Abstract
Background
Human leukocyte antigen (HLA) genes are characterized by high levels of polymorphism and linkage disequilibrium (LD), important characteristics to study the genetic background of human populations and their genetic structure. Here, we analyse the allele distribution and LD extent of HLA class I and II in São Miguel Island population (Azores archipelago, Portugal).
Findings
The sample set was composed of 106 healthy blood donors living in São Miguel Island obtained from the anonymized Azorean DNA bank. HLA class I (-A, -B and -Cw) and class II (-DRB1, -DQB1, -DPA1 and -DPB1) genotyping was performed by PCR-SSP Olerup SSP™ (GenoVision Inc.), according to the manufacturer's instructions.
Genetic diversity values, based on the 7 loci, ranged from 0.821 both for HLA-DPA1 and -DQB1 to 0.934 for HLA-B, with a mean value of 0.846. Analysis of 5 HLA-A-Cw-B-DRB1-DQB1 haplotypes revealed that A*01-Cw*07-B*08-DRB1*03-DQB1*02 is the most frequent in São Miguel (7.9%) followed by A*24-B*08-Cw*07-DRB1*03-DQB1*02 (3.8%). In addition, even though the reports of high LD for HLA markers in worldwide populations, São Miguel islanders do not have extensive LD (average D' = 0.285).
Conclusions
In summary, the results demonstrate high variability of HLA in São Miguel Island population as well as absence of genetic structure and extensive LD. The data here presented suggest that in São Miguel islanders autoimmune diseases studies will necessarily encompass a more focused analysis of HLA extended haplotypes as well as the evaluation of other non-HLA candidate genes.
Background
The Azores is a Portuguese archipelago composed of nine islands distributed by three geographical groups: the Eastern (São Miguel and Santa Maria), the Central (Terceira, Pico, Faial, São Jorge and Graciosa) and the Western (Flores and Corvo). The Portuguese explorers, who discovered the archipelago in 1427, only started the settlement in 1439 through a long and difficult process. Historical data report a contribution from people with genetic backgrounds other than Portuguese, including Flemish, Spanish, French, Italian, German, Scottish, Jewish, and also from Moorish prisoners and black slaves from Guinea, Cape Verde and São Tomé [1]. São Miguel is the largest island of the Azores and is composed of 131,609 inhabitants (2001 Census, Portugal National Institute of Statistics). Several studies have been performed to characterize the genetic pool of the Azoreans [2-10]. These studies report a high genetic variability and heterogeneity of the Azorean population, explained by the settling history of the islands, where a major contribution of mainland Portugal individuals is evident. Moreover, the data revealed absence of population structure, even though the archipelago's geographical discontinuity and demographic disproportionality. Currently, this knowledge is being fundamental for the design and development of pharmacogenetic research and genetic studies in common diseases, such as cardiovascular and autoimmune diseases.
The human leukocyte antigen (HLA) genes, a central component of the major histocompatibility complex (MHC) on 6p21.3, encode polymorphic class I, II and III molecules that play a major role in the immune response [11]. In addition, HLA loci are characterized by high levels of polymorphism and linkage disequilibrium (LD), important characteristics to study the genetic background of human populations, as well as their present-day genetic structure. Here, we analyse the allele frequency and LD extent of HLA class I and II, in order to identify its diversity and haplotype distribution and to gain further insight in the potential use of this genomic region for the study of autoimmune diseases in the São Miguel Island population.
Materials and methods
Population samples, genotyping and statistical analysis
The sample set was composed of 106 healthy blood donors living in São Miguel Island obtained from the anonymized Azorean DNA bank located at the Hospital of Divino Espírito Santo of Ponta Delgada, EPE, the main hospital in Azores [12]. HLA class I (-A, -Cw and -B) and class II (-DRB1, -DQB1, -DPA1 and -DPB1) genotyping was performed by PCR-SSP Olerup SSP™ (GenoVision Inc.), according to the manufacturer's instructions. After electrophoresis on a 4% agarose gel stained with SYBR® Green, PCR products were visualized, followed by HLA allele identification using the Helmberg-SCORE™ software version 3.320T (Olerup SSP AB, Saltsjöbaden, Sweden).
