Abstract
We demonstrate that the biological effect of an oligonucleotide is influenced by its route of cellular uptake. Utilizing a splice-switching antisense oligonucleotide (SSO) and a sensitive reporter assay involving correction of RNA splicing, we examined induction of luciferase in cells treated either with various concentrations of an unconjugated (“free”) SSO or an SSO conjugated to a bivalent RGD ligand that promotes binding to the αvβ3 integrin (RGD-SSO). Under conditions of equal accumulation in cells, the RGD-SSO consistently had a greater effect on luciferase induction than the unconjugated SSO. We determined that the RGD-SSO and the unconjugated SSO were internalized by distinct endocytotic pathways, suggesting that the route of internalization affects the magnitude of the biological response.
Introduction
There is currently great interest in the possible therapeutic uses of siRNA oligonucleotides and rekindled interest in antisense oligonucleotides for purposes of modulation of RNA splicing or antagonism of miRNA (Dean and Bennett, 2003; Sazani and Kole, 2003; Stein et al., 2005; Gewirtz, 2007; Castanotto and Rossi, 2009; Whitehead et al., 2009). However, these highly polar molecules do not readily cross biological membranes and enter cells; thus a great deal of effort has been devoted to improving the intracellular delivery of oligonucleotides (Song et al., 2005; Abes et al., 2007; Li and Szoka, 2007; Li et al., 2008; Meyer et al., 2008). A significant portion of this effort has simply dealt with increasing the total amount of oligonucleotide delivered, while other aspects include targeting to specific cell types, or improving release from endosomal compartments where oligonucleotides tend to initially accumulate. However, relatively little attention has been paid to the details of subcellular trafficking of oligonucleotides and how this might impact their biological effectiveness.
As a test system, we utilized αvβ3 integrin-expressing melanoma cells that contain a luciferase reporter gene interrupted by an abnormal intron that prevents expression of functional luciferase protein (Alam et al., 2008). Upon adequate delivery of an appropriate splice-switching antisense oligonucleotide (SSO) to the nucleus, the intron is spliced out and luciferase is expressed, thus providing a very sensitive positive readout of oligonucleotide delivery and subsequent biological effect (Sazani and Kole, 2003; Roberts et al., 2006). We compared the effects of an unconjugated or “free” SSO (termed 623) with those of a conjugate comprised of oligonucleotide 623 covalently linked to a cyclic RGD peptide that binds with high affinity to the αvβ3 integrin (RGD-623) (Alam et al., 2008). We show that the unconjugated SSO and the RGD-conjugated SSO differ in their ability to induce luciferase even when they are accumulated to equal levels in cells. We further demonstrate that the endocytotic uptake pathways for the RGD-SSO and the unconjugated SSO are substantially different, particularly in terms of their dependence on the actin cytoskeleton and on dynamin. Since the RGD-SSO and the unconjugated SSO are equally potent when delivered directly to the cytosol, our observations suggest that the endocytotic uptake pathway followed by an oligonucleotide can strongly influence its subsequent biological effects.
Materials and Methods
Cells and reagents
We utilized stable transfectants of human A375SM melanoma cells (A375SM-Luc705-B cells) containing a firefly luciferase gene cassette that includes an abnormal intron. The intranuclear presence of a “splice-switching oligonucleotide” (termed oligo 623) that targets an intronic splice site results in splicing out of the intron and allows expression of wild-type luciferase mRNA and protein (Alam et al., 2008). Cells were maintained in DMEM containing 100 μg/mL hygromycin B and 10% fetal bovine serum (FBS). Cytochalasin D, latrunculin A, methyl-β-cyclodextrin, β-cyclodextrin, and amiloride were purchased from Sigma (St. Louis, MO). A plasmid expressing a dominant negative (DN) version of dynamin (K44A) as a chimera with green fluorescent protein (GFP) was obtained from Dr. Joann Trejo, University of California, San Diego.
