Abstract
The GTF2IRD1 general transcription factor is a candidate for involvement in the varied cognitive and neurobehavioral symptoms of the microdeletion disorder, Williams–Beuren syndrome (WBS). We show that mice with heterozygous or homozygous disruption of Gtf2ird1 exhibit decreased fear and aggression and increased social behaviors. These findings are reminiscent of the hypersociability and diminished fear of strangers that are hallmarks of WBS. Other core features of WBS, such as increased anxiety and problems with spatial learning were not present in the targeted mice. Investigation of a possible neurochemical basis for the altered behaviors in these mice using high-performance liquid chromatography analysis showed increased levels of serotonin metabolites in several brain regions, including the amygdala, frontal cortex and parietal cortex. Serotonin levels have previously been implicated in fear and aggression, through modulation of the neural pathway connecting the prefrontal cortex and amygdala. These results suggest that hemizygosity for GTF2IRD1 may play a role in the complex behavioral phenotype seen in patients with WBS, either individually, or in combination with other genes, and that the GTF2I transcription factors may influence fear and social behavior through the alteration of neurochemical pathways.
Keywords: Behavior, fear, knockout mice, serotonin, social interaction, transcription factor
Williams–Beuren syndrome (WBS; OMIM 194050) is an autosomal dominant disorder, with a frequency of between in 1/7500 and 1/20 000 live births, that presents with a unique spectrum of physical and behavioral features (Greenberg 1990; Pober & Dykens 1996; Stromme et al. 2002). WBS is associated with mild to moderate mental retardation, but the uneven neurocognitive profile is characterized by significant deficits in visuospatial constructive cognition alongside relative strengths in language, auditory rote memory and facial recognition (Mervis & Klein-Tasman 2000; Mervis et al. 2000). The most striking aspect of the WBS phenotype is the distinctive behavioral profile, which is a unique combination of both friendliness and anxiety (Mervis & Klein-Tasman 2000; Pober & Dykens 1996).
The majority of individuals with WBS harbor a 1.55 Mb deletion spanning the same genes because of unequal meiotic recombination between flanking low copy repeats (Bayés et al. 2003). Individuals have been identified with smaller deletions, who do not exhibit all of the typical WBS cognitive and behavioral features, and based on their phenotypic descriptions, it appears that genes near the distal deletion breakpoint contribute much to the unique WBS cognitive and behavioral profile (Botta et al. 1999; Frangiskakis et al. 1996; Gagliardi et al. 2003; Heller et al. 2003; Hirota et al. 2003; Morris et al. 2003; Tassabehji et al. 1999, 2005). These candidate genes include a novel three-member family of general transcription factors characterized by multiple helix–loop–helix I-repeat domains (Hinsley et al. 2004; Roy 2001), two of which (GTF2I and GTF2IRD1) are always deleted and one of which (GTF2IRD2) is variably deleted in WBS (Tipney et al. 2004).
Both Gtf2i and Gtf2ird1 are widely expressed during the embryonic stages of mouse development (Enkhmandakh et al. 2004; Palmer et al. 2006). In adult mice, GTF2I is present exclusively in neurons, with the greatest expression levels observed in cerebellar Purkinje cells, hippocampal interneurons and the large neurons of the cerebral cortex, whereas GTF2IRD1 showed greatest expression in granular cell layer of the olfactory bulb, the Purkinje cells of the cerebellum and the neurons in the piriform cortex (Danoff et al. 2004; Palmer et al. 2006). These results suggest that the two proteins play nonredundant, differentially regulated roles, despite their similar structure.
A Gtf2ird1 insertional mutant was generated previously, but only limited cognitive testing was performed (Durkin et al. 2001; van Hagen et al. 2006; Tassabehji et al. 2005). To better understand the contribution GTF2IRD1 haploinsufficiency makes to the cognitive and behavioral phenotype of WBS, we generated a gene-targeted mouse model and subjected heterozygous and homozygous Gtf2ird1 mutant mice to a variety of neurobehavioral paradigms that evaluate different domains of central nervous system functioning. In addition, to further probe the mechanism by which haploinsufficiency for this gene might translate into altered behavior, we analyzed neurotransmitter levels in a variety of different brain areas.
Materials and methods
Generation of Gtf2ird1-targeted mice
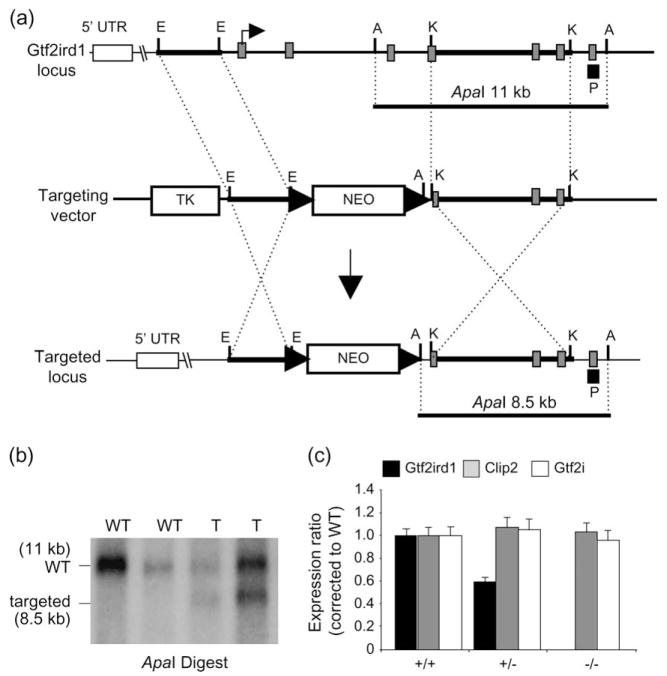
The murine Gtf2ird1 gene was disrupted using a conventional replacement targeting strategy. The targeting vector consisted of 2.7 kb short arm and a 5.8 kb long arm derived from RPCI-21-510M19 (PAC library derived from 129S6/SvEvTac mice) cloned into the EcoRI and KpnI sites, respectively, of the pKSLoxPNT cloning vector (Hanks et al. 1995). The resulting vector contained a neomycin-resistant gene (Neo), flanked by loxP sites, in the same transcriptional orientation as the Gtf2ird1 gene (Fig. 1a). Integration of the linearized vector into the Gtf2ird1 gene locus of R1 murine embryonic stem cells (Nagy et al. 1993) generated neomycin-resistant clones with the expected genomic fragments by Southern blot and polymerase chain reaction (PCR) analysis (Fig. 1b). The targeting resulted in the replacement of Gtf2ird1 exons 2, 3, 4 and part of 5 with the neomycin-resistant gene cassette transcribed by the PGK1 promoter (Fig. 1a). Mice carrying the targeted allele were generated by aggregation of targeted cells with ICR morula-stage embryos to obtain germline-transmitting chimeric mice (Nagy et al. 2002).
Figure 1. Targeted disruption of the Gtf2ird1 gene by homologous recombination.
(a) Structural organization of the murine Gtf2ird1 gene (top), of the targeting vector (middle) and the targeted Gtf2ird1 locus (bottom). Grey boxes indicate coding exons. Restriction enzymes are ApaI (A), EcoRI (E), and KpnI (K). Black box represents position of probe used to screen embryonic stem (ES) cells (P). The loxP sites are represented by arrows; NEO, neomycin-resistance cassette; TK, thymidine kinase gene. Bent arrow represents the predicted translation start site. (b) Southern blot analysis of Gtf2ird1-targeted ES cells. Genomic DNA was digested with ApaI, fractionated on agarose gel and hybridized with 32P-labeled probe. The 11-kb ApaI fragment corresponds to the WT locus and the 8.5-kb fragment to the targeted locus. (c) Real-time RT-PCR analysis of gene expression using brain cDNA from WT and targeted Gtf2ird1 mice. Gtf2ird1 and its two flanking genes (Gtf2i, Clip2) were assayed.
Chimeric males were mated with CD1 females to produce hybrid CD1x[ICRx129] Gtf2ird1 heterozygously targeted mice, and these mice were subsequently backcrossed to CD1 to generate animals for intercross breeding. Both Gtf2ird1+/− and Gtf2ird1−/− mice were viable and fertile and the mutant allele was transmitted at the expected Mendelian ratio. F1 heterozygous littermates were crossed to homozygosity in order to generate Gtf2ird1−/− mice. Genotyping was performed by PCR analysis of purified genomic DNA using the forward primer mIRD1-GF (5′-CGACCACCATAGGTTGAAGG-3′), in combination with the two reverse primers mIRD1-GR (5′-TGGGGAACTGTTTGAGAAGG-3′) and NEO-GR (5′-GGGGAACTTCCTGACTAGGG-3′). A 381 bp product is generated from the wild-type (WT) locus and a 350 bp product is generated from the targeted locus.
