Abstract
Adverse events linked to perturbations of cellular genes by vector insertion reported in gene therapy trials and animal models have prompted attempts to better understand the mechanisms directing viral vector integration. The integration profiles of vectors based on MLV, ASLV, SIV, and HIV have all been shown to be non-random, and novel vectors with a safer integration pattern have been sought. Recently we developed a producer cell line called CatPac that packages standard MoMLV vectors with FeLV gag, pol and env gene products. We now report the integration profile of this vector, asking if the FeLV integrase and capsid proteins could modify the MoMLV integration profile, potentially resulting in a less genotoxic pattern. We transduced rhesus macaque CD34+ hematopoietic progenitor cells with CatPac or standard MoMLV vectors, and determined their integration profile by LAM-PCR. We obtained 184 and 175 unique integration sites (IS) respectively for CatPac and standard MoMLV vectors, and these were compared to 10 000 in silico-generated random IS. The integration profile for CatPac vector was similar to MoMLV and equally non-random, with a propensity for integration near transcription start sites and in highly dense gene regions. We found an IS for CatPac vector localized 715 nucleotides upstream of LMO-2, the gene involved in the ALL developed by X-SCID patients treated via gene therapy using MoMLV vectors. In conclusion, we found that replacement of MoMLV env, gag, and pol gene products with FeLV did not alter the basic integration profile. Thus there appears to be no safety advantage for this packaging system. However, considering the stability and efficacy of CatPac vectors, further development is warranted, utilizing potentially safer vector backbones, for instance those with a SIN configuration.
Keywords: Gene Therapy, Retroviral Vector, Insertion Site Analysis, Viral Packaging Machinery, Feline Leukemia Virus
Introduction
Insertional mutagenesis after gene transfer in animal models1,2 as well as in X-SCID clinical trials3–5 has led numerous laboratories to focus on the safety of the virus-based vectors used to transfer and express genes of interest. A better understanding of the mechanisms that define the characteristics of the integration of virus-based vectors should allow the development of new vectors with increased biosafety. Taking advantage of the subgroup C Feline Leukemia Virus (FeLV-C) as a platform for efficient transduction of target human cell types, including CD34+ stem and progenitor cells, we recently developed an FeLV-C based packaging system named CatPac. HEK293 cells were engineered to stably package MoMLV vectors with FeLV 61E gag-pol gene products, in addition to FeLV-C Sarna env, resulting in a completely FeLV-derived packaging system6. In the current study we assessed the integration profile of CatPac vector in comparison to the same MoMLV vector packaged with MoMLV gag-pol and amphotropic env in order to elucidate the influence of the FeLV component packaging proteins on viral integration. Both primary vector sequences as well as integrase proteins have been implicated in genomic integration site selection for retroviruses, and thus we asked whether this hybrid vector might have a potentially less genotoxic integration pattern than standard MoMLV, due to the presence of the alternative FeLV integrase machinery. FeLV-C and MoMLV are both members of the mammalian C-type retrovirus family, but there is no prior information on the integration profile for either FeLV-C or hybrid MoMLV/FeLV packaging and vector systems.
Material and Methods
Rhesus macaque CD34+ cells were obtained as previously described7. Two rhesus macaques (RQ4984 & RQ4972) were treated with a combination of G-CSF and SCF and mobilized circulating hematopoietic progenitor and stem cells were collected by apheresis. PBMNCs contained in apheresis product were purified by density gradient centrifugation over lymphocyte separation media (LSM, MP Biomedicals) and CD34+ cells were positively immunoselected using the 12.8 IgM anti-CD34 biotinylated antibody and MACS streptavidin microbeads (Miltenyi Biotec). The purified CD34+ cells were stimulated for 48 hrs in DMEM supplemented with 10% FCS, SCF, Flt3-L, and TPO and then transduced twice on fibronectin-treated plates (Retronectin, Takara) with the CatPac or control MoMLV vector supernatants, using our standard in vitro transduction conditions8. The vectors contained the GFP marker gene, allowing assessment of transduction efficiency by flow cytometry. In both animals, the transduction efficiencies of CD34+ cells for CatPac and control MoMLV packaging