Oligonucleotides
Synthesis and chemical characterization of 2′-O-Me-phosphorothioate splice-switching oligonucleotide 623 (5′-GTT ATT CTT TAG AAT GGT GC-3′) and its cyclic RGD conjugate (RGD-623) were performed as described previously (Alam et al., 2008). In both cases, the oligonucleotide had a N-carboxy-(tetramethyl-rhodamine) (Tamra) fluorophore incorporated at the 3′ position. Versions of these materials containing 5 mismatches to the target splice site were also produced.
Cell uptake and luciferase assays
A375SM-Luc705-B cells were plated in 12-well plates at 1.5 × 105 cells per well in DMEM supplemented with 10% FBS. The following day, medium was changed to OPTI-MEM I (Invitrogen, Carlsbad, CA). Cells were treated with either free 623 oligonucleotide or RGD-623 conjugate, or with mismatched control oligonucleotides. Four hours after treatment, 1% FBS was added to each well. Twenty-four hours after oligonucleotide treatment, medium was replaced with DMEM containing 1% FBS, and at 24 hours afterward cell lysates were collected for determination of fluorescence intensity, luciferase activity, and total protein content. The luciferase assay used a kit (Promega, Madison, WI) and measurements were performed on a Monolight 2010 instrument (Analytical Luminescence Laboratory, San Diego, CA). Total cellular uptake of Tamra-labeled oligonucleotides was monitored using a Nanodrop microfluorimeter (Nanodrop Technologies, Wilmington, DE). After treatment with oligonucleotides, the cells were lysed in Reporter Lysis Buffer (Promega) and the Tamra fluorescence (excitation 560 nm and emission 583 nm) was quantitated based on a linear standard curve of unconjugated Tamra in buffer.
Confocal microscopy
Intracellular distribution of Tamra-labeled oligonucleotides in living cells was examined using an Olympus Confocal FV300 fluorescent microscope with 60× oil immersion objective, as previously described (Alam et al., 2008). Glass bottom microwell dishes (35 mm) were obtained from MatTek (Ashland, MA).
Results and Discussion
We explored the relationship between cell uptake and biological effect (luciferase induction in this case), first by performing careful measurements of these parameters over a wide range of oligonucleotide concentrations, and second by use of molecular and chemical inhibitors to perturb uptake and subcellular trafficking processes. Our observations indicate that the free oligonucleotide and the RGD=noligonucleotide conjugate are taken up via different endocytotic pathways leading to differing degrees of biological effectiveness.
Relationship between uptake and biological effect
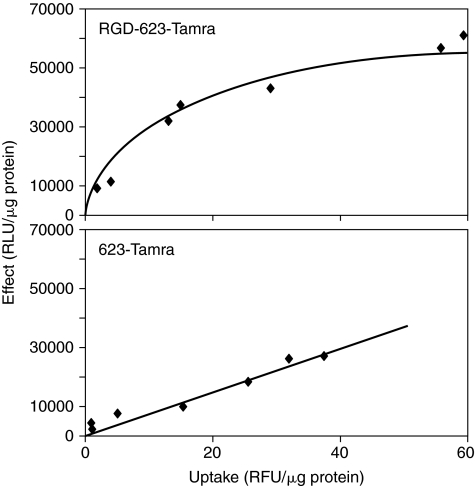
We used SSO 623 or its cyclic RGD conjugate, both labeled at the 3′ position with a Tamra fluorophore (Fig. 1). A375SM-Luc705-B cells were exposed to a wide range of concentrations of these molecules (0–n500 nM); thereafter, the cells were harvested and total cellular accumulation of oligonucleotide was monitored by fluorimetry, while luciferase induction was quantitated by luminometry. Results were normalized to cell protein content and the effect on luciferase induction was plotted vs. total cell uptake of fluorescent oligonucleotide. As seen in Figure 2, the plot of effect vs. uptake for unconjugated 623 was linear; in contrast, the plot for RGD-623 was hyperbolic. Further, the magnitude of the luciferase effect at a given level of uptake was substantially greater for the RGD-623 compared to unconjugated 623, especially at lower levels of uptake. For example, at 20 uptake units (RFU/μg protein) the luciferase induction was 15 units (RLU/μg protein) for 623 but 40 units for RGD-623. Similar experiments with any of the mismatched versions of 623 or RGD-623 did not result in luciferase induction (data not shown). It should be noted that the uptake studies described here track the accumulation and retention of the fluorophore; thus, it is possible that this measurement might not reflect accumulation of the oligonucleotide itself if the labels were to be cleaved. However, it has been our experience (Alam et al., 2008; Kang et al., 2008) and that of several other laboratories (Turner et al., 2005; Bendifallah et al., 2006; El-Andaloussi et al., 2006) that fluorophore labels placed at the 3′ position of stable oligonucleotides provide reasonably accurate tracking of oligonucleotides in cell culture experiments such as those described here.