Expression analysis
Total RNA was prepared from adult and neonate brain using TriRe-agent (Sigma-Aldrich Canada, Oakville, Ontario, Canada) following manufacturer’s instructions. Following DNAse treatment, 5 μg of RNA was converted to complementary DNA using the SuperScript™ First-Strand Synthesis System (Invitrogen Canada Inc., Burlington, Ontario, Canada) and random hexamer primers. Samples were diluted 1/100 with sterile water and used directly in real-time assays using the AB Prism 7900HT sequence detection system as described previously (Somerville et al. 2005). Gtf2ird1 and its flanking genes, Gtf2i and Cyln2 were tested. Each test gene was normalized to control genes Hprt1, Hmbs and Sdha. Normalized values for each gene were then pooled for each of the genotype groups (Gtf2ird1+/+, Gtf2ird1+/− and Gtf2ird1−/−). Comparative expression ratios (%) were calculated by dividing the pooled normalized values for each of the test genes in the Gtf2ird1+/− and Gtf2ird1−/− genotype groups by the normalized test gene values for the Gtf2ird1+/+ control group. Primers for real-time amplification from cDNA were as follows: Gtf2ird1 exon 2, mIRD1RTe2-F (5′-ACTGTGACATCCCCACCAAC-3′) and mIRD1RTe2-R (5′-GAGTCTAAGGCGGACACCAG-3′); Gtf2ird1 exon 9, mIRD1RTe9-F (5 ′-CGAGGCTGTGGAAATTGTG-3 ′) and mIRD1RTe9-R (5′-TGTGTCGCTCCTCCAGAATC-3′); Cyln2 exon 4, mCYLN2RTe4-F (5′-CAACAGAGGAGGCCACAGAG-3′) and mCYLN2RTe4-R (5′-CAAGGCCAAGAAGACCAAAC-3′); Gtf2i exon 30, m2IRTe30-F (5′-CAGGAAGATCACCATCAACC-3′) and m2IRTe30-R (5′-AGATCCTCCTCATGGAGCTG-3′).
General morphological analysis
Mice were routinely examined for obvious morphological or anatomical abnormalities. All testing was performed on mice produced from the intercross of hybrid CD1×[ICR×129] mice. For determination of body weight, adult male and female mice were weighed with the same scale (accuracy ±0.1 g). For determination of growth curves, mice were weighed twice per week from weaning (3 weeks) until 10 weeks at approximately the same time of day.
Behavioral experiments
For behavioral testing, adult mice between 3 and 9 months of age were used. All animals were group housed with access to food and water ad libitum and were on a 12 h light/dark cycle throughout the experiments. All the experiments were conducted during the light phase from 0900–1700 h. Four cohorts of Gtf2ird1+/− and Gtf2ird1−/− mice and their WT littermate controls were used to minimize the effect of multiple testing. Cohort 1: elevated plus maze, cube exploration and open field; cohort 2: resident intruder test and olfactory function test; cohort 3: Morris water maze test; cohort 4: cued and contextual fear conditioning. The mice were given minimum 7-day-interval between the tests. Prior to all experiments, mice were left undisturbed in the room for 30 min to allow acclimation. Where no effect of gender was found, male and female data were pooled.
Resident–intruder test
Aggression was assessed using the resident–intruder test in isolated male mice, essentially as previously described (Moy et al. 2004). Males (+/+, n = 17; +/−, n = 10; −/−, n = 18) were housed individually for at least 1 week before assessment, which was performed over three sessions in the same day. Intruders (unfamiliar socially housed C57BL/6J male mice, age and weight matched with each resident) were individually placed in the resident home cage for a 10-min test session and observation was started. The latency, duration and number of events were recorded as: aggressive behavior (contact between the resident and the intruder such as biting or wrestling and aggressive grooming of partner) or social interest behavior (following and sniffing of partner). A different intruder animal was used for each resident. Animals that did not attack the intruder were given an attack latency of 10 min. All behavioral events were video recorded and analyzed by Observer 5.0 software (Noldus Information Technology, Wageningen, the Netherlands).
A simple test of olfactory function in each test animal was conducted following the resident–intruder test. This was carried out as described previously, by measuring the time it took for each mouse to find food buried in bedding (Moy et al. 2004). All mice were first habituated to the food (Bud’s Best Cookies, Hoover, AL, USA) by the experimenter placing pieces of cookie in the home cage overnight. The next day, chow was removed from the cages and the mice were food deprived for 24 h. The test was conducted in a plastic cage 30 × 17 × 12 cm. The food was placed in the randomly chosen area (1 × 1 × 0.5 cm) and the entire bottom of the cage covered with standard bedding to a depth of 2.5 cm. Mice were then placed into the cage individually and the latency to find the food was recorded, with a maximum time of 15 min.
Elevated plus maze
The elevated plus maze was used to estimate the anxious state of the mice (Rodgers & Cole 1994). Testing was performed as described previously (Avgustinovich et al. 2004). All measurements were taken in a dimly lit experimental room to which the mice were acclimatized, and the maze was thoroughly cleaned between sessions. Over a 5-min test period, the following measures of plus-maze behavior were recorded: (1) time spent in the open arms, enclosed arms and on the central platform time, expressed as a percentage of total time; (2) number of open arms entries, enclosed arms entries and central platform entries, expressed as a percentage of total entries; (3) number of total entries; (4) number of head dips. Testing was performed on the following mice: +/+, n = 11 (9 males, 2 females); +/−, n = 12 (9 males, 3 females); −/−, n = 14 (10 males, 4 females).
Cube exploration test
The cube exploration test was used to assess approach anxiety as previously described (Avgustinovich et al. 2000). Briefly, a small cube (3 cm3) was carefully placed in the center of each home cage and measurements were recorded over a 5-min test period. All measurements were taken in a dimly lit experimental room to which the mice were acclimatized, and the novel object (cube) was thoroughly cleaned between tests.
Locomotor activity in the open field
Open-field activity assessments were carried out as described previously (Abramow-Newerly et al. 2006). The activity cage consisted of a cubical box (41 × 41 × 33 cm3) (model 7420/7430; Ugo Basile, Comerio, Italy) with a floor equipped with horizontal and vertical infrared sensors. The subject’s behavior was recorded using a computer event-recording program (Ethograph, observer 5.0 from Noldus Information Technology). Each mouse [+/+, n = 11 (9 males, 2 females); +/−, n = 12 (9 males, 3 females); −/−, n = 14 (10 males, 4 females)] was placed individually into the center of the activity cage for 5 min and the following behavioral measures recorded: (1) latency (seconds) to first escape from the center; (2) length of time of immobility periods (seconds); (3) time spent self grooming (seconds); (4) number of risk assessment behaviors involving the mouse stretching its body from corners/wall toward the center; (5) horizontal and (6) vertical activity. Exploratory activity and walking were recorded separately for the central and peripheral field of the open arena and ratio between central and peripheral activity was calculated. The arena was cleaned with 70% ethanol solution between the subjects.
Contextual and cued fear conditioning
Contextual and cued fear conditioning was carried out according to previously published protocols (Clapcote et al. 2005). Briefly, a fear conditioning apparatus (MED Associates Inc., Georgia, VT, USA) consisting of a test chamber (25 cm high × 30 cm wide × 25 cm deep) was cleaned prior to testing with 70% ethanol. Freezing activity was recorded using automated fear conditioning software (Actimetrics software, FREEZEFRAME v. 1.6e) and presented as a percentage of total time. Tests subjects [+/+, n = 9 (6 males, 3 females); +/−, n = 20 (12 males, 8 females); −/−, n = 6 (3 males, 3 females)] were removed from home cage and allowed to explore for 2 min. Conditioning consisted of a single pairing of an auditory cue (3600 Hz, 80 dB) with a foot shock (1 mA scrambled). The auditory cue was present 2 min after the training session started and was 30 seconds in duration. The foot shock was delivered continuously during the last 2 seconds of the auditory cue. The subject was removed from the chamber 30 seconds later and returned to its home cage. Approximately 24 h later, each subject was returned to the test chamber and monitored for 5 min. Two hours later, the context was altered and each subject was placed into the altered chamber and allowed 3 min for exploration, after which the auditory tone cue of 3 min was delivered.