Average gene diversity and estimation of the HLA haplotypes was carried out using Arlequin v3.0 [13]. Evaluation of standardized multiallelic disequilibrium coefficient, D', was performed by the Haploxt application from the GOLD software. Average D' values were calculated by a simple mathematical mean of all values obtained for each marker pair. Nei's FST genetic distance matrix was computed between pairs of populations by DISPAN [14] and used to construct a Neighbor-Joining (NJ) tree by PHYLIP 3.63 [15]. We employed TreeView 1.6.6 [16] to display tree phylogenies obtained from NJ. In order to obtain the best results concerning population comparisons a compromise between the number of populations and HLA loci was performed. Consequently, HLA-DPA1 and -DPB1 were excluded from analysis. FST values were based on allele frequencies obtained in an online database (HLA-Allele Project; http://www.allelefrequencies.net/), in 19 populations for 5 HLA loci: São Miguel, Terceira, Italy, France, Germany, Belgium, Turkey, Morocco, Japan, Mongolia Oold, Mongolia Tsaatan, Mongolia Khalkha, Basque, Ibiza, Majorca, Majorca Jewish, Chuetas, Minorca and Jordania. Along with FST values, 5 loci haplotypes were searched in the same database to further investigate the possible origins of the early settlers.
Results
The analysis of the HLA alleles in the São Miguel Island population (Table 1) revealed for the HLA-A locus a total of 16 different alleles, 13 HLA-Cw and 24 HLA-B alleles. Regarding HLA class II loci, we found 22 HLA-DPB1, 13 HLA-DRB1, 5 HLA-DQB1 and 6 HLA-DPA1 different alleles. HLA-B and HLA-DPB1 are the two loci with the highest numbers of alleles, suggesting higher diversity for these markers. The highest frequency observed, 0.462, was in HLA-DPA1 gene, which shows a low number of alleles. In contrast, the lowest frequency identified (0.5%) was present in HLA-A, -B and -DPB1 (Table 1). Genetic diversity values ranged from 0.821 both for HLA-DPA1 and -DQB1 to 0.934 for HLA-B, with a mean value of 0.846 (Table 2). Overall, HLA allele frequencies in São Miguel, mainland Portugal and other European populations demonstrated absence of statistically significant differences (GST = 0.03; data not shown). According to Wright [17] values of GST smaller than 0.05 indicate little genetic differentiation.
Table 1.
HLA class I and II allele frequencies in São Miguel population (the highest values are in bold).
| HLA class I (2n = 212) | HLA class II (2n = 212) | ||||||
| Alleles | Rel. Freq. | Alleles | Rel. Freq. | Alleles | Rel. Freq. | Alleles | Rel. Freq. |
| HLA-A | HLA-B | HLA-DPB1 | HLA-DRB1 | ||||
| A*01 | 0.151 | B*07 | 0.066 | DPB1*0101 | 0.057 | DRB1*01 | 0.085 |
| A*02 | 0.250 | B*08 | 0.137 | DPB1*0201 | 0.212 | DRB1*03 | 0.165 |
| A*03 | 0.094 | B*13 | 0.005 | DPB1*0202 | 0.014 | DRB1*04 | 0.123 |
| A*11 | 0.042 | B*14 | 0.071 | DPB1*0301 | 0.080 | DRB1*07 | 0.170 |
| A*23 | 0.019 | B*15 | 0.052 | DPB1*0401 | 0.316 | DRB1*08 | 0.028 |
| A*24 | 0.137 | B*18 | 0.052 | DPB1*0402 | 0.094 | DRB1*09 | 0.019 |
| A*25 | 0.005 | B*27 | 0.042 | DPB1*0501 | 0.014 | DRB1*10 | 0.019 |