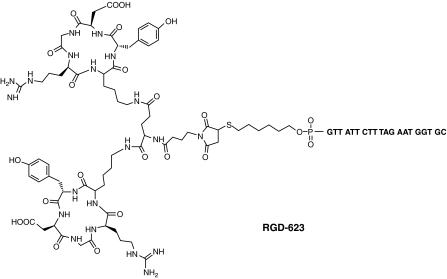
FIG. 1.
Diagram of the RGD-conjugated splice-shifting oligonucleotide (RGD-623). The RGD moiety is conjugated at the 5′ position, while a Tamra fluorophore is at the 3′ position. The oligonucleotide is a 2′-O-Me-phosphorothioate.
FIG. 2.
Uptake vs. effect. A375SM-Luc705-B cells (150,000/well) were treated with varying concentrations of 623-Tamra, or RGD-623-Tamra at 37°C for 4 hours in OPTI-MEM. At that point, 1% serum was added for 24 hours; the medium was then changed to 1% FBS/DMEM for another 24 hours. Cells were washed, lysed in reporter lysis buffer, freeze-thawed, vortexed, and centrifuged (14,000 rpm, 10 minutes); luciferase activity, protein concentration, and cell uptake of fluorescent oligonucleotides were measured as described in Materials and Methods. Luciferase activity was expressed as relative luminescent units (RLU)/μg protein, while cell uptake was expressed as relative fluorescent units (RFU)/μg protein. Results are the means of triplicate determinations. Graphs were produced with a curve-fitting program.
Evaluating the uptake pathway
The simple fluorometric measurement we report above provides information on the total amount of fluorophore-labeled oligonucleotide associated with the cell, but it is silent concerning the subcellular localization of the accumulated material. To address this issue, we turned to confocal fluorescence microscopy, and to use of chemical and molecular inhibitors of endocytosis. Cells employ several distinct endocytotic pathways including those involving clathrin-coated vesicles, caveolae, and various smooth vesicles (Kirkham and Parton, 2005; Mayor and Pagano, 2007).
Simple fluorescence microscopic observation of cells treated with 623-Tamra or RGD-623-Tamara did not reveal obvious differences (data not shown). In both cases the fluorescent oligonucleotide was primarily found in intracellular vesicles, similar to previous descriptions of the subcellular distribution of oligonucleotides and peptide=noligonucleotide conjugates observed by us and by others (Turner et al., 2005; Alam et al., 2008). Thus, we sought to probe possible differences in uptake and trafficking pathways by use of several putatively selective inhibitors of various endocytotic pathways (Parpal et al., 2001; Khalil et al., 2006). Prior to uptake experiments, we tested various doses of these agents for possible toxicity, and chose doses below those that permitted 90% cell survival (data not shown).
