Abstract
The self-renewal and multilineage differentiation of embryonic stem cells (ESC) is largely governed by transcription factors or repressors. Extensive efforts have focused on elucidating critical factors that control the differentiation of specific cell lineages, for instance, myeloid lineages in hematopoietic development. In this study, we found that Twist-2, a basic helix-loop-helix (bHLH) transcription factor, plays a critical role in inhibiting the differentiation of ESC. Murine ES cells, in which Twist-2 expression is silenced by lentivirally delivered shRNA, exhibit an enhanced formation of primary embryoid bodies (EB) and enhanced differentiation into mesodermally derived hematopoietic colonies. Furthermore, Twist-2 silenced (LV-siTwist-2) ESC display significantly increased generation of myeloid lineages (Gr-1+ and F4/80+ cells) during in vitro hematopoietic differentiation. Treatment with the Toll-like receptor (TLR) 4 ligand synergistically stimulates the generation of primary EB formation as well as of hematopoietic progenitors differentiated from LV-siTwist-2 ES cells. Thus, this study reveals the critical role of the transcriptional repressor Twist-2 in regulating the development of myeloid lineage in hematopoietic differentiation from ESC. This study also suggests a potential strategy for directional differentiation of ESC by inhibiting a transcriptional repressor.
Introduction
Mammalian embryonic stem cells (ESCs) are pluripotent cells derived from the early embryo blastocyst, which have the capability to differentiate into cells of the three embryonic germ layers. Given this unique ability, embryonic stem (ES) cells are a likely source for the cell types and tissues required for regenerative medicine-based therapies. One of the advantages of using ES cells is the ability to rapidly expand undifferentiated ES cells to large cell numbers prior to differentiation protocols (Smith et al., 1988; Thomson et al., 1998; Williams et al., 1988). However, a consistent bottleneck is the generally low yield of specific differentiation and purity of cell type generated (Docherty et al., 2007; Passier et al., 2008; Suter and Krause, 2008), especially with human embryonic stem cells (hESC) (Mountford, 2008). Recent reports using stromal cells from hematopoietic niches have increased hematopoietic induction from ES cells (Krassowska et al., 2006; Ledran et al., 2008). However, novel strategies enhancing the differentiation and yield of specific cell types from ES cells would help to successfully bring ES cell-based therapies into the clinic.
One obstacle to developing efficient in vitro differentiation methods is our limited understanding of the mechanisms that regulate ES cell differentiation. Although many groups have successfully used stimulatory factors to induce ES cell differentiation into multiple cell types (Copray et al., 2006; Dahl et al., 2008; Wang et al., 2008), the role of transcriptional repressors in ES cell differentiation is not well investigated (Beyer et al., 2007; Jankovic et al., 2007; Jhas et al., 2006; Szabo et al., 2008; Zhang et al., 2006). The Twist family (Twist-1, Twist-2) are comprised of basic helix-loop-helix (bHLH) transcription factors that inhibit the terminal differentiation of mesodermally derived tissues, including bone, muscle, and adipose tissue (Bialek et al., 2004; Hebrok et al., 1994; Lee et al., 2003; Rohwedel et al., 1995; Spicer et al., 1996). Recently, we found that Twist-2 is a key negative regulator of myeloid lineage development, as manifested by marked increases in mature myeloid populations of macrophage, neutrophils, and basophils in Twist-2-deficient mice (Sharabi et al., 2008). In this study, using murine ES cells, we found that the transcriptional repressor Twist-2 plays an important role in the inhibition of ESC differentiation into hematopoietic myeloid lineages. This study also suggests a strategy for directional differentiation of ESC by silencing a transcriptional repressor.
Materials and Methods
ES cell culture
Mouse ES D3 cells (ATCC CRL-1934) were cultured on 100-mm gelatin-coated tissue culture dishes with Knockout D-MEM (Invitrogen, Carlsbad, CA) supplemented with 15% fetal bovine serum (FBS) (StemCell Inc., Vancouver, Canada), 2 mM L-glutamine, 100 μM monothioglycerol (MTG), 100 U/mL penicillin, 10 μg/mL streptomycin, and 1000 U/mL LIF (ESGRO, Chemicon, Temecula, CA).
Lentivirus production
The LV-siGFP and LV-siTwist-2 lentiviral constructs were generated as previously described (Shen et al., 2004). Briefly, Twist-2 shRNA, or GFP shRNA hairpin sequences were inserted into the pTRIP vector driven by the H1 promoter. Vectors were verified by DNA sequencing. Recombinant lentiviral vectors were generated by cotransfection with packaging plasmids into 293T cells, and concentrated by ultracentrifugation as previously described (Schroers and Chen, 2004).
Western blotting analyses
siRNA oligonucleotide sequences targeting mouse Twist-2 were designed with the aid of an online program (www.dharmacon.com) as previously reported (Shen et al., 2004). To verify Twist-2 downregulation by siRNA oligos, Western blot analyses were performed. Briefly, 293T cells were cotranfected with one of synthetic 21 base-pair siRNA oligo duplexes (#A: AAGCGACGAGAUGGACAAUAA and #B: AACAAGAAAUCGAGCGAAGAU). and a FLAG-tagged Twist-1, or Twist-2 expression vector using Geneporter, following the manufacturer's protocol. The cells were harvested 48 h after cotransfection and subjected to SDS-PAGE. Following transfer to Hybond-P membrane (Amersham, Arlington Heights, IL), the samples were analyzed by Western blotting with anti-Flag (Sigma, St. Louis, MO), or anti-Actin (Santa Cruz, Santa Cruz, CA) antibodies, followed by detection with ECL-Plus reagent (Amersham).