systems were equivalent: for RQ4984 8.1 % and 8.8 % of GFP positive cells for CatPac and MoMLV vectors respectively, and for RQ4972 39.7 % and 20.3% respectively. Genomic DNA of transduced CD34+ cells was isolated using Qiagen DNeasy Blood & Tissue kit (#69506) and LAM-PCR was carried out as previously described9 with 100 ng of total genomic DNA. TasI was used as the restriction enzyme. Linear PCR was carried out with biotinylated primer LTRa (Biot 5′-TGCTTACCACAGATATCCTG-3′). The first exponential PCR was carried out with primer LCI (5′-GACCCGGGAGATCTGAAT-3′) and LTR-R1 primer (5′-CAGCTGTTCCATCTGTTC-3′), whereas the second exponential PCR was carried out with LCIII (5′-AGTGGCACAGCAGTTAGG-3′) and LTR-R2 primer (5′-GCTAGCTTGCCAAACCTA-3′). Sequences obtained were aligned by BLAT or BLAST to the rhesus macaque genome assembly (Mmul 1.0, Jan 2006). We obtained 200 and 187 valid integration sites (IS) for CatPac and MoMLV vectors respectively (Supplemental Information). Sites mapping to 2 or more genomic positions as well as sites mapping within repetitive genomic sequences were omitted to finally obtain 184 and 175 unique IS respectively for CatPac and MoMLV vectors. We also compared both profiles to in silico-randomly generated IS sets as previously described9. Briefly, 10 000 sets of 184 or 175 random IS were designed in silico as follows: An AATT (TasI) site in the genome was selected at random using a random number generator. The in silico IS was placed either upstream or downstream (p=0.5) of the AATT site, at a distance matching the size of one of the sequences obtained experimentally. The in silico IS was validated only when a BLAST alignment of the genomic sequence between the AATT and the IS returned a unique sequence in the genome. This operation was repeated 184 times and 175 times respectively for CatPac and MoMLV vectors to obtain a single matching random dataset. These control datasets were subjected to the same analyses as the experimental datasets, and the results were used to generate empiric p-values. We generated two groups of control genomic coordinates, one with 10 000 sets of 184 coordinates each to mimic the CatPac vector IS, and one with 10 000 sets of 175 coordinates each to mimic the MoMLV IS. For gene annotation, Ensembl release 54 (May 2009) comprising 38 146 predicted gene transcripts was used. The association between the CatPac and MoMLV vectors integration sites was tested using a Chi-square test.
Results and Discussion
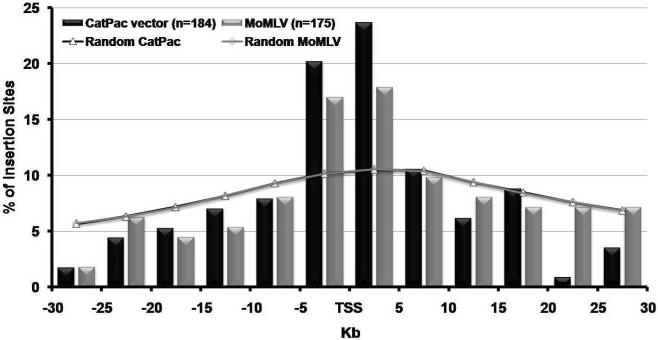
The results obtained for MoMLV are comparable to previously reported integration patterns for standard MoMLV derived vectors10,11. We confirmed the preference of MoMLV for integrating near the TSS, a feature also shared by CatPac vector. Like other retroviral vectors already studied such as MoMLV10,11, ASLV12, SIV10,13, and HIV12,14, CatPac vector exhibited a profile of integration significantly different from random integrations as can be seen in all the tables and figures presented in this report. As shown in Figure 1, out of the 114 CatPac vector and 112 MoMLV IS located in a window of 60 Kb centered on transcription start sites (TSS), 44% (CatPac vector) and 35% (MoMLV) occurred in a 10 Kb window centered on TSS, implicating a preference for integration near TSS. With regard to inter or intragenic insertions, Table 1 shows that approximately half of the vector integrations were found inside of a gene (49.5 and 54.3% for CatPac vector and MoMLV vectors respectively). One quarter of all integrations were found within a 30 Kb window upstream of a gene (Table 1). In these upstream regions CpG islands are motifs found in or in the vicinity of promoters. As shown in Table 2 integration of both vectors in or around (±1 Kb and ±5 Kb) CpG islands was significantly different from the random datasets. A quarter of the insertion sites were found ±5 Kb around CpG islands for CatPac (27.7 %) and MoMLV (25.7 %) vectors. However, this higher percentage probably represents the propensity of CatPac and MoMLV vectors to integrate close to the TSS, rather than a preference for CpG islands. Looking at integrations with regard to gene density, Table 2 shows that less than half of the integrations for both vectors were found in lower gene-density regions (0 to 10 genes / Mb) of the genome, and thus 53.8 % and 57.7 % of integration respectively for CatPac and MoMLV vectors in gene dense regions (11 genes/Mb and up). Although this integration profile is significantly different from in silico-generated integrations, there were no obvious differences in the integration pattern between CatPac and MoMLV vectors with respect to gene density.