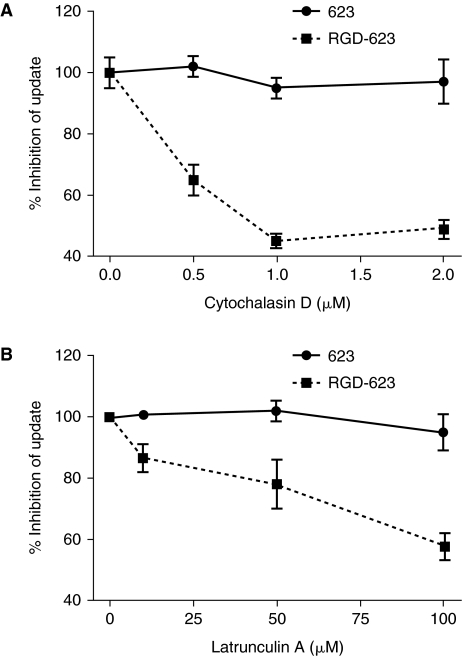
A notable difference was observed with regard to the effect of inhibitors of the actin cytoskeleton on cellular uptake of 623 vs. RGD-623. As seen in Figure 3A, uptake of RGD-623 was substantially reduced by treatment with cytochalasin D, whereas this agent had no effect on uptake of unconjugated 623. Use of latrunculin A, another actin inhibitor with a mechanism of action distinct from cytochalasin D (Aplin and Juliano, 1999; Uriarte et al., 2009), provided essentially the same result (Fig. 3B). Thus a substantial portion (~50%) of the uptake of RGD-623 was sensitive to disruption of actin filaments, while uptake of 623 was unaffected. While most endocytotic pathways involve actin, there have been reports of internalization and trafficking pathways that are not strongly actin-dependent (Damm et al., 2005). Similar studies were done with several endocytotic inhibitors that putatively have selective effects on clathrin-mediated endocytosis, lipid-raft-mediated endocytosis, and macropinocytosis (Parpal et al., 2001; Khalil et al., 2006), and the results are summarized in Table 1. To the extent that the agents used can be presumed to be selective, these studies suggest that clathrin-coated vesicles are not involved in uptake of 623 or RGD-623, nor in macropinocytosis. However, uptake of both RGD-623 and 623 seems to involve lipid-raft structures. We also examined the effect of some of the inhibitors on the induction of luciferase, as summarized in Table 2.
FIG. 3.
Effects of actin inhibitors. A375SM-Luc705-B cells (150,000/well) were preincubated with (A) cytochalasin D or (B) latrunculin A at the indicated concentrations for 1 hour at 37°C in OPTI-MEM containing 1% FBS. At that point, 623-Tamra (100 nM) or RGD-623-Tamra (100 nM) were added to the wells, and the cells were treated for 24 hours. The cells were washed with PBS, and lysed with 1× reporter lysis buffer (150 μL). The cell extracts were frozen and thawed, vortexed, and centrifuged. Relative fluorescent units (RFUs) were measured using a microfluorimeter. The uptake data was normalized on protein concentration. Results represent means and standard deviations of triplicate determinations. Uptake was considered as 100% where no inhibitor was added.
Table 1.
Effects of Inhibitors on Antisense Uptake
| Drug | Treatment | Endocytosis pathway putatively affected | Effect on 623-Tamra | Effect on RGD-623-Tamra |
|---|---|---|---|---|
| Control | None | None | 100% | 100% |
| Chlorpromazine | 2.5 μM | Clathrin-mediated endocytosis | None | None |
| Methyl-β-cyclodextrin | 1.0 mM | Lipid-raft-mediated endocytosis | 60% ± 2% | 55% ± 1% |
| β-Cyclodextrin | 2.5 mM | Lipid-raft-mediated endocytosis | 58% ± 2% | 62% ± 5% |
| Amiloride | 5 μM | Macropinocytosis | None | None |
A375SM-Luc705-B cells were treated with the indicated nontoxic concentration of drugs as well as with 100 nM of RGD-623-Tamra or 623-Tamra. After 20 hours, the cells were washed, lysed, and uptake of fluorescent oligonucleotide per microgram cell protein was determined as described in Materials and Methods. Results were normalized on uptake by the control cells as =100%. Statistically significant differences from control were observed only for the cyclodextrin-treated samples.
Table 2.