ES cell differentiation and hematopoietic colony assay
For primary EB formation assays, ES cells were seeded in plates at 500 cells/mL in 1% methylcellulose-based differentiation media that included Iscove modified Dulbecco medium (IMDM) containing 15% FBS, 2 mM L-glutamine, 150 μM MTG, 40 ng/mL murine Stem Cell Factor (mSCF) (StemCell Technologies, Vancouver, BC, Canada), and incubated for 10 days at 37°C in 5% CO2. EBs were viewed by light microscopy and each plate was scored. For secondary EB formation assays, primary EBs were digested with Collagenase (StemCell Technologies) 0.25% trypsin followed by passaging through a 20-gauge needle 2 to 10 times. The ES cells were then reseeded and cultured in the same culture conditions as described in the primary EB formulation assays.
In vitro hematopoietic differentiation of ES cells was performed using the two-step method in methylcellulose based media as described in reagent manufacture's protocol (StemCell Technologies). Briefly, ES cells were first differentiated into primary EBs as described above. Primary EBs were dissociated with Collagenase (StemCell Technologies) and cells were then replated in Iscove's MDM (IMDM) containing 1% methylcellulose, 15% FBS, 2 mM L-glutamine, insulin (10 μg/mL), transferrin (200 μg/mL), and the following colony-stimulating factors and cytokines: mSCF (10 μg/mL), murine IL-3 (10 μg/mL), human IL-6 (10 μg/mL), and 3 U/mL recombinant human erythropoietin. After 14 days of culture resultant colonies were counted and scored based upon morphology using an inverted microscope. For LPS (lipopolysaccharide) stimulation and colony analysis, 10 μg/mL LPS was added to ES cell cultures on day 0 of primary embryoid body formation and cells were cultured for 10 days as described. During secondary colony formation, 10 μg/mL LPS was added to cultures at day 0 and colonies were counted and scored at day 14. All hematopoietic colonies were subsequently harvested and furthermore analyzed by flow cytometry.
Real-time reverse-transcriptase polymerase chain reaction (RT-PCR)
Total RNA was isolated from ES cells using the RNeasy kit (Qiagen, Chatsworth, CA). First-strand cDNA was synthesized from 4 μg of total RNA using the Superscript II First-strand synthesis system (Invitrogen). Real-time quantitative RT-PCR was set up using Taqman Universal PCR Master Mix (Applied Biosystems, Bedford, MA) and analyzed on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Taqman gene expression assays for Twist-1 and Twist-2 were ordered from Applied Biosystems. All data was normalized to 18S rRNA internal controls. For semiquantitative analysis, primer pairs for pluripotent markers, Oct-3/4 (sense—TCT TTC CAC CAG GCC CCC GGC TC; antisense—TGC GGG CGG ACA TGG GGA GAT CC, annealing Tm = 60°C), Nanog (Sense—CAG GTG TTT GAG GGT AGC TC; antisense—CGG TTC ATC ATG GTA CAG TC, annealing Tm = 55°C) and β-actin (sense—GCT CGT CGT CGA CAA CGG AAG; antisense—CAA ACA TGA TCT GGG TCA TCT TCT C, annealing Tm = 58°C) as a internal control were used.
Flow cytometry and cell sorting
Single cell suspensions of ES cells or harvested hematopoietic colonies were stained for 30 min at 4°C with the following antibodies as indicated: phycoerythrin (PE)-conjugated anti-SSEA-1 (clone 480, Santa Cruz), PE-Cy7-conjugated anti-Sca-1 (D7, BD PharMingen, San Diego, CA), anti-Gr-1 (RB5-8C5, PharMingen), anti-CD11b (M1/70, BD PharMingen), PE-conjugated anti-CD45 (30-F11, BD PharMingen), allophycocyanin (APC)-conjugated anti-c-Kit (CD117) (2B8, BD PharMingen), anti-F4/80 (BM8, Caltag, Burlingame, CA). A panel of antibodies and matched isotype controls were used for staining mouse ES cells and colonies. Anti-CD16/CD32 Fcγ III/II receptor antibodies (BD Pharmingen) were routinely used to pretreat cells at 4°C for 30 min to block nonspecific Fcγ receptor binding in some experiments. Cell sorting was performed on a MoFlow (Dako, Carpenturia, CA) cell sorter and flow cytometric analysis was performed on a FACSaria (BD) with data analysis using FlowJo Software (TreeStar, Ashland, Or).
Statistical analysis
We used a two-tailed Student's t-test for statistical significance analysis (InStat 2.01 program, Graph Pad Software, San Diego, CA), and a 95% confidence limit was taken to be significant, defined as p < 0.05. Results are typically presented as means ± standard errors as indicated.