Morris water maze test
The Morris water maze apparatus and testing procedures were described previously (Clapcote et al. 2005). Briefly, on the first day, each mouse was given four visible platform trials (V) in the Morris water maze apparatus (117 cm diameter). Mice [+/+, n = 7 (four males, three females); +/−, n = 11 (seven males, three females); −/−, n = 12 (six males, six females)] were then subjected to 5 days of four training trials per day with the submerged platform in the same position (hidden phase). On the sixth day, the platform was moved to a different position, and the mice were subjected to 4 days of four training trials per day (reversal phase). A probe trial was administered 20 min after the last trial on the fifth day of the hidden phase and the fourth day of the reversal phase. Each subject was placed into the water diagonally opposite the target quadrant (T) and allowed 60 seconds to search the water, from which the platform had been removed. Behavioral variables were quantified with the aid of HVS Water 2020 (HVS Image Ltd, Twickenham, Middlesex, UK).
Neurochemical analyses
Mice were killed with a brief head-focused pulse of high-intensity microwave radiation (8 kW, 2.4 GHz), delivered by a 10 kW Muoromachi Brain Fixation System (Stoelting Co., Chicago, IL, USA) to rapidly and effectively fix the brain in situ. The fixed brains were dissected on ice to isolate the individual brain regions (amygdala, frontal cortex, parietal cortex and occipital cortex) and stored at −80°C until analysis. Tissues were processed as described previously, divided into aliquots of 50 μl, and stored at −80°C or analyzed immediately (Mount et al. 2004). Tissue pellets were retained for the determination of protein content. Six to nine individual tissue samples were obtained for each brain region.
Levels of serotonin (5-HT) and 5-hydroxyindoleacetic acid (5-HIAA) were quantified using high-performance liquid chromatography with electrochemical detection. Analyses were performed on a system consisting of a Thermo Separation Products (TSP, Piscataway, NJ, USA) P4000 pump, a TSP AS3000 autosampler with cooling unit, an ESA Coulochem II electrochemical detector (ESA 50ll Analytical Cell and 5020 Guard Cell; ESA Biosciences, Chelmsford, MA, USA) and a Spectra Physics 4290 Integrator (Spectra Physics, Irvine, CA, USA) connected to a PC running TSP WOW chromatography software. The mobile phase, an aqueous mixture of 0.098 M glacial acetic acid, 0.09 M sodium acetate (pH 3.7), 0.118 mM ethylenediaminetetra-acetic acid, 7% methanol and 0.8 mM octane sulphonate was delivered at a flow rate of 0.8 ml/min. Separation of the 100 μl samples was performed on an ACE 250 × 4.6 mm column with Ace C18, 5 μm stationary phase. Peak heights recorded at E2 were detected using electrode potentials as follows (Guard cell +450 mV: analytical cell E1 + 100 mV; E2 −400 mV). Quantification of monoamines was performed on 0.1 N percholoric acid extracts in a procedure involving two 30 min runs per sample. An appropriately diluted sample was run followed by a second run consisting of one-half sample and one-half standard cocktail (pure monoamines and metabolites in concentrations of 10 pg/μl). Monoamine and metabolite levels in pg/mg tissue wet weight were then calculated.
Statistical analysis
Data were expressed as mean ± SEM. Statistical analysis was performed using a two-way analysis of variance (ANOVA) to detect main effects or interacting effects of gender and genotype. Where no effect was found, male and female data were pooled. Body weight analysis was performed using a two-way ANOVA to detect a main effect or interaction, followed by Tukey’s post hoc analysis. Neurochemical analysis was performed using a two-tailed Student’s t-test for independent samples. For all other analyses, statistical analysis was performed using a one-way ANOVA to detect a main effect of genotype. Where stated, Tukey’s HSD post hoc analysis was used when ANOVAs yielded statistically significant main effect of genotype. A difference among genotypes in the Morris water maze was evaluated using repeated measures ANOVA. Probabilities of <0.05 were considered significant.
Results
Effects of Gtf2ird1 targeting on gene expression
To understand the function of GTF2IRD1, we generated a mouse model in which the first four coding exons of the gene have been deleted. We measured expression of Gtf2ird1 in brain tissue from mice heterozygous and homozygous for the targeted allele, using real-time PCR, and examined expression of the immediately flanking genes. A primer set within the deleted region of the gene (exon 2, containing the translation start site), showed a dose-dependent reduction in expression in the heterozygous mice and absence of expression in the Gtf2ird1−/− mice (Fig. 1c). Amplification with a primer set downstream of the replacement targeting (exon 9) showed the existence of aberrant transcripts consisting of the upstream, untranslated exon 1 spliced directly into exon 6 of Gtf2ird1. Depending on the initiation choice, translation of this transcript (lacking the first four coding exons) would produce a small, out-of-frame peptide, or an in-frame truncated protein lacking the leucine zipper necessary for dimerization, along with at least one of the I-repeats (Cheriyath & Roy 2000, 2001). Because of low expression of the GTF2IRD1 protein and the lack of a specific antibody against GTF2IRD1, we were unable to perform Western blot analysis.
Mice heterozygous and homozygous for the Gtf2ird1 deletion show mild growth retardation
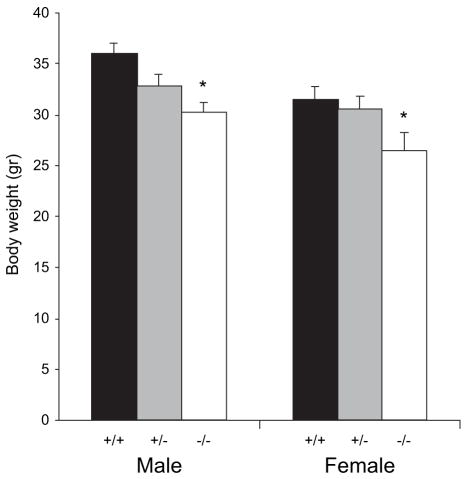
Two-way ANOVA showed that Gtf2ird1-targeted mice showed a significant main effect of gender (F1,114 = 9.20, P < 0.01) and phenotype (F2,114 = 7.43, P < 0.001) but no interaction of gender and phenotype (F2,114 = 1.49, P = 0.22). Post hoc analysis showed that male Gtf2ird1−/− mice showed significantly decreased body weights compared with WT males (P < 0.005). Adult female Gtf2ird1−/− mice were also found to be significantly smaller than WT females (P < 0.05) (Fig. 2). No significant difference was seen in the adult Gtf2ird1+/− male or female mice. Similar results were also seen in growth curve analysis in Gtf2ird1-targeted mice from 3 to 10 weeks of age (data not shown).
Figure 2. Gtf2ird1-targeted mice show mild growth retardation.
Body weight analysis of adult Gtf2ird1 targeted mice. Adult male (+/+, n = 23; +/−, n = 23; +/+, n = 14) and female (+/+, n = 24; +/−, n = 28; +/+, n = 8) mice were analyzed for changes in body weight (mean age, 20.7 ± 2.6 weeks). Both male and female Gtf2ird1−/− mice showed a significant decrease (P < 0.05) in body weight of approximately 15%. Although male and female Gtf2ird1+/− mice also showed a decrease in body weight, the decrease was only statistically significant in the Gtf2ird1−/− mice (*P < 0.05).