Figure 1.
Distribution of integration sites determined experimentally or in silico-generated in a 60 Kb window centered on Transcription Start Sites (TSS). The window of 60 Kb is subdivided into regions of 5 Kb and percentage of integration sites are reported for CatPac (black) and MoMLV (grey) vectors.
Table 1.
Summary of vector insertion site characteristics versus random in silico-generated integration datasets (n.s. = non significant).
| CatPac vector (n=184) | Matching Random | p-value | Control MoMLV (n=175) | Matching Random | p-value | p-value CatPac vector vs. MoMLV | |
|---|---|---|---|---|---|---|---|
| Sites inside of a gene (w/in exon or intron) | 49.5% | 32.9% | p < 0.0001 | 54.3% | 33.0% | p < 0.0001 | p ≤ 0.4181 (n.s.) |
| Sites inside of a gene and w/in exon | 11.0% | 3.2% | p ≤ 0.0031 | 3.2% | 3.2% | p ≤ 0.4793 (n.s.) | p ≤ 0.0709 (n.s.) |
| Sites inside of a gene and w/in intron | 89.0% | 96.8% | p ≤ 0.0031 | 96.8% | 96.7% | p ≤ 0.4793 (n.s.) | p ≤ 0.0709 (n.s.) |
| Sites w/in 30 kb upstream of 1 or more genes | 25.0% | 12.3% | p < 0.0001 | 26.9% | 12.3% | p < 0.0001 | p ≤ 0.7787 (n.s.) |
Table 2.
Distribution of vector insertion sites relative to gene density within 1 Mb window (±500 Kb around the integration site) and to CpG islands (n.s. = non significant).
| CatPac vector (n=184) | Matching Random | p-value | Control MoMLV (n=175) | Matching Random | p-value | p-value CatPac vector vs. MoMLV | ||
|---|---|---|---|---|---|---|---|---|
| Integration site contained w/in CpG island | 1.6% | 0.1% | p ≤ 0.0007 | 2.2% | 0.1% | p < 0.0001 | p ≤ 0.9466 (n.s.) | |
| Integration site contained w/in CpG island, or within 1 kb window on either side | 4.4% | 1.0% | p < 0.0001 | 5.2% | 1.0% | p < 0.0001 | p ≤ 0.5166 (n.s.) | |
| Integration site contained w/in CpG island, or within 5 kb window on either side | 27.7% | 4.9% | p < 0.0001 | 25.7% | 4.9% | p < 0.0001 | p ≤ 0.7571 (n.s.) | |
|
| ||||||||
| Insertion sites relative to gene density w/ in a 1Mb window | 0 – 10 genes | 46.2% | 79.5% | p < 0.0001 | 42.3% | 79.5% | p < 0.0001 | p ≤ 0.5227 (n.s.) |
| 11 – 20 genes | 25.5% | 15.4% | p ≤ 0.0007 | 33.1% | 15.4% | p < 0.0001 | p ≤ 0.1426 (n.s.) | |
| 21 genes and up | 28.3% | 5.1% | p < 0.0001 | 24.6% | 5.1% | p < 0.0001 | p ≤ 0.5013 (n.s.) | |
Theoretically, virus-based vectors can lead to tumor formation by insertional mutagenesis either by activating an oncogene or by disrupting a tumor suppressor gene15. Most, if not all, cases of malignant disease development following gene therapy for hematological disease have been associated with the activation of a proto-oncogene 2–5. We therefore looked at integration of CatPac and MoMLV vectors in the proximity of oncogenes (Table 3). We assessed both the proximity of proto-oncogenes to IS, and conversely the proximity of IS to proto-oncogenes. We defined proto-oncogenes based on the most recent version of the Sanger oncogene file (dated 2008-12-16, at http://www.sanger.ac.uk/genetics/CGP/Census/). Three hundred eighty four human entries were mapped to 368 unique Ensembl rhesus macaque gene identifiers. As shown in Table 3, with this size dataset, the MoMLV integrations were not notably different from randoms with regard to proximity to oncogenes. However, the CatPac vector integrations were more likely than the random datasets to be in proximity to oncogenes. The 12 CatPac vector IS in or within a 30 kb window of an oncogene were located: upstream of MLLT11, MUTYH, CYTSB, and LMO2; downstream of ARNT; in the intron of LASP1, Q4W6X8_MACMU, AKAP9, ERG, CBFB, and PCM1; and in the exon of SUFU. The 7 MoMLV IS were located: upstream of MLLT11 and TAL1; downstream of KRAS and STIL; in the intron of CDK6, PMS1, and CIITA. The only oncogene in proximity to both CatPac vector and MoMLV integrations was MLLT11, myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 11, also termed AF1q. Expression of this gene has been shown to be linked to poor prognosis in AML (reviewed in16) and has a function in apoptosis and drug resistance. We did not find any integrations in or near the MDS1-EVI1 gene complex, previously reported to be over-represented in engrafting human, murine, and rhesus macaque hematopoietic stem and progenitor cells transduced with MoMLV vectors17. Interestingly, in the 12 oncogenes targeted by the CatPac vector we found an integration 715 nucleotides upstream of LMO2, the gene activated in X-SCID trials and causing leukemia through a T cell clonal expansion3–5. LMO2 was also reported as an MLV insertion site in the ADA-SCID and CGD gene therapy trials, but so far, no clonal expansion has been observed18–20.
