuk University.
References
- 1.Gerd P, Thomas J. Seborrheic dermatitis. In: Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJ, editors. Fitzpatrick's dermatology in general medicine. 7th ed. New York: McGraw-Hill; 2008. pp. 1822–1830. [Google Scholar]
- 2.Comert A, Bekiroglu N, Gurbuz O, Ergun T. Efficacy of oral fluconazole in the treatment of seborrheic dermatitis: a placebo-controlled study. Am J Clin Dermatol. 2007;8:235–238. doi: 10.2165/00128071-200708040-00005. [DOI] [PubMed] [Google Scholar]
- 3.Miranda KC, de Araujo CR, Costa CR, Passos XS, de Fatima Lisboa Fernandes O, do Rosario Rodrigues Silva M. Antifungal activities of azole agents against the Malassezia species. Int J Antimicrob Agents. 2007;29:281–284. doi: 10.1016/j.ijantimicag.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 4.Drouhet E, Dompmartin D, Papachristou-Moraiti A, Ravisse P. Experimental dermatitis caused by Pityrosporum ovale and (or) Pityrosporum orbiculare in the guinea pig and the mouse. Sabouraudia. 1980;18:149–156. [PubMed] [Google Scholar]
- 5.Williamson P, Kligman AM. A new method for the quantitative investigation of cutaneous bacteria. J Invest Dermatol. 1965;45:498–503. doi: 10.1038/jid.1965.164. [DOI] [PubMed] [Google Scholar]
- 6.Leeming JP, Notman FH, Holland KT. The distribution and ecology of Malassezia furfur and cutaneous bacteria on human skin. J Appl Bacteriol. 1989;67:47–52. doi: 10.1111/j.1365-2672.1989.tb04953.x. [DOI] [PubMed] [Google Scholar]
- 7.Jang SJ, Choi YB, Ahn KJ. Malassezia species cultured from the lesions of Malassezia folliculitis. Korean J Med Mycol. 2003;8:55–62. [Google Scholar]
- 8.Leeming JP, Notman FH. Improved methods for isolation and enumeration of Malassezia furfur from human skin. J Clin Microbiol. 1987;25:2017–2019. doi: 10.1128/jcm.25.10.2017-2019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirhendi H, Makimura K, Zomorodian K, Yamada T, Sugita T, Yamaguchi H. A simple PCR-RFLP method for identification and differentiation of 11 Malassezia species. J Microbiol Methods. 2005;61:281–284. doi: 10.1016/j.mimet.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 10.Kim SM, Lim SH, Jung BR, Lee YW, Choe YB, Ahn KJ. The application of colony PCR in the molecular biological analysis of Malassezia yeasts. Korean J Med Mycol. 2007;12:180–188. [Google Scholar]
- 11.Ahn KJ. Taxonomy of the genus Malassezia. Korean J Med Mycol. 1998;3:81–88. [Google Scholar]
- 12.Lee YW, Lim SH, Ahn KJ. The application of 26S rDNA PCR-RFLP in the identification and classification of Malassezia yeast. Korean J Med Mycol. 2006;11:141–153. [Google Scholar]
- 13.Choe YB, Jang SJ, Yim SM, Ahn KJ. The quantitative study on the distribution of Malassezia yeasts on the normal skin of the young adults. Korean J Med Mycol. 2004;9:174–181. [Google Scholar]
- 14.Baillon EH. Traité de botanique médicale cryptogamique, suivi du tableau du droguier de la Faculté de médecine de Paris. Paris: Doin; 1889. [Google Scholar]
- 15.Gueho E, Simmons RB, Pruitt WR, Meyer SA, Ahearn DG. Association of Malassezia pachydermatis with systemic infections of humans. J Clin Microbiol. 1987;25:1789–1790. doi: 10.1128/jcm.25.9.1789-1790.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashbee HR, Ingham E, Holland KT, Cunliffe WJ. The carriage of Malassezia furfur serovars A, B and C in patients with pityriasis versicolor, seborrhoeic dermatitis and controls. Br J Dermatol. 1993;129:533–540. doi: 10.1111/j.1365-2133.1993.tb00480.x. [DOI] [PubMed] [Google Scholar]
- 17.Mayser P, Haze P, Papavassilis C, Pickel M, Gruender K, Gueho E. Differentiation of Malassezia species: selectivity of cremophor EL, castor oil and ricinoleic acid for M. furfur. Br J Dermatol. 1997;137:208–213. doi: 10.1046/j.1365-2133.1997.18071890.x. [DOI] [PubMed] [Google Scholar]