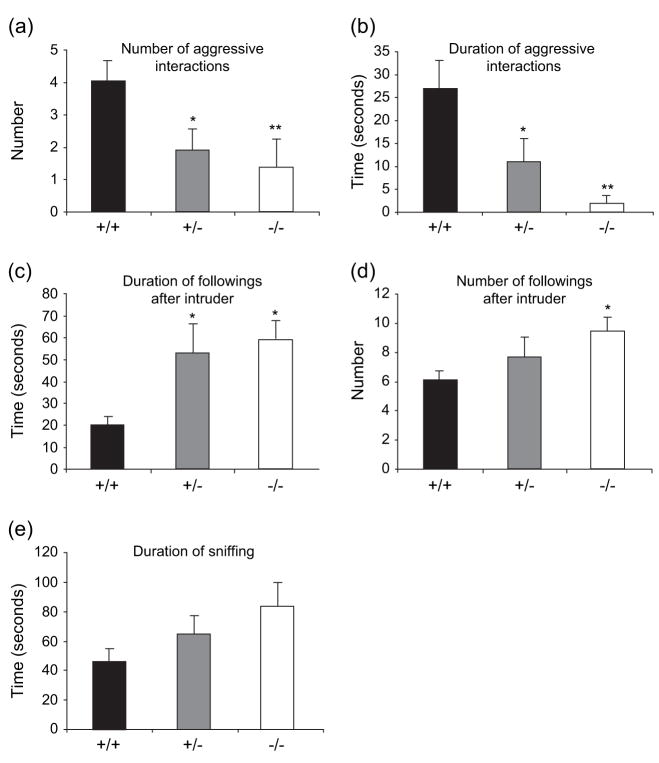
Gtf2ird1-targeted mice are less aggressive and engage in more social interactions
Gtf2ird1-targeted mice showed a significant main effect of genotype (F2,42 = 4.17, P < 0.05) on the number of aggressive interactions and duration of aggressive interaction (F2,42 = 10.2, P < 0.001). Tukey’s post hoc analysis showed that Gtf2ird1+/− (P < 0.05) and Gtf2ird1−/− (P < 0.01) mice showed a significant decrease in the number of aggressive interactions (Fig. 3a) and a reduction in the time spent engaging in aggressive interactions (Fig. 3b) with an unknown intruder mouse compared with WT mice. In tests to measure changes in social interactions, there was a main effect of genotype on the duration of (F2,42 = 5.09, P < 0.05) and number of followings after (F2,42 = 4,39, P < 0.05) the intruder. Tukey’s post hoc analysis showed that Gtf2ird1+/− and Gtf2ird1−/− mice followed the intruder more often (P < 0.05; Fig. 3d) and the following was of longer duration (P < 0.05; Fig. 3c). Although Gtf2ird1+/− and Gtf2ird1−/− mice spent longer sniffing the intruder compared with WT mice (Fig. 3e), no main effect of genotype was seen for the duration of sniffing (F2,42 = 2.42, P = 0.10). In the olfactory function test, the time required for finding unfamiliar buried food was not significantly different between the genotypes (WT, 354 ± 84.7 seconds; Gtf2ird1+/−, 314.6 ± 22.7 seconds; Gtf2ird1−/−, 279 ± 58.2 seconds), showing that the Gtf2ird1-targeted mice had normal olfactory function.
Figure 3. The resident-intruder test detects decreased aggression and increased social interaction in Gtf2ird1+/− and Gtf2ird1−/− mice.
Both the Gtf2ird1+/− and the Gtf2ird1−/− mice showed a significant decrease in the number of aggressive interactions (a) and length of these aggressive interactions (b) as compared with WT mice. Both the Gtf2ird1+/− and the Gtf2ird1−/− mice also showed a significant increase in the number of social interactions including the time spent following the intruder (c) compared with WT mice. Gtf2ird1−/− mice also displayed a significant increase in the number of followings (d) and an increased, but not significant time spent sniffing the intruder (e) compared with WT mice. A similar trend in noted for Gtf2ird1+/− mice although this change was not statistically significant. Values are mean ± SEM (+/+, n = 17; +/−, n = 10; −/−, n = 18) (*P < 0.05, **P < 0.01).
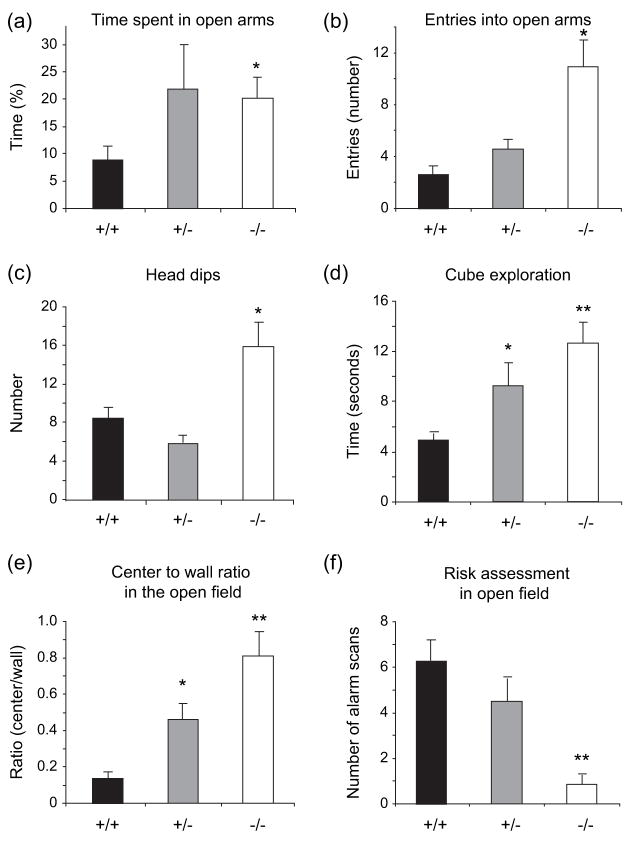
Gtf2ird1-targeted mice have decreased anxiety
We evaluated state anxiety in the elevated plus maze and open-field tests, and found that anxiety was significantly reduced in the mutant mice. There was a main effect of genotype on the time spent in the open arms of the elevated plus maze (F2,33 = 3.454, P < 0.05). Post hoc analysis showed that Gtf2ird1−/− mice displayed a significant increase in the amount of time spent in the open arms of the elevated plus maze compared with WT mice (P < 0.05), indicative of reduced anxiety involving avoidable anxiety-provoking stimuli (Fig. 4a). A similar, but not significant trend was seen for the heterozygous mice. A significant increase was also seen in Gtf2ird1-targeted mice in entries into the open arm (F2,34 = 6.43, P < 0.005) and head dips (F2,34 = 9.243, P < 0.001). Post hoc pair-wise comparison also showed significant increases in entries into the open arm (P < 0.05) and head dips (P < 0.05) for the Gtf2ird1−/− mice (Fig. 4b,c). In the novel object test, there was a highly significant main effect of genotype on the time spent exploring the novel object (F2,21 = 23.6, P < 0.001; Fig. 4d). Post hoc analysis showed that both Gtf2ird1+/− mice (P < 0.05) and Gtf2ird1−/− mice (P < 0.001) spent a significantly increased amount of time exploring the novel object.
Figure 4. Anxiety is decreased in Gtf2ird1−/− mice.
In the elevated plus maze, significant increases in the percentage of time spent in open arms (a) entries into the open arms (b) and head dips (c) were seen in Gtf2ird1−/− mice compared with WT mice. In the novel object test, both Gtf2ird1+/− and Gtf2ird1−/− mice showed a significant increase in the time spent exploring an unfamiliar object (cube) (d). In the open field, both Gtf2ird1+/− and Gtf2ird1−/− mice showed a significant increase in the center to wall ratio (e) and Gtf2ird1−/− mice showed a significant decrease in risk assessment (alarm scanning) (f). Values are mean ± SEM (+/+, n = 11; +/−, n = 12; −/−, n = 14) (*P < 0.05, **P < 0.01).
These results could not be attributed to increased activity because neither Gtf2ird1+/− nor Gtf2ird1−/− mice showed any significant difference in total horizontal or vertical locomotor activity when compared with WT mice in the open-field test (data not shown). Gtf2ird1-targeted mice did show a highly significant increase in the center-to-wall ratio (F2,24 = 15.9, P < 0.001; Fig 4e) and a decrease in risk assessment (alarm scanning) (F2,24 = 9.04, P < 0.001; Fig 4f). Post hoc analysis showed that both Gtf2ird1+/− mice (P < 0.01) and Gtf2ird1−/− mice (P < 0.001) had an increase in center-to-wall activity and that Gtf2ird1−/− mice (P < 0.001) had a decrease in risk assessment in the open field.
Gtf2ird1-targeted mice have deficits in cued but not contextual fear conditioning
In contextual fear conditioning, although a significant overall increase in freezing compared with baseline was observed after 24 h (F5,64 = 6.61, P < 0.001), the level of freezing to context in Gtf2ird1-targeted mice did not differ significantly from that of WT littermates (F2,32 = 3.12, P > 0.05) (Fig. 5). In cued tests, there was no significant change in amount of freezing observed in the new context compared the baseline (F5,64 = 0.88, P > 0.05) or between genotypes (F2,32 = 0.78, P > 0.05) before the presentation of the auditory cue (pre-CS). Upon presentation of the auditory cue (post-CS), mice showed a significant overall increase in the amount of freezing (F5,64 = 12.32, P < 0.001), however, freezing in Gtf2ird1-targeted mice was significantly less than in WT littermates (F2,32 = 4.05, P < 0.05). Post hoc analysis showed that both Gtf2ird1+/− mice (P < 0.01) and Gtf2ird1−/− mice (P < 0.05) displayed significantly less freezing compared with WT mice (Fig. 5).
Figure 5. Gtf2ird1-targeted mice have deficits in cued but not contextual fear conditioning.