Results
Generation of stable Twist-2 silenced ES cells
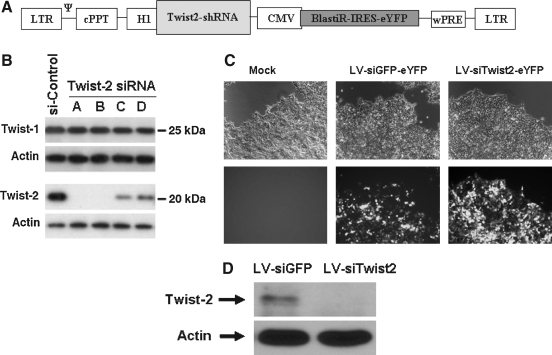
To investigate the role of Twist-2 in embryonic stem cells we generated a lentiviral construct expressing Twist-2 shRNA followed by a blasticidin resistance gene and eYFP as a marker (Fig. 1A). To select siRNA sequences that can efficiently silence target gene expression, four different siRNA sequences targeting Twist-2 were designed using the Dharmacon siDesign Center. These siRNAs were screened for specificity and potency of knockdown by western blotting in a transient transfection assay using FLAG-tagged Twist-2 expression constructs in 293T cells (Fig. 1B). The siRNAs in lanes A and B demonstrated highly potent and specific knockdown of Twist-2 without affecting the levels of Twist-1. It has been reported that target gene expression can be effectively downregulated in undifferentiated murine ES cells using a lentiviral delivery system (Zaehres, H., and Daley, G.Q., 2006). Lentivirus containing the shRNA expression construct was produced as described in the Materials and Methods section. We observed high-intensity YFP expression in 293T cells transduced with lentivirus containing Twist-2 shRNA (LV-siTwist-2) and lentivirus containing GFP shRNA (LV-siGFP) compared to mock control (Fig. 1C). Furthermore, we confirmed that, in contrast to the cells infected with LV-siGFP, endogenous Twist-2 expression was efficiently downregulated in 293T cells transduced with lentivirus containing Twist-2 shRNA. Actin expression, as an internal control was similar in both groups (Fig. 1D). Thus, we concluded that our lentiviral vector containing Twist-2 shRNA sequence (LV-siTwist2) expressed from the H1-RNA promoter can efficiently silence endogenous Twist-2 expression.
FIG. 1.
Generation of lentivirus expressing Twist-2 shRNA and verification of Twist-2 knockdown efficiency. (A) A schematic diagram of lentiviral construct containing Twist-2 shRNA sequence driven by an H1 promoter followed by the eYFP marker driven by a CMV promoter. (B) Western blotting for FLAG-tagged Twist-1 and Twist-2 expression in 293T cell lysates cotransfected with indicated siRNA oligos. Actin was used as a loading control. Results representative of three independent experiments. (C) Bright-field and fluorescent microscopy of 293T cells transduced with indicated lentiviral vectors. Strong fluorescence was observed at 48 h in cells transduced with constructs harboring eYFP marker. Original magnification; × 20. (D) Western blotting for endogenous Twist-2 expression in 293T cells transduced with lentivirus encoding GFP shRNA and Twist-2 shRNA. Results representative of three independent experiments.
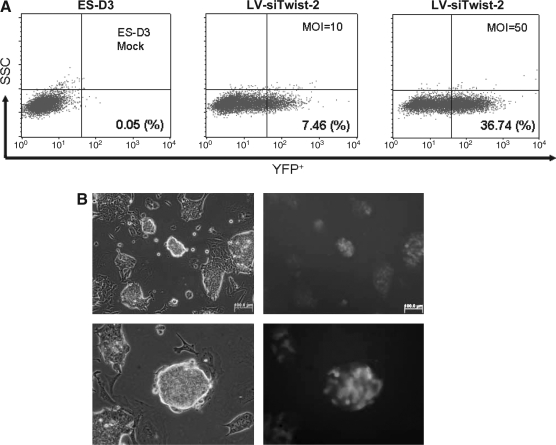
Next we generated a stable Twist-2 silenced murine embryonic stem cell line and a control GFP shRNA expressing embryonic stem cell line in parallel. First, we tested whether these lentiviral vectors can efficiently and stably drive transgene expression in murine ES cells. Undifferentiated ES cells were cultured on gelatine-coated plates and then transduced with lentivirus encoding Twist-2 shRNA (LV-siTwist-2 ES) at MOIs of 10 and 50. Cells were cultured for 48 h in the presence of leukemia inhibitory factor (LIF) and expanded. The transduction efficiency was evaluated by monitoring YFP expression by FACS analyses. We found that at 72 h after transduction approximately 7% of ES cells detectably expressed YFP when transduced at an MOI of 10. When cells were transduced at an MOI of 50, 36% of ES cells were shown to be YFP+ (Fig. 2A). The cytotoxic effects of lentiviral transduction were also examined by analyzing the number of living cells using Trypan blue staining. At 72 h after transduction at an MOI of 50, less than 5% displayed cytotoxic effects relating to lentiviral transduction (data not shown). To propagate YFP+ homogenous cells, after 48 h, resulting YFP high expressing cells were sorted and maintained in culture as described in the Materials and Methods section. ES cell colonies from both the Twist-2 shRNA (LV-siTwist2 ES) and GFP shRNA (LV-siGFP ES) cells were established and maintained proper morphology with bright expression of the YFP marker (Fig. 2B). These cells were maintained in parallel to ensure equal passage number.
FIG. 2.
Generation of stable Twist-2 shRNA expressing embryonic stem cells. (A) Flow cytometry scatter plots of YFP positive ES-D3 cells transduced with lentivirus at indicated MOIs of 10 and 50. Representative gates in dot plots were used to sort YFPhigh ES cells for culture. (B) Bright-field (left panels) and fluorescent images (right panels) of sorted Twist-2 shRNA ES cells cultured on gelatin coated plates. Low magnification (upper images) and high magnification (lower images).