Table 3.
Proximity of vector insertions to oncogenes (n.s. = non significant).
| CatPac vector (n=184) | Matching Random | p-value | Control MoMLV (n=175) | Matching Random | p-value | p-value CatPac vector vs. MoMLV | |
|---|---|---|---|---|---|---|---|
| Number of oncogenes w/ in 30 kb of one or more integration sites | 12 | 2.8 | p < 0.0001 | 7 | 2.7 | p ≤ 0.0201 | p ≤ 0.3525 (n.s.) |
| Number of genes w/ in 30 kb of one or more integration sites | 288 | 132.6 | p < 0.0001 | 268 | 126.4 | p < 0.0001 | p ≤ 0.4181 (n.s.) |
| Genes that are oncogenes w/ in 30 Kb of integration site | 4.2% | 2.2% | p ≤ 0.0705 (n.s.) | 2.6% | 2.1% | p ≤ 0.3216 (n.s.) | p ≤ 0.4385 (n.s.) |
|
| |||||||
| Number of integration sites w/in 30 kb of one or more oncogenes | 12 | 2.8 | p < 0.0001 | 6 | 2.7 | p ≤ 0.0562 (n.s.) | p ≤ 0.2711 (n.s.) |
| Number of integration sites w/i 30 kb of one or more genes | 150 | 96.5 | p < 0.0001 | 156 | 91.9 | p < 0.0001 | p ≤ 0.0593 (n.s.) |
| Sites w/ in 30 Kb of a gene that is an oncogene | 8.0% | 2.9% | p ≤ 0.0075 | 3.9% | 2.9% | p ≤ 0.2836 (n.s.) | p ≤ 0.1933 (n.s.) |
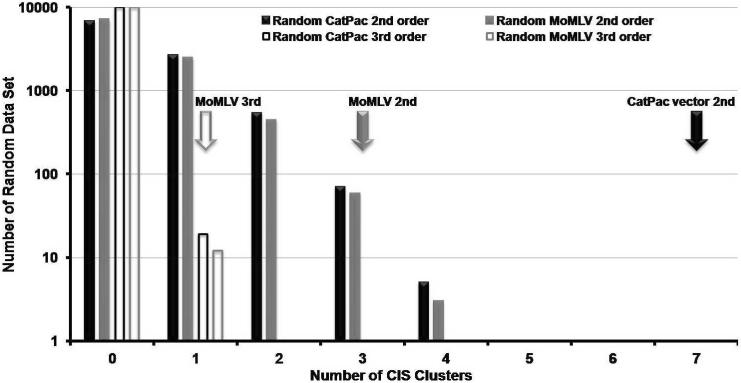
In order to analyze if there was a significant clustering of vector insertions we used the definition of common integration sites (CIS) reported by Suzuki and coworkers21. As shown in Figure 2, there is a difference between experimental data obtained for CatPac vector and random integrations. We found 7 second order CIS for CatPac vector, a higher value than the frequency of second order CIS in the matching random datasets. None of the CIS identified were localized within 200 Kb of a known proto-oncogene.
Figure 2.