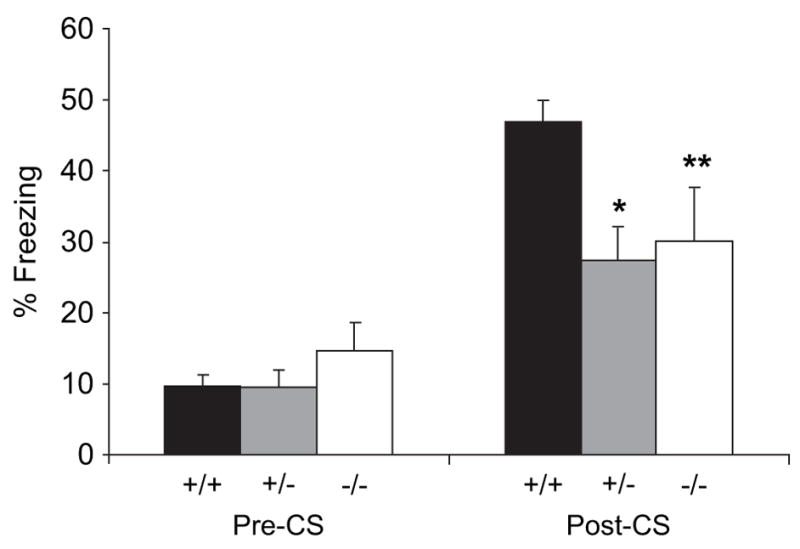
In cued tests, there was no significant change in amount of freezing observed between genotypes in the altered context compared the baseline (pre-CS). Upon presentation of the auditory cue (post-CS), freezing in Gtf2ird1-targeted mice was decreased with both Gtf2ird1+/− and Gtf2ird1−/− mice displaying significantly less freezing than WT littermates. In contextual testing, Gtf2ird1-targeted mice did not differ significantly from WT littermates (data not shown). (*P < 0.01, **P < 0.05).
Gtf2ird1-targeted mice have normal spatial learning and memory
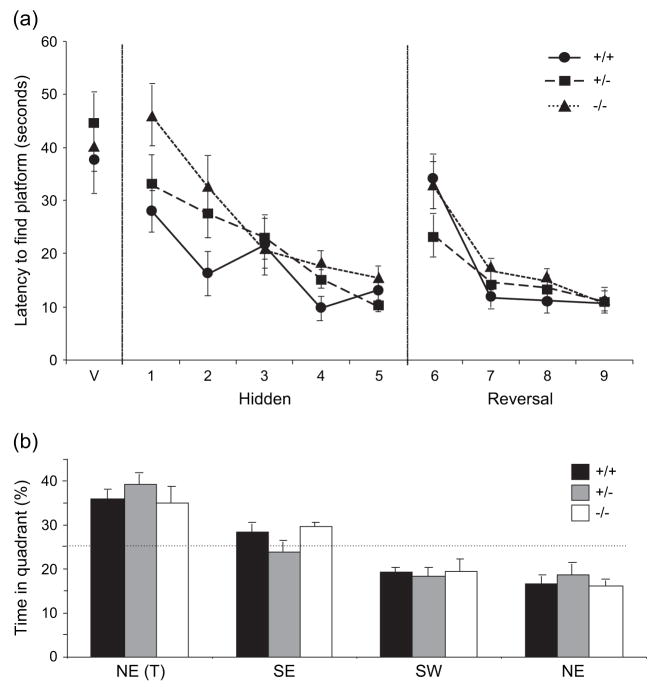
Neither Gtf2ird1+/− nor Gtf2ird1−/− mice showed a significant difference in their performance (latency, path length or swim speed) compared with WT littermates during either the acquisition or the reversal phase of the Morris water maze test (latency Fig. 6a, path length and swim speed data not shown). The increase in latency on the first day of reversal training is indicative of place learning. Further to this, no main effect of genotype was observed between WT littermates and Gtf2ird1-targeted mice in the probe test following the hidden phase for percentage of time spent in the target quadrant (F2,81 = 0.174, P = 0.84) with all genotypes showing a significant preference for the target quadrant (F3,81 = 37.0, P < 0.0001; Fig 6b). Similar results were seen for the probe test following the reversal phase (data not shown).
Figure 6. Gtf2ird1-targeted mice perform normally in the Morris water maze evaluation of visuospatial processing.
Neither Gtf2ird1+/− nor Gtf2ird1−/− mice showed a significant difference in their performance compared to WT littermates (P > 0.05) during either the visible (V), hidden (acquisition) or the reversal phase (a). No significant differences were observed in either probe test (test administered at the end of the hidden platform trial is shown) (b). Values are mean ± SEM (+/+, n = 7; +/−, n = 11; −/−, n = 12).
Gtf2ird1 homozygous mutant mice show altered 5-HT metabolite levels in the brain
We analyzed 5-HT and 5-HIAA levels based on the wealth of literature linking 5-HT metabolism with anxiety and aggression – phenotypes that are significantly altered in our mice. As the amygdala and cortical areas are known to be functionally active areas within the serotonergic system, we chose these areas for analysis. Levels of 5-HT and 5-HIAA neurotransmitter were evaluated across functionally relevant brain regions using a two-tailed Student’s t-test for independent samples. Levels of 5-HT were unchanged in the Gtf2ird1−/− mice as compared with WT mice in all brain regions tested (Table 1). Levels of 5-HIAA were significantly elevated in the amygdala (P < 0.05), frontal cortex (P < 0.05) and parietal cortex (P < 0.05) of Gtf2ird1−/− mice compared with WT animals (Table 2). Increases in 5-HIAA were also seen in the occipital cortex although this increase did not reach statistical significance.
Table 1.
Mean ± SEM concentration (pg/mg) of 5-HT in indicated brain areas of Gtf2ird1−/− and WT mice
| 5-HT | Amygdala | Frontal cortex | Parietal cortex | Occipital cortex |
|---|---|---|---|---|
| +/+ | 966 ± 69 (n = 7) | 832 ± 51 (n = 7) | 736 ± 57 (n = 6) | 830 ± 96 (n = 6) |
| −/− | 1120 ± 63 (n = 8) | 886 ± 58 (n = 9) | 780 ± 45 (n = 9) | 800 ± 81 (n = 8) |
Results were analyzed by two-tailed Student’s t-test for independent samples.
Table 2.
Mean ± SEM concentration (pg/mg) of 5-HIAA levels are increased in the amygdala, frontal cortex and parietal cortex of Gtf2ird1−/− mice compared with WT mice
| 5-HIAA | Amygdala | Frontal cortex | Parietal cortex | Occipital cortex |
|---|---|---|---|---|
| +/+ | 415 ± 18 (n = 7) | 234 ± 16 (n = 7) | 366 ± 29 (n = 6) | 321 ± 33 (n = 6) |
| −/− | 509 ± 32* (n = 8) | 297 ± 17* (n = 9) | 469 ± 41* (n = 9) | 370 ± 25 (n = 8) |
Results were analyzed by two-tailed Student’s t-test for independent samples (*P < 0.05).
Discussion
Individuals with WBS display a spectrum of clinical, neurobehavioral and cognitive abnormalities, but to date only a single gene (elastin) has been unequivocally implicated in any aspect of the disorder, namely the cardiovascular abnormalities (Curran et al. 1993). This is, at least in part, because of the paucity and phenotypic heterogeneity of individuals with smaller deletions of the region, making genotype–phenotype correlation difficult. In addition, it is possible, perhaps likely, that there are combinatorial consequences of multiple gene deletion. As a result of these limitations, mouse models have become a logical route to understanding the role of specific genes in the complex WBS phenotype. Here, we have shown that mice either heterozygously or homozygously disrupted for the Gtf2ird1 transcription factor exhibit some behavioral features of WBS that have not been reported in previous mouse models (Crackower et al. 2003; Fujiwara et al. 2006; van Hagen et al. 2006; Hoogenraad et al. 2002; Li et al. 1998; Meng et al. 2002; Tassabehji et al. 2005; Zhao et al. 2005). The distinctive behavioral profile seen in people with WBS is one of the defining features of WBS, and insight into the genetic basis of aspects of this unique phenotype will be important not only for understanding the molecular basis of WBS, but of normal human behavior.
Perhaps the most intriguing finding in mice with disruption of Gtf2ird1 was the decrease in aggressive behavior toward, and increased social interest in, unfamiliar mice. This was paired with a significantly blunted natural fear response. People with WBS almost universally exhibit overfriendliness with inappropriate social boundaries and lack of normal risk assessment, and frequently approach and/or initiate social interactions with strangers (Doyle et al. 2004; Klein-Tasman & Mervis 2003). The altered behaviors seen in the Gtf2ird1 mutants are intriguingly reminiscent of these hallmark features of WBS.