- 18.Crespo Erchiga V, Ojeda Martos A, Vera Casano A, Crespo Erchiga A, Sanchez Fajardo F. Malassezia globosa as the causative agent of pityriasis versicolor. Br J Dermatol. 2000;143:799–803. doi: 10.1046/j.1365-2133.2000.03779.x. [DOI] [PubMed] [Google Scholar]
- 19.Ashbee HR. Recent developments in the immunology and biology of Malassezia species. FEMS Immunol Med Microbiol. 2006;47:14–23. doi: 10.1111/j.1574-695X.2006.00057.x. [DOI] [PubMed] [Google Scholar]
- 20.Gueho E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996;69:337–355. doi: 10.1007/BF00399623. [DOI] [PubMed] [Google Scholar]
- 21.Sugita T, Takashima M, Shinoda T, Suto H, Unno T, Tsuboi R, et al. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J Clin Microbiol. 2002;40:1363–1367. doi: 10.1128/JCM.40.4.1363-1367.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugita T, Takashima M, Kodama M, Tsuboi R, Nishikawa A. Description of a new yeast species, Malassezia japonica, and its detection in patients with atopic dermatitis and healthy subjects. J Clin Microbiol. 2003;41:4695–4699. doi: 10.1128/JCM.41.10.4695-4699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugita T, Tajima M, Takashima M, Amaya M, Saito M, Tsuboi R, et al. A new yeast, Malassezia yamatoensis, isolated from a patient with seborrheic dermatitis, and its distribution in patients and healthy subjects. Microbiol Immunol. 2004;48:579–583. doi: 10.1111/j.1348-0421.2004.tb03554.x. [DOI] [PubMed] [Google Scholar]
- 24.Hirai A, Kano R, Makimura K, Duarte ER, Hamdan JS, Lachance MA, et al. Malassezia nana sp. nov., a novel lipid-dependent yeast species isolated from animals. Int J Syst Evol Microbiol. 2004;54:623–627. doi: 10.1099/ijs.0.02776-0. [DOI] [PubMed] [Google Scholar]
- 25.Cabanes FJ, Theelen B, Castella G, Boekhout T. Two new lipid-dependent Malassezia species from domestic animals. FEMS Yeast Res. 2007;7:1064–1076. doi: 10.1111/j.1567-1364.2007.00217.x. [DOI] [PubMed] [Google Scholar]
- 26.Nakabayashi A, Sei Y, Guillot J. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med Mycol. 2000;38:337–341. doi: 10.1080/mmy.38.5.337.341. [DOI] [PubMed] [Google Scholar]
- 27.Ahn KJ. Malassezia species cultured from the lesions of pityriasis versicolor. Korean J Dermatol. 1997;35:736–743. [Google Scholar]
- 28.Ginarte M, Fabeiro JM, Toribio J. Confluent and reticulated papillomatosis (Gougerot-Carteaud) successfully treated with tacalcitol. J Dermatolog Treat. 2002;13:27–30. doi: 10.1080/09546630252775216. [DOI] [PubMed] [Google Scholar]
- 29.Yesudian P, Kamalam S, Razack A. Confluent and reticulated papillomatosis (Gougerot-Carteaud). An abnormal host reaction to Malassezzia furfur. Acta Derm Venereol. 1973;53:381–384. [PubMed] [Google Scholar]
- 30.Chowdhary A, Randhawa HS, Sharma S, Brandt ME, Kumar S. Malassezia furfur in a case of onychomycosis: colonizer or etiologic agent? Med Mycol. 2005;43:87–90. doi: 10.1080/13693780400006070. [DOI] [PubMed] [Google Scholar]
- 31.Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL., Jr Skin diseases associated with Malassezia species. J Am Acad Dermatol. 2004;51:785–798. doi: 10.1016/j.jaad.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 32.Curvale-Fauchet N, Botterel F, Legrand P, Guillot J, Bretagne S. Frequency of intravascular catheter colonization by Malassezia spp. in adult patients. Mycoses. 2004;47:491–494. doi: 10.1111/j.1439-0507.2004.01047.x. [DOI] [PubMed] [Google Scholar]
- 33.Devlin RK. Invasive fungal infections caused by Candida and Malassezia species in the neonatal intensive care unit. Adv Neonatal Care. 2006;6:68–77. doi: 10.1016/j.adnc.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Tajima M, Sugita T, Nishikawa A, Tsuboi R. Molecular analysis of Malassezia microflora in seborrheic dermatitis patients: comparison with other diseases and healthy subjects. J Invest Dermatol. 2008;128:345–351. doi: 10.1038/sj.jid.5701017. [DOI] [PubMed] [Google Scholar]