| A*26 | 0.009 | B*35 | 0.061 | DPB1*0601 | 0.005 | DRB1*11 | 0.118 |
| A*29 | 0.066 | B*37 | 0.014 | DPB1*0901 | 0.005 | DRB1*12 | 0.009 |
| A*30 | 0.033 | B*38 | 0.014 | DPB1*1001 | 0.028 | DRB1*13 | 0.146 |
| A*31 | 0.024 | B*39 | 0.009 | DPB1*1101 | 0.024 | DRB1*14 | 0.019 |
| A*32 | 0.061 | B*40 | 0.028 | DPB1*1301 | 0.052 | DRB1*15 | 0.075 |
| A*33 | 0.028 | B*41 | 0.024 | DPB1*1401 | 0.014 | DRB1*16 | 0.024 |
| A*66 | 0.005 | B*44 | 0.156 | DPB1*1501 | 0.005 | HLA-DQB1 | |
| A*68 | 0.071 | B*45 | 0.009 | DPB1*1601 | 0.005 | DQB1*02 | 0.302 |
| A*80 | 0.005 | B*47 | 0.005 | DPB1*1701 | 0.038 | DQB1*03 | 0.321 |
| HLA-Cw | B*49 | 0.052 | DPB1*1901 | 0.014 | DQB1*04 | 0.028 | |
| Cw*01 | 0.024 | B*50 | 0.033 | DPB1*2501 | 0.005 | DQB1*05 | 0.151 |
| Cw*02 | 0.066 | B*51 | 0.066 | DPB1*3901 | 0.005 | DQB1*06 | 0.198 |
| Cw*03 | 0.075 | B*53 | 0.024 | DPB1*5101 | 0.005 | ||
| Cw*04 | 0.104 | B*55 | 0.019 | DPB1*6601 | 0.005 | ||
| Cw*05 | 0.071 | B*57 | 0.042 | DPB1*7801 | 0.005 | ||
| Cw*06 | 0.090 | B*58 | 0.014 | HLA-DPA1 | |||
| Cw*07 | 0.311 | B*78 | 0.005 | DPA1*01 | 0.462 | ||
| Cw*08 | 0.052 | DPA1*0103 | 0.255 | ||||
| Cw*12 | 0.047 | DPA1*0105 | 0.005 | ||||
| Cw*14 | 0.019 | DPA1*0201 | 0.226 | ||||
| Cw*15 | 0.047 | DPA1*0202 | 0.042 | ||||
| Cw*16 | 0.071 | DPA1*0301 | 0.009 | ||||
| Cw*17 | 0.024 | ||||||
Table 2.
Gene diversity (GD) and linkage disequilibrium (D') values for 7 HLA loci in São Miguel Island population.
| Gene diversity | Linkage disequilibrium | ||
|---|---|---|---|
| HLA loci | GD | HLA loci pair | D' |
| A | 0.877 | A-Cw | 0.301 |
| A-B | 0.317 | ||
| A-DRB1 | 0.231 | ||
| A-DQB1 | 0.207 | ||
| A-DPA1 | 0.172 | ||
| A-DPB1 | 0.175 | ||
| B | 0.934 | B-Cw | 0.571 |
| B-DRB1 | 0.341 | ||
| B-DQB1 | 0.258 | ||
| B-DPA1 | 0.221 | ||
| B-DPB1 | 0.205 | ||
| Cw | 0.839 | Cw-DRB1 | 0.356 |
| Cw-DQB1 | 0.253 | ||
| Cw-DPA1 | 0.275 | ||
| Cw-DPB1 | 0.164 | ||
| DPA1 | 0.821 | DPA1-DPB1 | 0.398 |
| DPA1-DQB1 | 0.163 | ||
| DPA1-DRB1 | 0.270 | ||
| DPB1 | 0.906 | DPB1-DQB1 | 0.191 |
| DPB1-DRB1 | 0.213 | ||
| DQB1 | 0.821 | DQB1-DRB1 | 0.712 |
| DRB1 | 0.877 | -- | -- |
| Average GD | 0.846 | Average D' | 0.285 |
In bold are the highest and the smallest values.
Considering the 7 HLA loci, haplotype determination demonstrates a total of 176 different haplotypes corresponding to an 83.0% discriminatory power. Analysis of 5 HLA-A-Cw-B -DRB1-DQB1 haplotypes was also performed (see Additional file 1 for details). The results indicate that A*01-Cw*07-B*08-DRB1*03-DQB1*02 is the most frequent in São Miguel (7.9%), followed by A*24-Cw*07-B*08-DRB1*03-DQB1*02 (3.8%). Both A*02-Cw*05-B*44-DRB1*04-DQB1*03 and A*29-Cw*16-B*44-DRB1*07-DQB1*02 are present at a frequency of 1.9%. A total of 157 haplotypes were matched against worldwide populations (HLA-Allele Project; http://www.allelefrequencies.net/). The results showed that the second most frequent haplotype, above described, appears only on Tunisia. Moreover, just 9 haplotypes (Haplotype number - HN - 1, 29, 37, 42, 84, 85, 101, 104 and 112; see Additional file 1 for details) were identified in this database.