Effects of Inhibitors on Luciferase Induction
| Drug | Treatment | Endocytosis pathway putatively affected | Effect on 623-Tamra | Effect on RGD-623-Tamra |
|---|---|---|---|---|
| Control | None | None | 100% | 100% |
| Chlorpromazine | 2.5 μM | Clathrin-mediated endocytosis | 89% ± 9% | 97% ± 8% |
| Amiloride | 5 μM | Macropinocytosis | 110% ± 14% | 95% ± 5% |
A375SM-Luc705-B cells were treated with the indicated nontoxic concentration of drugs as well as with 100 nM of RGD-623-Tamra or 623-Tamra. After 20 hours, the medium was changed and the cells were placed in culture medium with 1% serum. After an additional 48 hours, the cells were harvested and analyzed for luciferase activity and total protein (RLU/μg) as described in Materials and Methods. Results were normalized on the control cells as =100%. Cyclodextrins were not included because they proved toxic during the lengthier incubations required in these experiments. There were no statistically significant differences between the chloropromazine- or amiloride-treated samples and the controls.
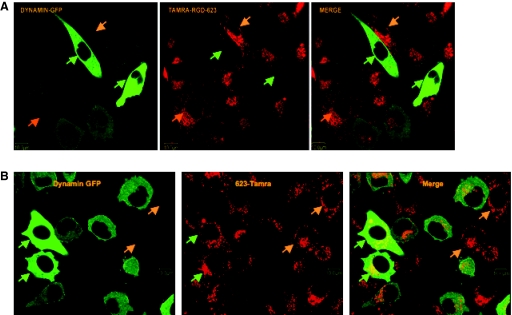
Dynamin is a small GTPase that plays a key role in pinching off membrane vesicles (Mayor and Pagano, 2007). Most endocytotic pathways depend on the action of dynamin, but there have been reports of dynamin-independent endocytosis (Sabharanjak et al., 2002; Massol et al., 2004). We examined the potential role of dynamin in oligonucleotide uptake in this system by transfecting cells with plasmids coding for a chimeric protein comprised of a DN form of dynamin linked to GFP. We then treated these cells with Tamra-labeled RGD-623 or 623 and observed the extent and subcellular distribution of the accumulated fluorescent oligonucleotides. As seen in Figure 4A, expression of high levels of DN-dynamin/GFP almost completely blocked the accumulation of RGD-623 in intracellular vesicles; cells with lower levels of expression of the chimeric protein displayed a partial inhibition. Interestingly, as seen in Figure 4B, high levels of expression of DN-dynamin/GFP failed to inhibit accumulation of 623 into intracellular vesicles. This suggests that the uptake pathway for unconjugated 623 oligonucleotide is largely independent of dynamin function.
FIG. 4.
Effects of dynamin. A375SM-Luc705-B cells (1 million) were transfected at 90% confluence with 2 μg of DN dynamin-GFP plasmid using an Amaxa Nucleoporation system. Cells were then cultured in DMEM plus 10% FBS for 24 hours to allow protein expression. The cells were then treated with 100 nM of 623-Tamra or 50 nM of RGD-623-Tamra for 4 hours. Cells were then trypsinized and seeded in 35-mm glass bottom microwell dishes previously coated with 5 μg/cm2 of fibronectin for 1 hour at room temperature. Confocal fluorescent images were obtained as described in Materials and Methods. Cells expressing high levels of DN dynamin-GFP are marked by green arrows while untransfected cells are marked by orange arrows and cells with weaker dynamin-GFP expression are not marked. (A) RGD-623-Tamra. (B) 623-Tamra. Results are typical of multiple microscopic fields observed.
Thus it seems clear that the endocytotic uptake pathways for RGD-623 and unconjugated 623 are distinct, with RGD-623 trafficking at least in part via a lipid raft-dependent, actin-dependent, dynamin-dependent pathway, while 623 utilizes a lipid raft-dependent, actin-independent, dynamin-insensitive pathway. These discrete uptake and trafficking processes may underlie the different pharmacological effectiveness of these 2 molecules when they are accumulated in cells at equal total levels. A portion of the uptake of RGD-623 seems to take place via the same actin-independent pathway as 623; presumably, this is some form of nonspecific pinocytosis (Mayor and Pagano, 2007).