In an apparent direct contrast to people with WBS, the majority of whom have either generalized anxiety disorder or simple non-social phobias (Dykens 2003; Mervis & Klein-Tasman 2000), Gtf2ird1−/− mice displayed decreased anxiety when tested in the elevated plus maze and open field (Fig. 4). Although this was somewhat unexpected, it could be that alteration in the expression of GTF2IRD1 affects the anxiety state of both humans and Gtf2ird1-targeted mice, albeit in different ways. Alternatively, it is possible that these tests do not examine the same behavioral response as that seen in people with WBS. The presentation of a novel object normally elicits fear response, or neophobia, in animals. However, Gtf2ird1-targeted mice showed an enhanced interest in the home cage cube exploration test (Fig. 4d), in part confirming their less fearful state observed in the plus maze and open field. However, it is also possible that the apparent lack of fear (approach/avoidance behavior) exhibited by these mice is masking any potential anxiety that would normally be elicited by the elevated plus maze or open field. Additional testing, including the administration of anxiogenic drugs before testing in the elevated plus maze may help distinguish between lack of fear response and reduced anxiety-observed behavior of Gtf2ird1-targeted mice.
The neural mechanisms regulating social behavior and aggression are still being elucidated, but models of human aggression specifically implicate the amygdala and paralimbic prefrontal regions (Davidson et al. 2000), with lesions of the amygdala in non-human primates resulting in impaired or inappropriate social function (Amaral 2002; Prather et al. 2001). The molecular mechanisms governing the perception of, and reaction to, danger and threatening situations have also been linked to the amygdala, with the orbitofrontal cortex hypothesized to play a key role in modulating limbic reactivity to threat (Davidson et al. 2000; Izquierdo et al. 2005). Recent functional neuroimaging studies of people with WBS showed reduced activation of the amygdala when processing images of threatening faces, suggesting an underlying dysfunction (Meyer-Lindenberg et al. 2005). Amygdala function is often measured using associative fear conditioned learning and memory, specifically with a cue, such as a tone. Impaired cued fear conditioning was noted in both the Gtf2ird1+/− and Gtf2ird1−/− mice although no difference was detected in contextual fear conditioning. Biochemically, oxytocin has been firmly established as central mediator of social behavior through its action in the amygdala, and stathmin, a molecule highly expressed in this region, has also been implicated in both innate and learned fear (Shumyatsky et al. 2005; Winslow & Insel 2002). Mice lacking GDI1, which encodes a protein controlling the activity of the small guanosine triphosphatase of the Rab family in vesicle fusion and intracellular trafficking, also exhibit decreased aggression and altered social behavior, but the biological basis for this remains unknown (D’Adamo et al. 2002). It will be interesting to investigate these and other molecules and genes that have been implicated in fear and social response, in the Gtf2ird1 mutant mice.
It is known that 5-HT plays an important role in emotional disorders with decreased 5-HT levels shown to cause an increase in aggressive behavior in rodents (Vergnes et al. 1986) as well as depression in humans (Ogilvie et al. 1996), while administration of a 5-HT(1B) receptor agonist reduced aggression in rats (De Almeida et al. 2006). Alteration of 5-HT(1A) and 5-HT(2A) receptor density and binding have also been linked to changes in aggression in rodents (Caramaschi et al. 2007; Schiller et al. 2006). Gtf2ird1−\− mice showed a significant increase of the 5-HT metabolite 5-HIAA in the frontal and parietal cortices and the amygdala, although the tissue 5-HT content was not significantly increased in any of the regions tested. These observations suggest an alteration in postsynaptic 5-HT turnover rather than an overall increase in 5-HT production. Further experiments, including studies of 5-HT receptor density and binding, are needed to determine the mechanism by which serotonergic pathways are altered in the Gtf2ird1-targeted mice.
The increased sociability seen in the Gtf2ird1 mutant mice suggests that this gene plays an important role in the regulation of normal social interaction in rodents, possibly in pathways that influence transcription of the molecules mentioned above. Interestingly, although almost all individuals with WBS exhibit the same cognitive profile, three children with smaller than normal deletions of the WBS region, leaving genes at the distal end intact, did not exhibit hypersociability (Doyle et al. 2004; van Hagen et al. 2006; Tassabehji et al. 2005). Recently, an individual was identified with a unique deletion that extends out of the WBS region toward the telomere, resulting in hemizygosity for GTF2IRD1 and GTF2I, but no other genes from the common WBS deletion region (Edelmann et al. 2006). This patient exhibited inappropriate friendliness toward strangers, even though her phenotype was compounded by a diagnosis of autism. It seems likely, therefore, that in humans, the WBS behavioral profile may be the product of the combinatorial effect of hemizygosity for both GTF2IRD1 and GTF2I. Mouse models with multiple gene deletions will go some way to elucidate this and will be very helpful in studying the interplay between different genes within the WBS deletion.
Even homozygous disruption of Gtf2ird1 was not sufficient to produce deficits in learning tasks that rely heavily on the hippocampus, such as the Morris water maze test of spatial learning and memory and contextual fear conditioning. This was supported by electrophysiological recordings of the CA1 region of the hippocampus, which showed normal basal synaptic activity and long-term potentiation (data not shown). Reports of two patients with unusual deletions of 7q11.23 support the role of other genes in spatial learning. One individual with a deletion that removed GTF2IRD1, but left GTF2I intact, did not exhibit as severe visual spatial impairment as people with WBS (Tassabehji et al. 2005), whereas a second individual with a deletion that removed both GTF2IRD1 and GTF2I, but none of the other commonly deleted genes (Edelmann et al. 2006), showed weakness in visuospatial skills equivalent to that seen in people with WBS. Our findings, together with the published reports, suggest that GTF2IRD1 and GTF2I may both play an important role in proper visuospatial cognition, but that their effect may only be evident when both are in the heterozygous state.
Growth deficiency has long been associated with WBS, with mean adult height corresponding to the third percentile in both sexes (Pankau et al. 1992). Consistent with this, mild growth deficiencies were observed in Gtf2ird1-deficient mice, as was recently reported for homozygous Gtf2ird1 mutants (Tassabehji et al. 2005). Similar results were seen in a mouse deficient for another gene from the WBS deletion, Cyln2 (Hoogenraad et al. 2002), raising the possibility that the growth deficiency seen in individuals with WBS results from additive hemizygosity for Cyln2 and Gtf2ird1.
Craniofacial abnormalities were also reported in Gtf2ird1 mutants, including a proportion of homozygous mice with severely misaligned jaws (Tassabehji et al. 2005). We did not see any obvious craniofacial abnormalities in our homozygous mice although a careful quantitative analysis may be required to identify subtle abnormalities. A recent report of a second Gtf2ird1 null mouse generated using fusion of a LacZ cassette into exon 2 of the gene, also failed to detect any overt craniofacial abnormalities (Palmer et al. 2006). The difference in phenotype penetrance may be because of the influence of other genes involved in the pathways regulating craniofacial development. Our mice are maintained on a mixed, predominantly outbred genetic background, whereas the insertional mutants were on a mixed inbred background (C57BL/6 × CBA/J). The penetrance of craniofacial anomalies in mouse models of Smith–Magenis syndrome was recently shown to be highly dependent on genetic background (Yan et al. 2007).
Disruption of only a single copy of Gtf2ird1 in mice results in decreased aggression and natural fear response, and increased social interaction combined with impaired amygdala-based learning. These alterations closely resemble phenotypes observed in WBS and suggest that the haploin-sufficiency of GTF2IRD1 contributes to the physical and behavioral deficits associated with this disorder. The Gtf2ird1 mutant mice present an opportunity to identify downstream genes and pathways that are essential for proper development and maintenance of certain aspects of human behavior. These mice provide the basis for manipulations not possible in humans, for example, the global analysis of gene expression in the amygdala before and after behavioral testing, and should prove a valuable model for an intriguing human disorder.
Acknowledgments
The authors thank Dr Zhengping Jia for his help with measurement of basal synaptic activity and long-term potentiation in hippocampal slices, and Dr Andras Nagy and members of his laboratory for their help with embryonic stem cell targeting. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to L.R.O. and J.C.R. and by a Premier’s Research Excellence Award to L.R.O. J.C.R. holds a Canada Research Chair, S.J.C. holds a postdoctoral fellowship from the Royal Society of London (UK), T.L. holds a postdoctoral fellowship from the CIHR and E.J.Y. holds a graduate scholarship from the University of Toronto.