Linkage disequilibrium was based on the calculation of standardized multiallelic disequilibrium coefficient, D'. The range values are 0.163 for HLA markers DPA1-DQB1 and 0.712 for DQB1-DRB1 (Table 2). This wide variation averages 0.285 for the 7 loci. Curiously, the genetically closest markers (DPA1-DPB1, 0.011 Mb; D' = 0.398) do not present the highest value of D' (DQB1-DRB1, 0.081 Mb; D' = 0.712). A poor correlation between distance (Mb) and D' is observed, although there is a decrease of LD values over physical distance increase, as expected.
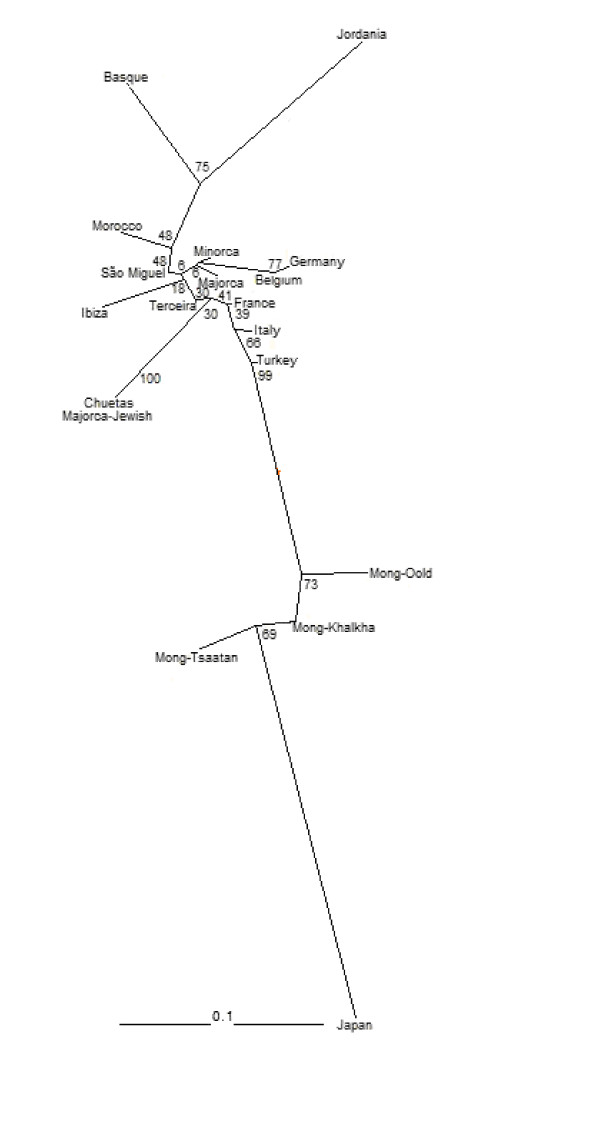
In order to obtain a graphical view of the genetic similarity between São Miguel (106 individuals, 5 HLA loci) and other populations, we computed Nei's genetic distances and depicted them in Figure 1. Interestingly, São Miguel is closer to Morocco population than to Terceira, another Azorean island. Nevertheless, in general, both populations cluster within the Europeans.
Figure 1.
Neighbor-Joining tree comparing 5 HLA loci in 19 populations. São Miguel clusters with Europeans and Moroccans.