Testing intrinsic efficacy and the role of the actin-dependent component of uptake
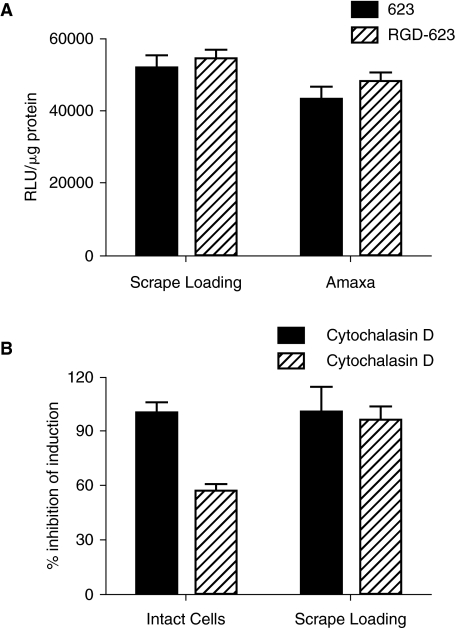
An alternative possibility is that RGD-623 and 623 have some innate difference in efficacy. To rule this out, we compared the effects of 623 and RGD-623 when administered to intact cells (and thus taken up by endocytosis) vs. effects when the oligonucleotides were delivered directly to the cytoplasm by scrape-loading or electroporation (Ostap et al., 2003; Touyz et al., 2005). As seen in Figure 5A, RGD-623 and 623 were equally effective in inducing luciferase when delivered directly to the cytosol. These observations suggest that there is no difference in the intrinsic efficacy of 623 and RGD-623.
FIG. 5.
Endocytosis vs. direct delivery to the cytosol. (A) RGD-623-Tamra or 623-Tamra (50 nM) was directly delivered to the cytosol of A375SM-Luc705-B cells by electroporation (Amaxa Nucleoporation system) or scrape-loading (see B below). The cells were incubated in DMEM plus 1% FBS for 24 hours and then the cells were harvested and analyzed for protein content and luciferase activity. Results are the means and standard deviations of 3 determinations and are expressed as RLU/μg protein. (B) Cells were treated with 100-nM RGD-623-Tamra in the presence or absence of 2-μM cytochalasin D. In one set of experiments, the intact cells were simply incubated with the oligonucleotide and inhibitor in DMEM plus 1% FBS. In the other case, the cells were “scrape-loaded” in the presence of oligonucleotide and inhibitor by dislodging them from the substratum with a spatula. The cells were then allowed to re-attach. After 24 hours, the cells were harvested, lysed, and analyzed for protein content and luciferase activity as described in Materials and Methods. Results are means and standard deviations of triplicate determinations and are expressed as RLU/μg protein. The hatched bar represents cells treated with cytochalasin D, while the solid bar represents control cells. For the intact cell case, the difference between control- and cytochalasin D-treated cells is significant at the P < 0.01 level.
We also tested the effect of cytochalasin D on luciferase induction when RGD-623 was administered to intact cells or delivered via scrape-loading. As seen in Figure 5B, cytochalasin D had no effect on luciferase induction in the scrape-loaded cells but it significantly reduced luciferase induction when the RGD-623 was accumulated by normal endocytotic processes. This indicates, first that the concentrations of cytochalasin D used are not toxic to the cells' transcription, splicing, and translation machinery, and second that the actin-dependent uptake process makes a substantial contribution to the pharmacological effect of the RGD-623. Thus the observed differences in the ability of 623 and RGD-623 to induce luciferase may be due to their distinct pathways of internalization and subcellular trafficking, with the dynamin-dependent, actin-dependent pathway utilized by RGD-623 being particularly effective.
Discussion
We have identified an actin-dependent, dynamin-dependent pathway for uptake of the RGD-623 oligonucleotide conjugate in A375 cells, and an unusual actin-independent, dynamin-independent pathway for uptake of the unconjugated 623 oligonucleotide. At this point, it is not known whether this latter unusual pathway is operant in other cell types or for other chemical forms of “free” monomeric or duplexed oligonucleotides. If this pathway were widely used by many cell types, it might have significant implications for the pharmacology of “free” antisense and siRNA molecules.