Occurrence of Common Integration Sites (CIS) in CD34+ cells transduced with CatPac or MoMLV vectors. According to the definition of Suzuki and coworkers21, a second order CIS is defined as 2 or more IS located within a 30 kb window; a third order CIS as 3 or more IS within a 50 kb window; and a fourth order CIS as 4 or more IS within a 100 kb window. The black solid arrow indicates the number of second order CIS found in the experimental dataset obtained for CatPac vector (7). There was no third order CIS found in the CatPac experimental dataset. The solid grey and empty grey arrows respectively indicate the number of second (3) and third (1) order CIS found in the experimental dataset for MoMLV. These numbers can be compared to the CIS obtained for the 10 000 independent random datasets generated in silico to match the CatPac and MoMLV datasets. The X-axis gives the number of CIS and the Y-axis gives the number of random datasets, out of 10 000, that included second order (solid boxes) or third order (empty boxes) CIS. Black and grey boxes respectively represent data obtained for random sets matching CatPac and MoMLV vectors. There was no fourth order CIS found in any set (random or experimental).
These investigations contribute to knowledge regarding the determinants of retrovirus integration into the genome. At least two vector-related elements are involved: the integrase/capsid proteins and/or the primary sequence of the vector backbone itself. The relative contributions of each element in determining integration preferences remain under intense investigation, but it appears that the integrase core region is critical. Swapping the integrase between even closely related viruses, such as HIV and FIV, results in a change in the integration pattern determined primarily by the integrase, along with a contribution from gag-encoded capsid proteins22,23. The path of entry into the cell, determined by the env gene product, appears to have little impact on integration preferences24. In a previous study, pseudotyping of a standard MoMLV retroviral vector with the FeLV-C env protein only resulted in more efficient transduction of primitive CD34+ cells compared to a standard GALV env pseudotype, but no change in integration profile, based on a limited number of integration sites retrieved from myeloid cells following in vivo engraftment25. In the present study, we went on to study the impact of using FeLV gag and pol to package an MoMLV backbone and compared it to an MoMLV backbone packaged with standard MoMLV components, to determine if the integrase and capsid proteins of FeLV would change the integration pattern of MoMLV vector backbones. Utilization of FeLV gag, pol, and env components did not alter the integration profile of MoMLV vectors, in contrast to the prior results comparing the impact of swapping integrase between HIV and FIV viruses. Both standard MoMLV and CatPac vectors integrated within genes and in areas of high gene density, near TSS. Other studies have indicated that vector backbone sequences are also important determinants of integration patterns. The pre-integration complex contains viral integrase and capsid proteins, reverse-transcribed vector DNA, and host cell factors, all implicated in both site-selection and the actual integration process26–28. There are no large-scale integration profiles published for FeLV, however CIS identification in tumors induced by FeLV infection indicate activation of an overlapping but not identical set of genes by FeLV as compared to MoMLV29. Swapping the U3 region of the MoLV LTR with the same region of FeLV results in a partial shift in CIS in tumors30. However, the integration profile of CIS in tumors may relate more to the enhancer activity of a specific U3 region in a target cell type as opposed to actual integration characteristics, since analysis of tumor CIS relies to a large degree on in vivo selection of clones with activating insertions, from an initial highly polyclonal pool of cells.
In conclusion, retroviral vectors packaged using the CatPac system integrate into the genome in a non-random fashion, with a general profile very similar to vectors packaged using standard MoMLV gag and pol gene products, unfortunately characterized by a preference for integration within genes and near TSS. Although the study of a larger number of integration sites might uncover subtle differences between FeLV and MoMLV integrase and capsid determinants of integration, it is clear from the data we obtained in this pilot study that their general profiles are similar and characterized by the same risks. Therefore, while CatPac vector may have advantages in terms of ease of high titer vector production and potential improved efficiency of gene transfer to some target cell populations, the integration profile we have characterized does not suggest any increased safety of these vectors compared to standard retrovirus packaged with MoMLV components.
Supplementary Material
Acknowledgments
Jean-Yves Métais would like to dedicate this manuscript to his grandmother Marie Pradier born Sautereau.
This research was supported in part by the Intramural Research Programs of the National Heart, Lung, and Blood Institute and the National Human Genome Research Institute, National Institutes of Health.
Raymond T. Doty and Janis L. Abkowitz are the inventors on a patent filed for the CatPac producer cell line which may have potential for commercial use (patent application serial number 61/058,148). Upon request CatPac packaging cells will be supplied to academic investigators. The authors have no other potential conflicts of interest to declare.