Characterization of Twist-2-silenced (LV-siTwist-2) ES cells
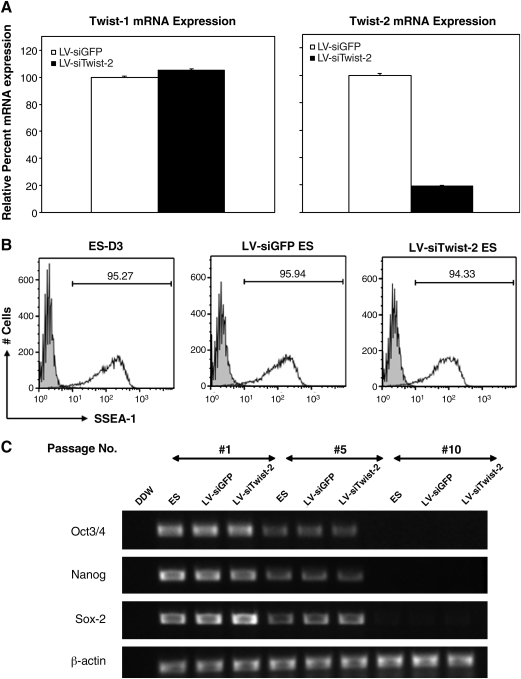
Next we verified knockdown of Twist-2 in the ES cell line. ES cells from the Twist-2 shRNA expressing cells (LV-siTwsit-2 ES) were cultured on gelatin-coated plates with medium containing LIF, harvested, and mRNA was isolated using the RNAeasy Quiagen Kit. Real-time quantitative RT-PCR was subsequently performed to analyze expression of Twist-2. We observed an approximately 80% reduction in Twist-2 mRNA in the LV-siTwist-2 ES cells compared to the LV-siGFP ES cells (Fig. 3A). However, Twist-1 mRNA levels were unchanged, indicating specific and potent knockdown of Twist-2 in the LV-siTwist-2 ES cells (Fig. 3A).
FIG. 3.
Twist-2 silenced ES cells maintain an undifferentiated state with high expression of SSEA-1. (A) Real-time quantitative RT-PCR for Twist-1 and Twist-2 expression in mRNA isolated from indicated ES cells. Significant knockdown of Twist-2 mRNA is observed the stable Twist-2 shRNA, LV-siTwist-2 ES cells, whereas Twist-1 mRNA levels are unchanged compared to the control GFP shRNA, LV-siGFP ES cells. Data represent the means ± SD of triplicate well from representative of three independent experiments. (B) Flow cytometry of SSEA-1 expression on the surface of sorted YFP+ transduced ES cells or parental ES cells cultured in the presence of LIF. Experiment was repeated twice with similar results. (C) Semiquantitative RT-PCR analyses showing the comparable expression levels of pluripotent marker genes (Oct-3/4, Nanog and Sox-2) at the passage #1 of transduced ES cells grown from sorted YFP+ transduced ES cells and parental ES cells in the presence of LIF. Both LV-siTwist-2 and LV-siGFP ES cells gradually lost the expression of pluripotent marker genes in the in vitro differentiation culture without LIF at the comparable rate (passages #5 and 10). β-Actin as used as an internal control.
Because Twist-1 and Twist-2 are expressed at relatively high levels in ES cells (data not shown) and known to play important roles in embryogenesis and mesoderm development, knockdown of one of these important transcription factors could impact the ability to culture these cells in an undifferentiated state. To investigate this possibility, we cultured the LV-siTwist-2 ES cells for multiple passages and stained for Stage-Specific Embryonic Antigen-1 (SSEA-1), a marker of undifferentiated ES cells. We found that levels of SSEA-1 expression on LV-siTwist-2 ES cells in the ES culture with LIF (leukemia inhibitory factor) were similar to that of LV-siGFP ES cells and untreated ES cells as controls (Fig. 3B). Besides examining SSEA-1 expression levels on the cell surface, we used semiquantitative RT-PCR analyses to examine the expression levels of pluripotent marker genes in transduced ES cells. Figure 3C shows that expression levels of pluripotent marker genes (Oct-3/4, Nanog, and Sox-2) in transduced ES cells grown from sorted YFP+ LV-siTwist-2 ES cells after many rounds of cell division were comparable to those in LV-siGFP ES and parental ES cells in the ES culture with LIF (passage #1). Moreover, we observed that both LV-siTwist-2 and LV-siGFP ES cells spontaneously lost the expression of pluripotent marker genes in the in vitro differentiation culture without LIF at a comparable rate (passages #5 and #10). These data indicate that lentiviral transduction and knockdown of Twist-2 did not apparently affect the maintenance of these ES cells and their differentiation in vitro.
Increased primary embryoid body formation in the LV-siTwist-2 ES cells
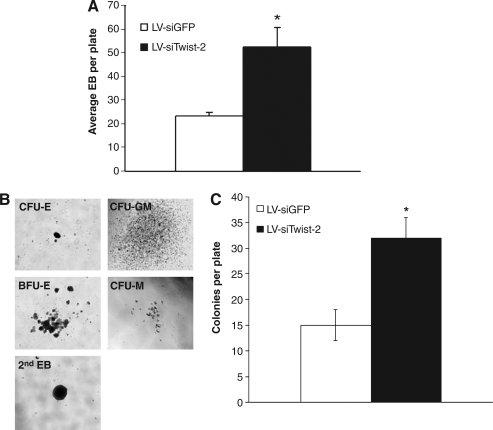
Twist-2 is known to inhibit the terminal differentiation of mesodermally derived tissues (Bialek et al., 2004; Hebrok et al., 1994; Lee et al., 2003; Rohwedel et al., 1995; Spicer et al., 1996). Our recent study found that Twist-2 is a key negative regulator of myeloid lineage development (Sharabi et al., 2008). We therefore set out to examine whether loss of this inhibitor in the LV-siTwist-2 ES cells would impact or enhance differentiation of mesodermally derived cell types, specifically hematopoietic lineages. To differentiate hematopoietic cells from ES cells we first cultured ES cells in methylcellulose to form primary embryoid bodies (EBs) (Dang et al., 2002). Surprisingly, we observed that formation of primary embryoid bodies was significantly enhanced in the LV-siTwist-2 ES cells compared to the LV-siGFP ES cells (Fig. 4A, p < 0.05, two-tail t-test). To examine the effects of growth factors influencing the formation of primary EB, we also compared the efficiency of EB formation derived from LV-siTwist-2 ES cells and LV-siGFP ES cells in the absence of growth factors. We observed that a sufficient number of primary EB failed to develop in the absence of both growth factors and serum. There were no significant differences in generation of primary EB compared to LV-siGFP ES cells as a control (data not shown). Thus, given the same number of plated ES cells, the LV-siTwist-2 ES cells generated more primary embryoid bodies in the presence of growth factors, suggesting an enhanced generation of primary EB formation caused by loss of Twist-2 in ES cells under the growth factor stimulation.