Many recent studies have sought to increase the pharmacological effectiveness of antisense and siRNA oligonucleotides by increasing their total cellular uptake. While this is no doubt valid, our current studies suggest that it may also be worthwhile to consider the pathway of uptake. By targeting oligonucleotides to specific cell-surface receptors, it may be possible both to increase cellular uptake and, at least to some degree, to steer the path of intracellular trafficking in a beneficial manner. At this point, little is known about the properties of various endosomal compartments in terms of releasing oligonucleotides to the cytosol or nucleus; this may prove to be an interesting area of investigation.
Acknowledgments
This work was supported by a grant from the NIH [PO1 GM 26599] to R.L.J. The authors thank Osamu Nakagawa Ph.D. for assistance with Figure 1.
Author Disclosure Statement
None of the authors has any commercial associations that may result in a conflict of interest situation.
References
- ABES S. MOULTON H. TURNER J. CLAIR P. RICHARD J.P. IVERSEN P. GAIT M.J. LEBLEU B. Peptide-based delivery of nucleic acids: design, mechanism of uptake and applications to splice-correcting oligonucleotides. Biochem. Soc. Trans. 2007;35:53–55. doi: 10.1042/BST0350053. [DOI] [PubMed] [Google Scholar]
- ALAM M.R. DIXIT V. KANG H. LI Z.B. CHEN X. TREJO J. FISHER M. JULIANO R.L. Intracellular delivery of an anionic antisense oligonucleotide via receptor-mediated endocytosis. Nucleic Acids Res. 2008;36:2764–2776. doi: 10.1093/nar/gkn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APLIN A.E. JULIANO R.L. Integrin and cytoskeletal regulation of growth factor signaling to the MAP kinase pathway. J. Cell. Sci. 1999;112(Pt 5):695–706. doi: 10.1242/jcs.112.5.695. [DOI] [PubMed] [Google Scholar]
- BENDIFALLAH N. RASMUSSEN F.W. ZACHAR V. EBBESEN P. NIELSEN P.E. KOPPELHUS U. Evaluation of cell-penetrating peptides (CPPs) as vehicles for intracellular delivery of antisense peptide nucleic acid (PNA) Bioconjug. Chem. 2006;17:750–758. doi: 10.1021/bc050283q. [DOI] [PubMed] [Google Scholar]
- CASTANOTTO D. ROSSI J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAMM E.M. PELKMANS L. KARTENBECK J. MEZZACASA A. KURZCHALIA T. HELENIUS A. Clathrin- and caveolin-1-independent endocytosis: entry of simian virus 40 into cells devoid of caveolae. J. Cell Biol. 2005;168:477–488. doi: 10.1083/jcb.200407113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEAN N.M. BENNETT C.F. Antisense oligonucleotide-based therapeutics for cancer. Oncogene. 2003;22:9087–9096. doi: 10.1038/sj.onc.1207231. [DOI] [PubMed] [Google Scholar]
- EL-ANDALOUSSI S. JOHANSSON H.J. LUNDBERG P. LANGEL U. Induction of splice correction by cell-penetrating peptide nucleic acids. J. Gene Med. 2006;8:1262–1273. doi: 10.1002/jgm.950. [DOI] [PubMed] [Google Scholar]
- GEWIRTZ A.M. On future's doorstep: RNA interference and the pharmacopeia of tomorrow. J. Clin. Invest. 2007;117:3612–3614. doi: 10.1172/JCI34274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANG H. ALAM M.R. DIXIT V. FISHER M. JULIANO R.L. Cellular delivery and biological activity of antisense oligonucleotides conjugated to a targeted protein carrier. Bioconjug. Chem. 2008;19:2182–2188. doi: 10.1021/bc800270w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHALIL I.A. KOGURE K. AKITA H. HARASHIMA H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol. Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- KIRKHAM M. PARTON R.G. Clathrin-independent endocytosis: new insights into caveolae and non-caveolar lipid raft carriers. Biochim. Biophys. Acta. 2005;1745:273–286. doi: 10.1016/j.bbamcr.2005.06.002. [DOI] [PubMed] [Google Scholar]