Discussion
Extensive studies have been performed in several geographical areas to characterize the diversity of HLA genetic markers. These evaluations allow a better knowledge of the population structure considering non-neutral markers, as well as an understanding of the influence of evolutionary processes in the overall signature of a population. These genetic data are crucial for the comprehension of the molecular ethiology and epidemiology of common diseases. In general, the data here presented corroborate previous works [3,6-10], where Azoreans including São Miguel islanders show high values of genetic diversity when compared to mainland Portugal and other European populations. This may be a direct consequence of the Azorean settlement, with a major contribution of mainland Portuguese (~60%) and, to a lesser extent, Flemish, Spanish, French, Italians, Germans, Scottish, Jews, Moors and blacks from Guinea, Cabo Verde and São Tomé. Previous studies of HLA markers in mainland Portugal (3 loci, -A, -B and -DRB1, [18]) and in Azores (6 loci, -A, -Cw, -B, -DRB1, -DQA1 and -DQB1, [5]) demonstrate values of average diversity of 0.92 in both populations. The results obtained in the present study, based in 7 loci, showed a smaller value (0.84). This may be explained by the fact that Spinola et al. [5] used a high-resolution methodology to genotype HLA. Because alleles A*0101 and A*0102 are not considered the same allele (A*01), this methodology allows the identification of a higher number of different alleles. Nonetheless, the data show no significant differences between allele frequencies in São Miguel and Terceira islands. Considering HLA alleles distribution, the presence of -A*30 and -A*80, commonly found in sub-Saharan populations [19-21], in São Miguel validates historic records of slave settlers. In addition, the presence of alleles -B*35, -B*57 and -B*15 suggest a direct contribution of Moorish prisoners in Azores [22-24]. Nevertheless, the influence of early Portuguese settlers can not be ruled out since allele frequencies are similar. In general, these results are corroborated by the NJ tree (Figure 1), where São Miguel shows influence of both African and European populations.
Linkage disequilibrium is considered a good measure of population structure. According to Sanchez-Mazas [25] HLA-DPB1, located on the centromeric side of the HLA chromosomal region, does not show high values of LD with the other HLA loci. Interestingly, in the present study, the lowest values of D' observed are related with this marker. This result is explained by the high recombination region involving one or several hotspots, which separates HLA-DPB1 from the rest of the other HLA loci. Abecasis et al. [26] discuss that a value of D' = 0.33, which corresponds to a 10-fold increase in the required sample size, is commonly taken as the minimum usable amount of LD. Considering the 21 possible HLA loci combinations, 17 demonstrated values inferior to 0.33, and only 2 (Cw-B and DQB1-DRB1) showed values significantly higher (0.571 and 0.712, respectively). The HLA data reported by Meyer et al. [27] indicate a significant LD between all HLA loci in around 40 worldwide studied populations. The present research did not indicate large D' values and corroborates the results obtained by Service et al. [28] and Branco et al. [9,10], where the Azoreans have the lowest values of LD when compared with isolated and outbred populations.
HLA diversity in human populations is an important aspect of disease epidemiology, especially autoimmune disorders, such as type I diabetes, ankylosing spondylitis and celiac disease. According to Bakker et al. [29], the association of HLA alleles and/or haplotypes with disease susceptibility may be confounded by the presence of population stratification in neighboring HLA and non-HLA genomic regions. The high variability of HLA, and the absence of genetic structure and extensive LD, here demonstrated, suggest that autoimmune diseases studies in São Miguel islanders will necessarily encompass a more focused analysis of HLA extended haplotypes, as well as the evaluation of other non-HLA candidate genes.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PRP and CCB, contributed equally, by performing the experiments, statistical analysis and drafting the manuscript. CTG and RC genotyped individuals from the patients sample and provide technical help, respectively. LMV provided scientific orientation and revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplemental data to Results. Details each haplotype found in the São Miguel Island considering 5 HLA loci (A*-Cw*B*-DRB1*-DQB1) as well as their relative frequency.
Contributor Information
Paula R Pacheco, Email: paularpacheco@hdes.pt.
Claudia C Branco, Email: claudiacbranco@hdes.pt.
Cidália T Gomes, Email: g.cidalia@gmail.com.
Rita Cabral, Email: ritacabral@hdes.pt.
Luisa Mota-Vieira, Email: lmotavieira@hdes.pt.
Acknowledgements
This work was supported by grants from the Direcção Regional da Ciência e Tecnologia, Azores (Ref. M1.2.1/I/003/2005 and M1.2.1/I/002/2008), and Fundação para a Ciência e a Tecnologia of Portugal, FCT, through the POCI-2010 program (POCI/SAL-MMO/58413/2004). PRP has a FCT grant (Ref. SFRH/BD/27453/2006).