FIG. 4.
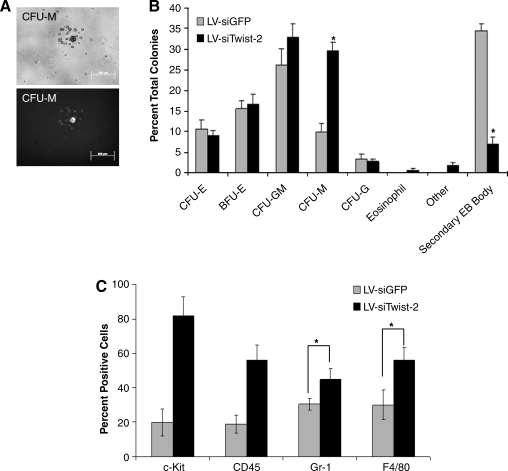
Embryoid body formation upon primary and secondary differentiation is significantly enhanced in Twist-2 silenced ES cells. (A) Results from primary embryoid body (EB) colony counting assay. The stable LV-siTwist-2 ES cells generates a significantly increased frequency of primary embryoid bodies with the size of larger than 100 μm (n = 8 per group, 60 × 15 mm plates). Data are shown as means ± standard error mean (SEM) of two independent experiments. *p < 0.05, two-tail t-test; LV-siTwist-2 versus LV-siGFP ES cells. (B) Representative images for different colonies, colony forming unit (CFU)-erythroid (E), CFU-GM (granulocyte-macrophage), burst forming unit (BFU)-E, CFU-M (macrophage), and secondary EB (2nd EB) at day 14 of secondary hematopoietic differentiation derived from LV-siTwist-2 ES cells. (C) Results of total colonies counts at day 14 of secondary hematopoietic differentiation. The stable LV-siTwist-2 ES cell generated a significantly increased frequency of secondary hematopoietic colonies (n = 8 per group, 60 × 15 mm plates). Data are shown as means ± standard error mean (SEM) of two independent experiments. *p < 0.05, two-tail t-test; LV-siTwist-2 versus LV-siGFP ES cells.
Because the hematopoietic system is derived from the mesoderm, we performed directed secondary hematopoietic differentiation using the cells from primary EB to investigate if there were any changes in mesodermal differentiation. First, the cells from the primary EB were dissociated, counted, and replated in a methylcellulose semisolid media with additional growth factors and cytokines supporting multilineage hematopoietic differentiation. Secondary colonies were cultured for 14 days and counted and scored for colony type based on colony morphology (Fig. 4B). Interestingly, we found an increased frequency of secondary hematopoietic colonies derived from the LV-siTwist-2 ES cells compared to the LV-siGFP ES cells (Fig. 4C, p < 0.05, Two-tail t-test). Thus, these results show that the Twist-2 silenced ES cells significantly increased not only primary EB formation, but also mesodermal differentiation of hematopoietic lineages, indicating that Twist-2 expression suppresses the differentiation of hematopoietic progenitors derived from ES cells.
Enhanced myeloid lineage differentiation during hematopoietic differentiation of Twist-2 silenced ES cells
To determine the types of hematopoietic colonies produced from Twist-2 silenced ES cells, we screened and scored the colony types according to their respective morphologies. First, to verify that hematopoietic colonies counted were all derived from transduced ES cells we imaged differentiated colonies under the fluorescence microscope and observed detectable levels of YFP fluorescence (Fig. 5A). Upon scoring the colonies we observed a skewed differentiation towards myeloid colonies of the CFU (colony forming unit)-M (macrophage) type (Fig. 5B). Importantly, this increase in the relative percentage of CFU-M colonies was associated with a dramatic decrease in the percentage of secondary EB remaining in culture (Fig. 5B). Secondary EBs are thought to be derived from the cells of primary EB, which do not undergo further differentiation (Chan et al., 2003). Thus, this suggests that silencing Twist-2 enhances the susceptibility or decreases the threshold for hematopoietic differentiation of primary EB cells.
FIG. 5.
Twist-2 silenced ES cells are dramatically skewed toward myeloid lineages during hematopoietic differentiation. (A) Representative images of the hematopoietic CFU-M colony derived from ES cells transduced with LV-siTwist-2 after 14 days hematopoietic differentiation; bright-field (upper image) and fluorescence (lower). (B) Results of hematopoietic colony scoring assay. The Twist-2 silenced, LV-siTwist ES cells generates a significantly skewed percentage of macrophage colonies (CFU-M), but reduced numbers of secondary EBs formation as well compared to a control, LV-siGFP ES cells. Each different hematopoietic colony based on the morphology was counted in each group (n = 6 per group, 60-mm plates) and calculated a percentage in total colonies of each group. Data are presented as a percentage means ± standard error mean (SEM) of colonies from three individual experiments. *p < 0.05, two-tail t-test; LV-siTwist-2 versus LV-siGFP. (C) Results from flow cytometry analyses showing positive hematopoietic colonies with indicated markers (c-Kit, CD45, Gr-1, and F4/80) at day 14 hematopoietic differentiation derived from LV-siGFP, LV-siTwist-2 ES cells. Hematopoietic colonies were harvested and stained with markers according to the Materials and Methods section. All data are shown as means percentages of double-positive cells (YFP+) ± standard error mean (SEM) of three independent experiments. *p < 0.05, two-tail t-test; LV-siTwist-2 vs. LV-siGFP.