- LI S.D. CHEN Y.C. HACKETT M.J. HUANG L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol. Ther. 2008;16:163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI W. SZOKA F.C., Jr. Lipid-based nanoparticles for nucleic acid delivery. Pharm. Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- MASSOL R.H. LARSEN J.E. FUJINAGA Y. LENCER W.I. KIRCHHAUSEN T. Cholera toxin toxicity does not require functional Arf6- and dynamin-dependent endocytic pathways. Mol. Biol. Cell. 2004;15:3631–3641. doi: 10.1091/mbc.E04-04-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAYOR S. PAGANO R.E. Pathways of clathrin-independent endocytosis. Nat. Rev. Mol. Cell Biol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEYER M. PHILIPP A. OSKUEE R. SCHMIDT C. WAGNER E. Breathing life into polycations: functionalization with pH-responsive endosomolytic peptides and polyethylene glycol enables siRNA delivery. J. Am. Chem. Soc. 2008;130:3272–3273. doi: 10.1021/ja710344v. [DOI] [PubMed] [Google Scholar]
- OSTAP E.M. MAUPIN P. DOBERSTEIN S.K. BAINES I.C. KORN E.D. POLLARD T.D. Dynamic localization of myosin-I to endocytic structures in Acanthamoeba. Cell Motil. Cytoskeleton. 2003;54:29–40. doi: 10.1002/cm.10081. [DOI] [PubMed] [Google Scholar]
- PARPAL S. KARLSSON M. THORN H. STRÅLFORS P. Cholesterol depletion disrupts caveolae and insulin receptor signaling for metabolic control via insulin receptor substrate-1, but not for mitogen-activated protein kinase control. J. Biol. Chem. 2001;276:9670–9678. doi: 10.1074/jbc.M007454200. [DOI] [PubMed] [Google Scholar]
- ROBERTS J. PALMA E. SAZANI P. ØRUM H. CHO M. KOLE R. Efficient and persistent splice switching by systemically delivered LNA oligonucleotides in mice. Mol. Ther. 2006;14:471–475. doi: 10.1016/j.ymthe.2006.05.017. [DOI] [PubMed] [Google Scholar]
- SABHARANJAK S. SHARMA P. PARTON R.G. MAYOR S. GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev. Cell. 2002;2:411–423. doi: 10.1016/s1534-5807(02)00145-4. [DOI] [PubMed] [Google Scholar]
- SAZANI P. KOLE R. Therapeutic potential of antisense oligonucleotides as modulators of alternative splicing. J. Clin. Invest. 2003;112:481–486. doi: 10.1172/JCI19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SONG E. ZHU P. LEE S.K. CHOWDHURY D. KUSSMAN S. DYKXHOORN D.M. FENG Y. PALLISER D. WEINER D.B. SHANKAR P. MARASCO W.A. LIEBERMAN J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- STEIN C.A. BENIMETSKAYA L. MANI S. Antisense strategies for oncogene inactivation. Semin. Oncol. 2005;32:563–572. doi: 10.1053/j.seminoncol.2005.09.003. [DOI] [PubMed] [Google Scholar]
- TOUYZ R.M. YAO G. QUINN M.T. PAGANO P.J. SCHIFFRIN E.L. p47phox associates with the cytoskeleton through cortactin in human vascular smooth muscle cells: role in NAD(P)H oxidase regulation by angiotensin II. Arterioscler. Thromb. Vasc. Biol. 2005;25:512–518. doi: 10.1161/01.ATV.0000154141.66879.98. [DOI] [PubMed] [Google Scholar]
- TURNER J.J. ARZUMANOV A.A. GAIT M.J. Synthesis, cellular uptake and HIV-1 Tat-dependent trans-activation inhibition activity of oligonucleotide analogues disulphide-conjugated to cell-penetrating peptides. Nucleic Acids Res. 2005;33:27–42. doi: 10.1093/nar/gki142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- URIARTE S.M. JOG N.R. LUERMAN G.C. BHIMANI S. WARD R.A. MCLEISH K.R. Counterregulation of clathrin-mediated endocytosis by the actin and microtubular cytoskeleton in human neutrophils. Am. J. Physiol., Cell Physiol. 2009;296:C857–C867. doi: 10.1152/ajpcell.00454.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITEHEAD K.A. LANGER R. ANDERSON D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]