References
- Guill JH. A history of the Azores islands. California, Division of Golden Shield International Publications Cooperation; 1993. [Google Scholar]
- Santos C, Lima M, Montiel R, Angles N, Pires L, Abade A, Aluja MP. Genetic structure and origin of peopling in the Azores islands (Portugal): The view from mtDNA. Ann Hum Genet. 2003;67:433–456. doi: 10.1046/j.1469-1809.2003.00031.x. [DOI] [PubMed] [Google Scholar]
- Pacheco PR, Branco CC, Cabral R, Costa S, Araújo AL, Peixoto BR, Mendonça P, Mota-Vieira L. The Y-chromosomal heritage of the Azores Islands population. Ann Hum Genet. 2005;69:145–156. doi: 10.1046/j.1469-1809.2004.00147.x. [DOI] [PubMed] [Google Scholar]
- Montiel R, Bettencourt C, Silva C, Santos C, Prata MJ, Lima M. Analysis of Y-chromosome variability and its comparison with mtDNA variability reveals different demographic histories between islands in the Azores Archipelago (Portugal) Ann Hum Genet. 2005;69:135–144. doi: 10.1046/j.1469-1809.2004.00146.x. [DOI] [PubMed] [Google Scholar]
- Spínola H, Brehm A, Bettencourt B, Middleton D, Bruges-Armas J. HLA class I and II polymorphisms in Azores show different settlements in Oriental and Central islands. Tissue Antigens. 2005;66:217–230. doi: 10.1111/j.1399-0039.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- Branco CC, Palla R, Lino S, Pacheco PR, Cabral R, de Fez L, Peixoto BR, Mota-Vieira L. Assessment of the Azorean ancestry by Alu insertion polymorphisms. Am J Hum Biol. 2006;18:223–226. doi: 10.1002/ajhb.20492. [DOI] [PubMed] [Google Scholar]
- Branco CC, São-Bento M, Gomes CT, Cabral R, Pacheco PR, Mota-Vieira L. Azores Islands: genetic origin, gene flow and diversity patterns. Ann Hum Biol. 2008;35:65–74. doi: 10.1080/03014460701793782. [DOI] [PubMed] [Google Scholar]
- Branco CC, Pacheco PR, Cabral R, Vicente AM, Mota-Vieira L. Genetic signature of the São Miguel Island population (Azores) assessed by 21 microsatellite loci. Am J Hum Biol. 2008;20:118–120. doi: 10.1002/ajhb.20692. [DOI] [PubMed] [Google Scholar]
- Branco CC, Cabrol E, São Bento M, Gomes CT, Cabral R, Vicente AM, Pacheco PR, Mota-Vieira L. Evaluation of linkage disequilibrium on the Xq13.3 region: comparison between the Azores Islands and mainland Portugal. Am J Hum Biol. 2008;20:364–366. doi: 10.1002/ajhb.20734. [DOI] [PubMed] [Google Scholar]
- Branco CC, Pacheco PR, Cabrol E, Cabral R, Vicente AM, Mota Vieira L. Linkage disequilibrium and diversity for three genomic regions in Azoreans and mainland Portuguese. Genet Mol Biol. 2009. in press . [DOI] [PMC free article] [PubMed]
- van Oosterhout C. A new theory of MHC evolution: beyond selection on the immune genes. Proc Biol Sci. 2009;276:657–665. doi: 10.1098/rspb.2008.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota-Vieira L, Pacheco PR, Almeida ML, Cabral R, Carvalho J, Branco CC, de Fez L, Peixoto BR, Araújo AL, Mendonça P. In: Progress in Forensic Genetics, Proceedings of the 21st International Congress of Forensic Genetics, 13 - 17 September Ponta Delgada. Amorim A, Côrte-Real F, Morling N, editor. Vol. 1288. 2005. Human DNA bank in São Miguel island (Azores), a resource for genetic diversity studies; pp. 388–390. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin: A software for population genetics data analysis. University of Geneva, Genetics and Biometry Laboratory. Geneva; 2000. [Google Scholar]
- Ota T. Institute of Molecular Evolutionary Genetics. The Pennsylvania State University, USA; 1993. DISPAN: Genetic distance and phylogenetic analysis. [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 35c. Distributed by the author. Department of Genetics, University of Washington, Seattle, WA; 1993. [Google Scholar]