To further characterize and identify these cell populations we performed FACS analysis of secondary hematopoietic colonies after mature development at 14 days. We stained single-cell suspensions of secondary hematopoietic colonies for the stem cell and hematopoietic markers Sca-1, c-Kit, and CD45, as well as markers of differentiated progeny Mac-1, F4/80, and Gr-1 (Fig. 5C). At the early time point (day 7) of hematopoietic differentiation we observed increases in the hematopoietic stem cell markers CD45 in cells derived from the Twist-2 silenced ES cell line (data not shown). At the later time point (day 14) we observed a marked increase in the percentage of Twist-2 silenced cells stained positive for the hematopoietic and myeloid progenitor marker c-Kit and hematopoietic marker CD45 (Fig. 5C). Sca-1−c-Kit+ positive hematopoietic progenitors differentiated from LV-siTwist-2 ES cells were dramatically increased compared to a control LV-siGFP (data not shown). We also observed increases in the percentages of Twist-2 silenced cells staining positive for F4/80, which is a macrophage-restricted cell surface glycoprotein and Gr-1, which is known as a marker of differentiated monocytes and granulocytes (Fig. 5C). These results indicate that silencing of Twist-2 in ES cells enhances the differentiation and generation of F4/80+ macrophage population as well as Gr-1+ myeloid cells. However, in contrast to the increasing percentage of myeloid cells, the percentage of erythroid Ter-119-positive cells differentiated from LV-siTwist-2 ES cells was slightly decreased when compared to LV-siGFP ES cells (data not shown), suggesting that differentiation is skewed away from erythroid lineages and toward myeloid lineages in the Twist-2 silenced ES cell line. Taken together, these results support our hypothesis that silencing Twist-2 in ES cells is an effective strategy for enhancing the yield and differentiation of mesodermally derived hematopoietic lineages from ES cells.
TLR ligands further promote primary EB formation and hematopoietic differentiation of Twist-2-silenced ES cells
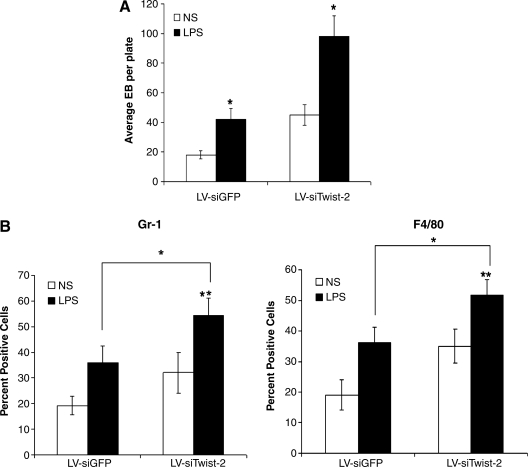
Twist-2 is an inducible feedback regulator of TLR-mediated transcription of NF-κB (Sosic et al., 2003). Our recent study found that Twist-2 functions as a key negative regulator of myeloid lineage differentiation and that Twist-2-silenced cells are hypersensitive to stimulation with LPS (Sharabi et al., 2008). Furthermore, we found that TLRs such as TLR4 were constitutively expressed on murine ES cells (Lee et al., 2009). It was also reported that TLR ligands stimulate the generation of hematopoietic progenitors for rapid replenishment of the innate immune system during infection (Nagai et al., 2006). Hence, we tested the hypothesis that stimulation with TLR ligands may further promote the myeloid lineage differentiation of Twist2-silenced ES cells. Figure 6A shows that stimulation with LPS significantly promoted the formation of primary EB derived from both LV-siGFP and LV-siTwist-2 ES cells. Furthermore, LPS stimulation consistently enhanced the myeloid differentiation, as manifested by increased generation of F4/80+ macrophage and Gr-1+ myeloid cells derived from both LV-siGFP and LV-siTwist-2 ES cells (Fig. 6B). LPS stimulation more effectively promoted the formation of EBs, as well as myeloid cell differentiation of LV-siTwist-2 ES cells compared to LV-siGFP ES cells (Fig. 6A and B). Taken together, these data suggest a synergistic effect of TLR signaling and silencing of Twist-2 in promoting the differentiation of myeloid lineages from ES cells.
FIG. 6.
Enhanced generation of myeloid cells differentiated from Twist-2 silenced ES cells in response to stimulation with TLR agonists. (A) Enhanced primary EB formation of Twist-2-silenced ES cells by LPS stimulation. Primary embryoid body (EB) formation assays were performed by counting EB (n = 6 per group, 60 × 15-mm plates) after 10 days culture with, or without LPS (10 μg/mL) stimulation. NS, no stimulation. Data are shown as means ± standard error mean (SEM) of threee independent experiments. *p < 0.05, two-tail t-test; LV-siTwist-2 versus LV-siGFP. (B) Enhanced myeloid differentiation of Twist-2-silenced ES cells by LPS stimulation. Increased Gr-1+ or F4/80+ myeloid cells differentiated from LV-siTwist-2 ES cells after 14-day culture in the presence of LPS (10 μg/mL) stimulation, or without stimulation. Data presented as means ± standard error mean (SEM) of three independent experiments. *p < 0.05, two-tail t-test; LV-siGFP versus LV-siTwist-2; **p < 0.05, LPS versus NS.
Discussion
ES cells have great potential as a source for regenerative medicine-based therapies in the future. However, to maximize clinical efficacy, further strategies have to be developed to enhance the yield and purity of the specific cell types and tissues derived from ES cells. Here we used siRNA technology to enhance hematopoietic differentiation from ES cells by silencing Twist-2, a critical regulator of mesodermal development during embryogenesis. We demonstrate that Twist-2 silenced ES cells generate an increased yield of primary embryoid bodies as well as secondary mesodermally derived hematopoietic colonies. We show that silencing Twist-2 in ES cells skews hematopoietic differentiation toward specific myeloid lineages, raising the possibility that specific cell types can also be targeted for increased purity using this strategy. Finally, we demonstrate a synergistic effect of TLR signaling and silencing of Twist-2 in promoting the differentiation of myeloid lineages from ES cells.