References
- Abramow-Newerly W, Lipina T, Abramow-Newerly M, Kim D, Bechard AR, Xie G, Clapcote SJ, Roder JC. Methods to rapidly and accurately screen a large number of ENU mutagenized mice for abnormal motor phenotypes. Amyotroph Lateral Scler. 2006;7:112–118. doi: 10.1080/14660820500443000. [DOI] [PubMed] [Google Scholar]
- Amaral DG. The primate amygdala and the neurobiology of social behavior: implications for understanding social anxiety. Biol Psychiatry. 2002;51:11–17. doi: 10.1016/s0006-3223(01)01307-5. [DOI] [PubMed] [Google Scholar]
- Avgustinovich DF, Lipina TV, Bondar NP, Alekseyenko OV, Kudryavtseva NN. Features of the genetically defined anxiety in mice. Behav Genet. 2000;30:101–109. doi: 10.1023/a:1001999020138. [DOI] [PubMed] [Google Scholar]
- Avgustinovich DF, Alekseenko OV, Bakshtanovskaia IV, Koriakina LA, Lipina TV, Tenditnik MV, Bondar NP, Kovalenko IL, Kudriavtseva NN. Dynamic changes of brain serotonergic and dopaminergic activities during development of anxious depression: experimental study. Usp Fiziol Nauk. 2004;35:19–40. [PubMed] [Google Scholar]
- Bayés M, Magano LF, Rivera N, Flores R, Pérez Jurado LA. Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta A, Novelli G, Mari A, Novelli A, Sabani M, Korenberg J, Osborne LR, Digilio MC, Giannotti A, Dallapiccola B. Detection of an atypical 7q11.23 deletion in Williams syndrome patients which does not include the STX1A and FZD3 genes. J Med Genet. 1999;36:478–480. [PMC free article] [PubMed] [Google Scholar]
- Caramaschi D, de Boer SF, Koolhaas JM. Differential role of the 5-HT1A receptor in aggressive and non-aggressive mice: an across-strain comparison. Physiol Behav. 2007;90:590–601. doi: 10.1016/j.physbeh.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Cheriyath V, Roy AL. Alternatively spliced isoforms of TFII-I. Complex formation, nuclear translocation, and differential gene regulation. J Biol Chem. 2000;275:26300–26308. doi: 10.1074/jbc.M002980200. [DOI] [PubMed] [Google Scholar]
- Cheriyath V, Roy AL. Structure-function analysis of TFII-I. Roles of the N-terminal end, basic region, and I-repeats. J Biol Chem. 2001;276:8377–8383. doi: 10.1074/jbc.M008411200. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Lazar NL, Bechard AR, Roder JC. Effects of the rd1 mutation and host strain on hippocampal learning in mice. Behav Genet. 2005;35:591–601. doi: 10.1007/s10519-005-5634-5. [DOI] [PubMed] [Google Scholar]
- Crackower MA, Kolas NK, Noguchi J, Sarao R, Kikuchi K, Kaneko H, Kobayashi E, Kawai Y, Kozieradzki I, Landers R, Mo R, Hui CC, Nieves E, Cohen PE, Osborne LR, Wada T, Kunieda T, Moens PB, Penninger JM. Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science. 2003;300:1291–1295. doi: 10.1126/science.1083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran ME, Atkinson DL, Ewart AK, Morris CA, Leppert MF, Keating MT. The elastin gene is disrupted by a translocation associated with supravalvular aortic stenosis. Cell. 1993;73:159–168. doi: 10.1016/0092-8674(93)90168-p. [DOI] [PubMed] [Google Scholar]
- D’Adamo P, Welzl H, Papadimitriou S, Raffaele di Barletta M, Tiveron C, Tatangelo L, Pozzi L, Chapman PF, Knevett SG, Ramsay MF, Valtorta F, Leoni C, Menegon A, Wolfer DP, Lipp HP, Toniolo D. Deletion of the mental retardation gene Gdi1 impairs associative memory and alters social behavior in mice. Hum Mol Genet. 2002;11:2567–2580. doi: 10.1093/hmg/11.21.2567. [DOI] [PubMed] [Google Scholar]
- Danoff SK, Taylor HE, Blackshaw S, Desiderio S. TFII-I, a candidate gene for Williams syndrome cognitive profile: parallels between regional expression in mouse brain and human phenotype. Neuroscience. 2004;123:931–938. doi: 10.1016/j.neuroscience.2003.08.038. [DOI] [PubMed] [Google Scholar]
- Davidson RJ, Putnam KM, Larson CL. Dysfunction in the neural circuitry of emotion regulation – a possible prelude to violence. Science. 2000;289:591–594. doi: 10.1126/science.289.5479.591. [DOI] [PubMed] [Google Scholar]
- De Almeida RM, Rosa MM, Santos DM, Saft DM, Benini Q, Miczek KA. 5-HT(1B) receptors, ventral orbitofrontal cortex, and aggressive behavior in mice. Psychopharmacology (Berl) 2006;185:441–450. doi: 10.1007/s00213-006-0333-3. [DOI] [PubMed] [Google Scholar]
- Doyle TF, Bellugi U, Korenberg JR, Graham J. “Everybody in the world is my friend” hypersociability in young children with Williams syndrome. Am J Med Genet A. 2004;124:263–273. doi: 10.1002/ajmg.a.20416. [DOI] [PubMed] [Google Scholar]
- Durkin ME, Keck-Waggoner CL, Popescu NC, Thorgeirsson SS. Integration of a c-myc transgene results in disruption of the mouse Gtf2ird1 gene, the homologue of the human GTF2IRD1 gene hemizygously deleted in Williams-Beuren syndrome. Genomics. 2001;73:20–27. doi: 10.1006/geno.2001.6507. [DOI] [PubMed] [Google Scholar]
- Dykens EM. Anxiety, fears, and phobias in persons with Williams syndrome. Dev Neuropsychol. 2003;23:291–316. doi: 10.1080/87565641.2003.9651896. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Prosnitz A, Pardo S, Bhatt J, Cohen N, Lauriat T, Ouchanov L, Jimenez Gonzalez P, Manghi ER, Bondy P, Esquivel M, Monge S, Fallas M, Splendore A, Francke U, Burton BK, McInnes LA. An atypical deletion of the Williams-Beuren Syndrome interval implicates genes associated with defective visuospatial processing and autism. J Med Genet. 2006;44:136–143. doi: 10.1136/jmg.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkhmandakh B, Bitchevaia N, Ruddle F, Bayarsaihan D. The early embryonic expression of TFII-I during mouse preimplantation development. Gene Expr Patterns. 2004;4:25–28. doi: 10.1016/s1567-133x(03)00155-8. [DOI] [PubMed] [Google Scholar]
- Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, Ensing GJ, Everett LA, Green ED, Proschel C, Gutowski NJ, Noble M, Atkinson DL, Odelberg SJ, Keating MT. LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell. 1996;86:59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Mishima T, Kofuji T, Chiba T, Tanaka K, Yamamoto A, Akagawa K. Analysis of knock-out mice to determine the role of HPC-1/syntaxin 1A in expressing synaptic plasticity. J Neurosci. 2006;26:5767–5776. doi: 10.1523/JNEUROSCI.0289-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi C, Bonaglia MC, Selicorni A, Borgatti R, Giorda R. Unusual cognitive and behavioural profile in a Williams syndrome patient with atypical 7q11.23 deletion. J Med Genet. 2003;40:526–530. doi: 10.1136/jmg.40.7.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg F. Williams syndrome professional symposium. Am J Med Genet. 1990;37 (Suppl 6):85–88. [Google Scholar]
- van Hagen JM, van der Geest JN, van der Giessen RS, Lagersvan Haselen GC, Eussen HJ, Gille JJ, Govaerts LC, Wouters CH, de Coo IF, Hoogenraad CC, Koekkoek SK, Frens MA, van Camp N, van der Linden A, Jansweijer MC, Thorgeirsson SS, De Zeeuw CI. Contribution of CYLN2 and GTF2IRD1 to neurological and cognitive symptoms in Williams Syndrome. Neurobiol Dis. 2006;26:112–124. doi: 10.1016/j.nbd.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- Heller R, Rauch A, Luttgen S, Schroder B, Winterpacht A. Partial deletion of the critical 1.5 Mb interval in Williams–Beuren syndrome. J Med Genet. 2003;40:e99. doi: 10.1136/jmg.40.8.e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsley TA, Cunliffe P, Tipney HJ, Brass A, Tassabehji M. Comparison of TFII-I gene family members deleted in Williams-Beuren syndrome. Protein Sci. 2004;13:2588–2599. doi: 10.1110/ps.04747604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota H, Matsuoka R, Chen XN, Salandanan LS, Lincoln A, Rose FE, Sunahara M, Osawa M, Bellugi U, Korenberg JR. Williams syndrome deficits in visual spatial processing linked to GTF2IRD1 and GTF2I on chromosome 7q11.23. Genet Med. 2003;5:311–321. doi: 10.1097/01.GIM.0000076975.10224.67. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Koekkoek B, Akhmanova A, Krugers H, Dortland B, Miedema M, van Alphen A, Kistler WM, Jaegle M, Koutsourakis M, Van Camp N, Verhoye M, van der Linden A, Kaverina I, Grosveld F, De Zeeuw CI, Galjart N. Targeted mutation of Cyln2 in the Williams syndrome critical region links CLIP-115 haploinsufficiency to neurodevelopmental abnormalities in mice. Nat Genet. 2002;32:116–127. doi: 10.1038/ng954. [DOI] [PubMed] [Google Scholar]
