Abstract
The N-glycan-dependent quality control of glycoprotein folding prevents endoplasmic to Golgi exit of folding intermediates, irreparably misfolded glycoproteins and incompletely assembled multimeric complexes. It also enhances folding efficiency by preventing aggregation and facilitating formation of proper disulfide bonds. The control mechanism essentially involves four components, resident lectin-chaperones that recognize monoglucosylated polymannose glycans, a lectin-associated oxidoreductase acting on monoglucosylated glycoproteins, a glucosyltransferase that creates monoglucosytlated epitopes in protein-linked glycans and a glucosidase that removes the glucose units added by the glucosyltransferase. This last enzyme is the only mechanism component sensing glycoprotein conformations as it creates monoglucosylated glycans exclusively in not properly folded species or in not completely assembled complexes. The glucosidase is a dimeric heterodimer composed of a catalytic subunit and an additional one that is partially responsible for the ER localization of the enzyme and for the enhancement of the deglucosylation rate as its mannose 6-phosphate receptor homologous domain presents the substrate to the catalytic site. This review deals with our present knowledge on the glucosyltransferase and the glucosidase.
Keywords: Glycoprotein folding, Quality control, Endoplasmic reticulum, Glucosyltransferase, Glucosidase II
The scenario
Nearly one third of proteins synthesized by eukaryotic cells enter the secretory pathway either co- or post-translationally. About 80 % of these proteins are N-glycosylated in the ER by the oligosaccharyltransferase (OST) complex at the sequon Asn-X-Ser/Thr (where X cannot be Pro). In most eukaryotic cells, OST transfers en bloc the Glc3Man9GlcNAc2 oligosaccharide from a dolichol pyrophosphate derivative (Fig 1). This common structure contrasts with the enormous glycan diversity found in mature glycoproteins, which results from the activity of several glycosidases and glycosyltransferases operating along the secretory pathway. Glycan processing begins in the ER immediately after the transfer reaction: glucosidase I (GI) hydrolyses the outermost glucose (residue n, Fig. 1), followed by glucosidase II (GII) that sequentially removes the remaining two glucose residues (residues l and n, Fig. 1). In addition, an ER α-mannosidase (ER α-mannosidase I) may eliminate one or two mannose residues from branches B and C (residues k and i, Fig. 1). The presence of monoglucosylated glycans was originally thought to be the exclusive result of partial trimming by GII. The finding of protein-linked monoglucosylated glycans in T. cruzi [1], in which the oligosaccharide transferred lacks glucose residues, indicated that glycans could be glucosylated in the ER once transferred. Glycoprotein glucosylation is present in most eukaryotic cells that transfer high mannose oligosaccharides, with the notable exception of S. cerevisiae. The enzyme responsible for the glucosylation reaction, named UDP-Glc:glycoprotein glucosyltransferase (UGGT), was firstly purified from rat liver [2]. UGGT employs UDP-Glc as glucose donor and high mannose glycoproteins as acceptors, in a reaction that requires relatively high calcium concentrations. The most striking property of UGGT is that acceptor glycoproteins must be displaying non native conformations to become good substrates [25]. The preference of UGGT for misfolded glycoproteins and the high specificity of calnexin (CNX) as a lectin for monoglucosylated glycans [3] prompted Ari Helenius and co-workers to propose a glycoprotein folding quality control (QC) mechanism (Fig. 2) [4]. The subsequent incorporation to this system of another lectin, calreticulin (CRT), highly homologous to CNX and of ERp57, a protein disulfide isomerase associated with CRT and CNX, did not modify its fundamentals. Briefly, trimming of the two outermost glucose residues by GI and GII triggers the binding of monoglucosylated glycoproteins to CNX and/or CRT, a phenomenon that can take place either co- or post-translationally depending on the particular substrate. This complex is disrupted upon GII removal of the innermost glucose (residue l, Fig. 1). At his stage, properly folded proteins may proceed to their final destinations. By contrast, deglucosylated misfolded species, folding intermediates or unassembled oligomers will be recognized by UGGT and the resulting reaction products will reassociate with the lectins (CNX/CRT). The complexes thus formed will be then retained in the ER. Deglucosylation-reglucosylation cycles driven by the opposite activities of GII and UGGT continues until the glycoprotein acquires its native fold or, alternatively, until it is marked for degradation by the ER associated degradation (ERAD) pathway. QC not only prevents the premature exit of immature glycoproteins, but also enhances the folding efficiency by inhibiting protein aggregation and allowing the action of ERp57. Although this model has been validated in numerous systems by different means, many issues remain open. For instance, it is unclear how QC and ERAD systems are able to discriminate between proteins undergoing a productive folding pathway from those irreparably misfolded that should be driven to degradation. In addition, recent observations suggest that UGGT may not be a mere folding sensor as it probably plays an active role in promoting the correct folding of certain glycoproteins. This review will focus on our present knowledge on two key enzymes of the QC, namely UGGT and GII.