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations: Variability within and among natural populations. Chicago: Chicago University Press; 1984. [Google Scholar]
- Spínola H, Middleton D, Brehm A. HLA genes in Portugal inferred from sequence-based typing: in the crossroad between Europe and Africa. Tissue Antigens. 2005;66:26–36. doi: 10.1111/j.1399-0039.2005.00430.x. [DOI] [PubMed] [Google Scholar]
- Arnaiz-Villena A, Elaiwa N, Silvera C, Rostom A, Moscoso J, Gómez-Casado E, Allende L, Varela P, Martínez-Laso J. The origin of Palestinians and their genetic relatedness with other Mediterranean populations. Hum Immunol. 2001;62:889–900. doi: 10.1016/S0198-8859(01)00288-9. [DOI] [PubMed] [Google Scholar]
- Arnaiz-Villena A, Gomez-Casado E, Martinez-Laso J. Population genetic relationships between Mediterranean populations determined by HLA allele distribution and a historic perspective. Tissue Antigens. 2002;60:111–121. doi: 10.1034/j.1399-0039.2002.600201.x. [DOI] [PubMed] [Google Scholar]
- Gómez-Casado E, del Moral P, Martínez-Laso J, García-Gómez A, Allende L, Silvera-Redondo C, Longas J, González-Hevilla M, Kandil M, Zamora J, Arnaiz-Villena A. HLA genes in Arabic-speaking Moroccans: close relatedness to Berbers and Iberians. Tissue Antigens. 2000;55:239–249. doi: 10.1034/j.1399-0039.2000.550307.x. [DOI] [PubMed] [Google Scholar]
- Muro M, Marín L, Torío A, Moya-Quiles MR, Minguela A, Rosique-Roman J, Sanchis MJ, Garcia-Calatayud MC, García-Alonso AM, Alvarez-López MR. HLA polymorphism in the Murcia population (Spain): in the cradle of the archaeologic Iberians. Hum Immunol. 2001;62:910–921. doi: 10.1016/S0198-8859(01)00290-7. [DOI] [PubMed] [Google Scholar]
- Modiano D, Luoni G, Petrarca V, Sodiomon Sirima B, De Luca M, Simporé J, Coluzzi M, Bodmer JG, Modiano G. HLA class I in three West African ethnic groups: genetic distances from sub-Saharan and Caucasoid populations. Tissue Antigens. 2001;57:128–137. doi: 10.1034/j.1399-0039.2001.057002128.x. [DOI] [PubMed] [Google Scholar]
- Bera O, Cesaire R, Quelvennec E, Quillivic F, de Chavigny V, Ribal C, Semana G. HLA class I and class II allele and haplotype diversity in Martinicans. Tissue Antigens. 2001;57:200–207. doi: 10.1034/j.1399-0039.2001.057003200.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Mazas A. An apportionment of human HLA diversity. Tissue Antigens. 2007;69:198–202. doi: 10.1111/j.1399-0039.2006.00802.x. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Noguchi E, Heinzmann A, Traherne JA, Bhattacharyya S, Leaves NI, Anderson GG, Zhang Y, Lench NJ, Carey A, Cardon LR, Moffatt MF, Cookson WO. Extent and distribution of linkage disequilibrium in three genomic regions. Am J Hum Genet. 2001;68:191–197. doi: 10.1086/316944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Single RM, Mack SJ, Erlich HA, Thomson G. Signatures of demographic history and natural selection in the human major histocompatibility complex loci. Genetics. 2006;173:2121–2142. doi: 10.1534/genetics.105.052837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Service S, DeYoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, Ospina J, Ruiz-Linares A, Macedo A, Palha JA, Heutink P, Aulchenko Y, Oostra B, van Duijn C, Jarvelin MR, Varilo T, Peddle L, Rahman P, Piras G, Monne M, Murray S, Galver L, Peltonen L, Sabatti C, Collins A, Freimer N. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M, Morrison J, Richardson A, Walsh EC, Gao X, Galver L, Hart J, Hafler DA, Pericak-Vance M, Todd JA, Daly MJ, Trowsdale J, Wijmenga C, Vyse TJ, Beck S, Murray SS, Carrington M, Gregory S, Deloukas P, Rioux JD. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data to Results. Details each haplotype found in the São Miguel Island considering 5 HLA loci (A*-Cw*B*-DRB1*-DQB1) as well as their relative frequency.