- Izquierdo A, Suda RK, Murray EA. Comparison of the effects of bilateral orbital prefrontal cortex lesions and amygdala lesions on emotional responses in rhesus monkeys. J Neurosci. 2005;25:8534–8542. doi: 10.1523/JNEUROSCI.1232-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Tasman BP, Mervis CB. Distinctive personality characteristics of 8-, 9-, and 10-year-olds with Williams syndrome. Dev Neuropsychol. 2003;23:269–290. doi: 10.1080/87565641.2003.9651895. [DOI] [PubMed] [Google Scholar]
- Li DY, Faury G, Taylor DG, Davis EC, Boyle WA, Mecham RP, Stenzel P, Boak B, Keating MT. Novel arterial pathology in mice and humans hemizygous for elastin. J Clin Invest. 1998;102:1783–1787. doi: 10.1172/JCI4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, Jia Z. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Mervis CB, Klein-Tasman BP. Williams syndrome: cognition, personality, and adaptive behavior. Ment Retard Dev Disabil Res Rev. 2000;6:148–158. doi: 10.1002/1098-2779(2000)6:2<148::AID-MRDD10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Mervis CB, Robinson BF, Bertrand J, Morris CA, Klein-Tasman BP, Armstrong SC. The Williams syndrome cognitive profile. Brain Cogn. 2000;44:604–628. doi: 10.1006/brcg.2000.1232. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Hariri AR, Munoz KE, Mervis CB, Mattay VS, Morris CA, Berman KF. Neural correlates of genetically abnormal social cognition in Williams syndrome. Nat Neurosci. 2005;8:991–993. doi: 10.1038/nn1494. [DOI] [PubMed] [Google Scholar]
- Morris CA, Mervis CB, Hobart HH, Gregg RG, Bertrand J, Ensing GJ, Sommer A, Moore CA, Hopkin RJ, Spallone PA, Keating MT, Osborne L, Kimberley KW, Stock AD. GTF2I hemizygosity implicated in mental retardation in Williams syndrome: genotype-phenotype analysis of five families with deletions in the Williams syndrome region. Am J Med Genet A. 2003;123:45–59. doi: 10.1002/ajmg.a.20496. [DOI] [PubMed] [Google Scholar]
- Mount HT, Martel JC, Fluit P, Wu Y, Gallo-Hendrikx E, Cosi C, Marien MR. Progressive sensorimotor impairment is not associated with reduced dopamine and high energy phosphate donors in a model of ataxia-telangiectasia. J Neurochem. 2004;88:1449–1454. doi: 10.1046/j.1471-4159.2003.02278.x. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertensten M, Vintersten K, Behringer R. Manipulating the Mouse Embryo; A Laboratory Manual. 3. Cold Spring Harbor Press; Cold Spring Harbor, New York: 2002. [Google Scholar]
- Ogilvie AD, Battersby S, Bubb VJ, Fink G, Harmar AJ, Goodwim GM, Smith CA. Polymorphism in serotonin transporter gene associated with susceptibility to major depression. Lancet. 1996;347:731–733. doi: 10.1016/s0140-6736(96)90079-3. [DOI] [PubMed] [Google Scholar]
- Palmer SJ, Tay ES, Santucci N, Cuc Bach TT, Hook J, Lemckert FA, Jamieson RV, Gunnning PW, Hardeman EC. Expression of Gtf2ird1, the Williams syndrome-associated gene, during mouse development. Gene Expr Patterns. 2006;7:396–404. doi: 10.1016/j.modgep.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Pankau R, Partsch CJ, Gosch A, Oppermann HC, Wessel A. Statural growth in Williams-Beuren syndrome. Eur J Pediatr. 1992;151:751–755. doi: 10.1007/BF01959084. [DOI] [PubMed] [Google Scholar]
- Pober BR, Dykens EM. Williams syndrome: an overview of medical, cognitive, and behavioral features. Child Adolesc Psychiatr Clin N Am. 1996;5:929–943. [Google Scholar]
- Prather MD, Lavenex P, Mauldin-Jourdain ML, Mason WA, Capitanio JP, Mendoza SP, Amaral DG. Increased social fear and decreased fear of objects in monkeys with neonatal amygdala lesions. Neuroscience. 2001;106:653–658. doi: 10.1016/s0306-4522(01)00445-6. [DOI] [PubMed] [Google Scholar]
- Rodgers RJ, Cole JC. Anxiolytic-like effect of (S)-WAY 100135, a 5-HT1A receptor antagonist, in the murine elevated plus-maze test. Eur J Pharmacol. 1994;261:321–325. doi: 10.1016/0014-2999(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Roy AL. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I. Gene. 2001;274:1–13. doi: 10.1016/s0378-1119(01)00625-4. [DOI] [PubMed] [Google Scholar]
- Schiller L, Donix M, Jahkel M, Oehler J. Serotonin 1A and 2A receptor densities, neurochemical and behavioural characteristics in two closely related mice strains after long-term isolation. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:492–503. doi: 10.1016/j.pnpbp.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Shumyatsky GP, Malleret G, Shin RM, Takizawa S, Tully K, Tsvetkov E, Zakharenko SS, Joseph J, Vronskaya S, Yin D, Schubart UK, Kandel ER, Bolshakov VY. stathmin, a gene enriched in the amygdala, controls both learned and innate fear. Cell. 2005;123:697–709. doi: 10.1016/j.cell.2005.08.038. [DOI] [PubMed] [Google Scholar]
- Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Perez-Jurado LA, Morris CA, Scherer SW, Osborne LR. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromme P, Bjornstad PG, Ramstad K. Prevalence estimation of Williams syndrome. J Child Neurol. 2002;17:269–271. doi: 10.1177/088307380201700406. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Metcalfe K, Karmiloff-Smith A, Carette MJ, Grant J, Dennis N, Reardon W, Splitt M, Read AP, Donnai D. Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am J Hum Genet. 1999;64:118–125. doi: 10.1086/302214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Hammond P, Karmiloff-Smith A, Thompson P, Thorgeirsson SS, Durkin ME, Popescu NC, Hutton T, Metcalfe K, Rucka A, Stewart H, Read AP, Maconochie M, Donnai D. GTF2IRD1 in craniofacial development of humans and mice. Science. 2005;310:1184–1187. doi: 10.1126/science.1116142. [DOI] [PubMed] [Google Scholar]
- Tipney HJ, Hinsley TA, Brass A, Metcalfe K, Donnai D, Tassabehji M. Isolation and characterisation of GTF2IRD2, a novel fusion gene and member of the TFII-I family of transcription factors, deleted in Williams-Beuren syndrome. Eur J Hum Genet. 2004;12:551–560. doi: 10.1038/sj.ejhg.5201174. [DOI] [PubMed] [Google Scholar]
- Vergnes M, Depaulis A, Boehrer A. Parachlorophenylalanine-induced serotonin depletion increases offensive but not defensive aggression in male rats. Physiol Behav. 1986;36:653–658. doi: 10.1016/0031-9384(86)90349-5. [DOI] [PubMed] [Google Scholar]
- Winslow JT, Insel TR. The social deficits of the oxytocin knockout mouse. Neuropeptides. 2002;36:221–229. doi: 10.1054/npep.2002.0909. [DOI] [PubMed] [Google Scholar]
- Yan J, Bi W, Lupski JR. Penetrance of craniofacial anomalies in mouse models of smith-magenis syndrome is modified by genomic sequence surrounding rai1: not all null alleles are alike. Am J Hum Genet. 2007;80:518–525. doi: 10.1086/512043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Aviles C, Abel RA, Almli CR, McQuillen P, Pleasure SJ. Hippocampal and visuospatial learning defects in mice with a deletion of frizzled 9, a gene in the Williams syndrome deletion interval. Development. 2005;132:2917–2927. doi: 10.1242/dev.01871. [DOI] [PubMed] [Google Scholar]