References
- 1.Li Z, Dullmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- 2.Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 5.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doty RT, Sabo KM, Chen J, Miller AD, Abkowitz JL. An all feline retroviral packaging system for transduction of human cells. Hum Gene Ther. doi: 10.1089/hum.2010.032. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donahue RE, Kirby MR, Metzger ME, Agricola BA, Sellers SE, Cullis HM. Peripheral blood CD34+ cells differ from bone marrow CD34+ cells in Thy-1 expression and cell cycle status in nonhuman primates mobilized or not mobilized with granulocyte colony-stimulating factor and/or stem cell factor. Blood. 1996;87:1644–1653. [PubMed] [Google Scholar]
- 8.Larochelle A, Choi U, Shou Y, Naumann N, Loktionova NA, Clevenger JR, et al. In vivo selection of hematopoietic progenitor cells and temozolomide dose intensification in rhesus macaques through lentiviral transduction with a drug resistance gene. J Clin Invest. 2009;119:1952–1963. doi: 10.1172/JCI37506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YJ, Kim YS, Larochelle A, Renaud G, Wolfsberg TG, Adler R, et al. Sustained high-level polyclonal hematopoietic marking and transgene expression 4 years after autologous transplantation of rhesus macaques with SIV lentiviral vector-transduced CD34+ cells. Blood. 2009;113:5434–5443. doi: 10.1182/blood-2008-10-185199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hematti P, Hong BK, Ferguson C, Adler R, Hanawa H, Sellers S, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crise B, Li Y, Yuan C, Morcock DR, Whitby D, Munroe DJ, et al. Simian immunodeficiency virus integration preference is similar to that of human immunodeficiency virus type 1. J Virol. 2005;79:12199–12204. doi: 10.1128/JVI.79.19.12199-12204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 15.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Gregory TK, Wald D, Chen Y, Vermaat JM, Xiong Y, Tse W. Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics. J Hematol Oncol. 2009;2:23. doi: 10.1186/1756-8722-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metais JY, Dunbar CE. The MDS1-EVI1 gene complex as a retrovirus integration site: impact on behavior of hematopoietic cells and implications for gene therapy. Mol Ther. 2008;16:439–449. doi: 10.1038/sj.mt.6300372. [DOI] [PubMed] [Google Scholar]
- 18.Aiuti A, Cassani B, Andolfi G, Mirolo M, Biasco L, Recchia A, et al. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J Clin Invest. 2007;117:2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassani B, Montini E, Maruggi G, Ambrosi A, Mirolo M, Selleri S, et al. Integration of retroviral vectors induces minor changes in the transcriptional activity of T cells from ADA-SCID patients treated with gene therapy. Blood. 2009 doi: 10.1182/blood-2009-02-202085. [DOI] [PubMed] [Google Scholar]
- 20.Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 22.Shibagaki Y, Chow SA. Central core domain of retroviral integrase is responsible for target site selection. J Biol Chem. 1997;272:8361–8369. doi: 10.1074/jbc.272.13.8361. [DOI] [PubMed] [Google Scholar]
- 23.Harper AL, Sudol M, Katzman M. An amino acid in the central catalytic domain of three retroviral integrases that affects target site selection in nonviral DNA. J Virol. 2003;77:3838–3845. doi: 10.1128/JVI.77.6.3838-3845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol Ther. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Lucas ML, Seidel NE, Porada CD, Quigley JG, Anderson SM, Malech HL, et al. Improved transduction of human sheep repopulating cells by retrovirus vectors pseudotyped with feline leukemia virus type C or RD114 envelopes. Blood. 2005;106:51–58. doi: 10.1182/blood-2004-11-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewinski MK, Bushman FD. Retroviral DNA integration--mechanism and consequences. Adv Genet. 2005;55:147–181. doi: 10.1016/S0065-2660(05)55005-3. [DOI] [PubMed] [Google Scholar]
- 27.Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, et al. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J Biol Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- 29.Fujino Y, Ohno K, Tsujimoto H. Molecular pathogenesis of feline leukemia virus-induced malignancies: insertional mutagenesis. Vet Immunol Immunopathol. 2008;123:138–143. doi: 10.1016/j.vetimm.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 30.Johnson C, Lobelle-Rich PA, Puetter A, Levy LS. Substitution of feline leukemia virus long terminal repeat sequences into murine leukemia virus alters the pattern of insertional activation and identifies new common insertion sites. J Virol. 2005;79:57–66. doi: 10.1128/JVI.79.1.57-66.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.