Twist-2 plays multiple key roles in regulating mesodermal development during embryogenesis (O'Rourke and Tam, 2002). Initially Twist is critical for proper embryonic patterning, gastrulation, and mesoderm development (Chen and Behringer, 1995; O'Rourke et al., 2002). During early development Twist is expressed in mesenchymal cells and mesodermal progenitors. However, in later stages of development expression of Twist actually inhibits the terminal differentiation of mesodermally derived tissues (Hebrok et al., 1994; Lee et al., 2003; Murray et al., 1992; Spicer et al., 1996). Indeed, constitutive expression of Twist was reported to inhibit muscle differentiation from murine embryonic stem cells (Rohwedel et al., 1995), and aberrant overexpression of Twist is sufficient to block differentiation of osteoblasts (Bialek et al., 2004). Furthermore, forced expression of Twist can even revert differentiated cells into a less differentiated state (Hjiantoniou et al., 2008). Conversely, loss of Twist expression is correlated with the rapid induction of terminal differentiation (Bialek et al., 2004). Thus, taken together, Twist acts as a critical inducer of mesodermal determination and development, but simultaneously functions to maintain cells in an undifferentiated mesodermal state. Generation of embryoid bodies is a key step in many embryonic stem cell differentiation protocols, and we used methylcellulose culture to induce EB formation because it has been shown to be efficient for mesodermal and hematopoietic differentiation (Kurosawa, 2007). Our finding of increased primary embryoid body formation in the Twist-2 silenced ES line is intriguing given the importance of Twist in mesodermal development. Primary EB are spherical structures that contain cells of the three germ layers: ectoderm, mesoderm, and endoderm. They resemble blastulas and undergo cellular differentiation that is similar in some regards to that which occurs during mammalian embryogenesis (Kurosawa, 2007). Twist is a powerful inducer and regulator of the epithelial to mesenchymal transition (EMT), which is critical for gastrulation and the development of mesoderm and endoderm (Yang et al., 2004). Silencing Twist-2 may thus disrupt the mechanisms regulating generation of EB and the three germ layers from ES cells and result in ES cells that are more prone to primary EB formation. One potential possibility is that in the absence of Twist-2 dysregulated differentiation of ectoderm results in an increase in EB with an altered composition of the three germ layers. Nevertheless, further studies are required to elucidate the role of Twist-2 in EB development from ES cells.
Twist-2 was recently found to have a critical role in controlling myeloid lineage differentiation (Sharabi et al., 2008). Moreover, Twist-2 is known to inhibit the terminal differentiation of mesodermally derived tissues, loss of Twist-2 is likely to enhance the differentiation of hematopoietic cells from mesodermal progenitors. The results of this study demonstrate the enhanced differentiation into the enhanced Sca-1−c-Kit+-positive hematopoietic progenitors and myeloid cells from ES cells by silencing Twist-2. This study suggests a novel strategy to enhance the yield of specific ES cell differentiation by removing specific inhibitors of cellular differentiation, such as Twist-2. Based upon our recent finding that TLRs such as TLR4 were constitutively expressed on murine ES cells (Lee et al., 2009), in this study we tested and demonstrated the stimulatory effect of TLR ligands on promoting the myeloid differentiation from ES cells, suggesting a synergistic effect of TLR signaling and Twist-2 silencing for enhanced myeloid differentiation. However, this strategy of silencing a transcriptional repressor for directed differentiation may be necessary, but not sufficient for directed differentiation to occur. Further investigation is clearly needed to determine the applicability of this strategy for directed differentiation by silencing various transcriptional repressors in both human and murine ES cells.
Acknowledgments
This work was supported by grants from the National Institute of Health (R01CA116677) and the Leukemia and Lymphoma Society SCOR.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Beyer K.S. Beauchamp R.L. Lee M.F., et al. Mediator subunit MED28 (Magicin) is a repressor of smooth muscle cell differentiation. J. Biol. Chem. 2007;282:32152–32157. doi: 10.1074/jbc.M706592200. [DOI] [PubMed] [Google Scholar]
- Bialek P. Kern B. Yang X., et al. A twist code determines the onset of osteoblast differentiation. Dev. Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Chan R.J. Johnson S.A. Li Y., et al. A definitive role of Shp-2 tyrosine phosphatase in mediating embryonic stem cell differentiation and hematopoiesis. Blood. 2003;102:2074–2080. doi: 10.1182/blood-2003-04-1171. [DOI] [PubMed] [Google Scholar]
- Chen Z.F. Behringer R.R. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Copray S. Balasubramaniyan V. Levenga J., et al. Olig2 overexpression induces the in vitro differentiation of neural stem cells into mature oligodendrocytes. Stem Cells. 2006;24:1001–1010. doi: 10.1634/stemcells.2005-0239. [DOI] [PubMed] [Google Scholar]
- Dahl L. Richter K. Hagglund A.C., et al. Lhx2 expression promotes self-renewal of a distinct multipotential hematopoietic progenitor cell in embryonic stem cell-derived embryoid bodies. PLoS ONE. 2008;3:e2025. doi: 10.1371/journal.pone.0002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang S.M. Kyba M. Perlingeiro R., et al. Efficiency of embryoid body formation and hematopoietic development from embryonic stem cells in different culture systems. Biotechnol. Bioeng. 2002;78:442–453. doi: 10.1002/bit.10220. [DOI] [PubMed] [Google Scholar]
- Docherty K. Bernardo A.S. Vallier L. Embryonic stem cell therapy for diabetes mellitus. Semin. Cell Dev. Biol. 2007;18:827–838. doi: 10.1016/j.semcdb.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Fehling H.J. Lacaud G. Kubo A., et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- Hebrok M. Wertz K. Fuchtbauer E.M. M-twist is an inhibitor of muscle differentiation. Dev. Biol. 1994;165:537–544. doi: 10.1006/dbio.1994.1273. [DOI] [PubMed] [Google Scholar]
- Hjiantoniou E. Anayasa M. Nicolaou P., et al. Twist induces reversal of myotube formation. Differentiation. 2008;76:182–192. doi: 10.1111/j.1432-0436.2007.00195.x. [DOI] [PubMed] [Google Scholar]
- Jankovic V. Ciarrocchi A. Boccuni P., et al. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:1260–1265. doi: 10.1073/pnas.0607894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhas S. Ciura S. Belanger-Jasmin S., et al. Hes6 inhibits astrocyte differentiation and promotes neurogenesis through different mechanisms. J. Neurosci. 2006;26:11061–11071. doi: 10.1523/JNEUROSCI.1358-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassowska A. Gordon-Keylock S. Samuel K., et al. Promotion of haematopoietic activity in embryonic stem cells by the aorta-gonad-mesonephros microenvironment. Exp. Cell Res. 2006;312:3595–3603. doi: 10.1016/j.yexcr.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J. Biosci. Bioeng. 2007;103:389–398. doi: 10.1263/jbb.103.389. [DOI] [PubMed] [Google Scholar]
- Ledran M.H. Krassowska A. Armstrong L., et al. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 2008;3:85–98. doi: 10.1016/j.stem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Lee S. Sharabi A. Huang X.F., et al. Embryonic stem cells and mammary luminal progenitors directly sense and respond to microbial products. Stem Cells. 2009;27:1604–1615. doi: 10.1002/stem.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S. Lee H.H. Park J., et al. Twist2, a novel ADD1/SREBP1c interacting protein, represses the transcriptional activity of ADD1/SREBP1c. Nucleic Acids Res. 2003;31:7165–7174. doi: 10.1093/nar/gkg934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountford J.C. Human embryonic stem cells: origins, characteristics and potential for regenerative therapy. Transfus. Med. 2008;18:1–12. doi: 10.1111/j.1365-3148.2007.00807.x. [DOI] [PubMed] [Google Scholar]
- Murray S.S. Glackin C.A. Winters K.A., et al. Expression of helix-loop-helix regulatory genes during differentiation of mouse osteoblastic cells. J. Bone Miner. Res. 1992;7:1131–1138. doi: 10.1002/jbmr.5650071004. [DOI] [PubMed] [Google Scholar]
- Nagai Y. Garrett K.P. Ohta S., et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke M.P. Soo K. Behringer R.R., et al. Twist plays an essential role in FGF and SHH signal transduction during mouse limb development. Dev. Biol. 2002;248:143–156. doi: 10.1006/dbio.2002.0730. [DOI] [PubMed] [Google Scholar]
- O'Rourke M.P. Tam P.P. Twist functions in mouse development. Int. J. Dev. Biol. 2002;46:401–413. [PubMed] [Google Scholar]
- Passier R. van Laake L.W. Mummery C.L. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–329. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- Rohwedel J. Horak V. Hebrok M., et al. M-twist expression inhibits mouse embryonic stem cell-derived myogenic differentiation in vitro. Exp Cell Res. 1995;220:92–100. doi: 10.1006/excr.1995.1295. [DOI] [PubMed] [Google Scholar]
- Schroers R. Chen S.Y. Lentiviral transduction of human dendritic cells. Methods Mol. Biol. 2004;246:451–459. doi: 10.1385/1-59259-650-9:451. [DOI] [PubMed] [Google Scholar]
- Sharabi A.B. Aldrich M. Sosic D., et al. Twist-2 controls myeloid lineage development and function. PLoS Biol. 2008;6:e316. doi: 10.1371/journal.pbio.0060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. Evel-Kabler K. Strube R., et al. Silencing of SOCS1 enhances antigen presentation by dendritic cells and antigen-specific anti-tumor immunity. Nat. Biotechnol. 2004;22:1546–1553. doi: 10.1038/nbt1035. [DOI] [PubMed] [Google Scholar]
- Smith A.G. Heath J.K. Donaldson D.D., et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Sosic D. Richardson J.A. Yu K., et al. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Spicer D.B. Rhee J. Cheung W.L., et al. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- Suter D.M. Krause K.H. Neural commitment of embryonic stem cells: molecules, pathways and potential for cell therapy. J. Pathol. 2008;215:355–368. doi: 10.1002/path.2380. [DOI] [PubMed] [Google Scholar]
- Szabo E. Qiu Y. Baksh S., et al. Calreticulin inhibits commitment to adipocyte differentiation. J. Cell Biol. 2008;182:103–116. doi: 10.1083/jcb.200712078. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Thomson J.A. Itskovitz-Eldor J. Shapiro S.S., et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wang Y.Y. Deng X. Xu L., et al. Bcl2 enhances induced hematopoietic differentiation of murine embryonic stem cells. Exp. Hematol. 2008;36:128–139. doi: 10.1016/j.exphem.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.L. Hilton D.J. Pease S., et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Yang J. Mani S.A. Donaher J.L., et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Zaehres H. Daley G.Q. Transgene expression and RNA interference in embryonic stem cells. Methods Enzymol. 2006;420:49–64. doi: 10.1016/S0076-6879(06)20004-1. [DOI] [PubMed] [Google Scholar]
- Zhang J. Grindley J.C. Yin T., et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]