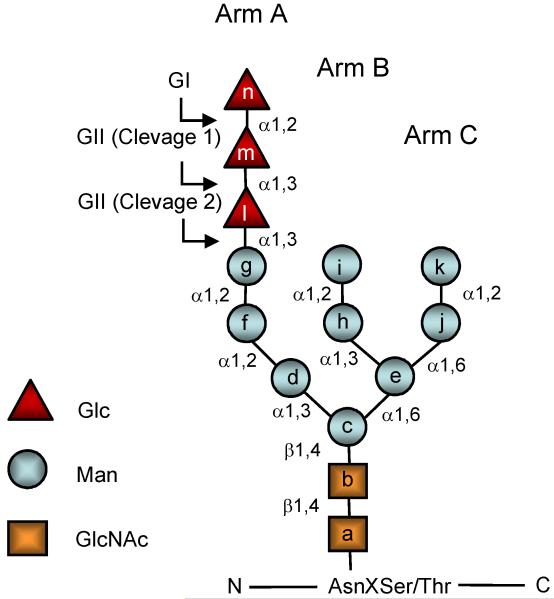
Fig. 1.
Structure of glycans. Lettering (a-n) follows the order of addition of monosaccharides in the synthesis of Glc3Man9GlcNAc2-P-P-dolichol. GI removes residue n and GII residues l and m. UGGT adds residue l to residue g.
Fig. 2.
Model proposed for the quality control of glycoprotein folding. Proteins entering the ER are N-glycosylated by the oligosaccharyltransferase (OST) as they emerge from the translocon (Sec61). Two glucoses are removed by the sequential action of GI and GII to generate monoglucosylated species that are recognized by CNX and/or CRT (only CNX is shown), that are associated with ERp57. The complex formed by the lectins and folding intermediates/misfolded glycoproteins dissociates upon removal of the last glucose by GII and is reformed by UGGT activity. Once glycoproteins have acquired their native conformations, either free or complexed with the lectins, GII hydrolyses the remaining glucose residue and releases the glycoproteins from the lectin anchors. These species are nor recognized by UGGT and are transported to the Golgi. Glycoproteins unable to properly fold are retrotranslocated to the cytosol where they are deglycosylated by the proteasome. One or more mannose residues may be removed by the ER α–mannosidase I (α-Man) during the whole folding process.
The UDP-Glc:glycoprotein glucoyltransferase (UGGT)
UGGT is a large monomeric protein composed of approximately 1600 amino acid residues that displays an ER retention/retrieval sequence at its C-terminus (HDEL in Yarrowia lipolytica [5], PDEL in Schizosaccharomyces pombe [6], HGEL in Drosophila melanogaster [7], HEEL in rat [8], REEL in human [9]). Consequently, UGGT is mainly found in the ER and the ER-Golgi intermediate compartment (ERGIC) [10], and is one of the few soluble glycosyltransferases of the secretory pathway described so far. Interestingly, the UGGT from T. cruzi lacks an ER retention/retrieval sequence, and thence it is unknown how its ER localization is achieved [11]. UGGT is present in most organisms that transfer high mannose oligosaccharides. By contrast, organisms that transfer extremely short versions of the glycan such as Giardia lamblia and Plasmodium falciparum, lack UGGT [12]. This enzyme transfers a single glucose residue from UDP-Glc to the terminal mannose of branch A of high mannose oligosaccharides (residue g, Fig. 1). As the reaction preserves the configuration of the glucose anomeric carbon, a covalent bound glucose-enzyme intermediate is expected to occur. UGGT is conformed by at least two structural domains [13]. The N-terminal domain comprises 80% of the molecule and has no homology to other known proteins. This domain is thought to be responsible for the recognition of non-native conformers, although this has not been conclusively established yet [14]. The C-terminal or catalytic domain comprises around 20 % of the protein, binds [β-32P]5N3UDP-Glc and displays significant similarity to members of glycosyltransferase family 8 [8]. This domain displays the conserved motif DQDXXN, where the triad DQD may serve to coordinate a divalent cation necessary for UDP-Glc binding. UGGT C-terminal domains from different species share a significant similarity (60–70%), but much lower values occur between N-terminal ones. For instance, S. pombe and D. melanogaster N-terminal domains show a poor similarity (16.3 %), but chimeras combining the C and N domains from both species were active in vivo, suggesting that their N-terminal domains share similar structural features [13]. Humans have two homologous genes coding for for UGGT that share 55 % identity, hUGGT1 and hUGGT2, but only the former appeared to be enzymatically active [9]. ER stress triggered by tunicamycin or the ionophore A23187 induces the expression of hUGGT1 but not hUGGT2. The biological function of hUGGT2 is unknown, but a chimera protein consisting of N-terminal domain of hUGGT1 and the C-terminal domain of hUGGT2 is active, suggesting that hUGGT2 has evolved to fulfill an alternative biological function [14].
UGGT substrate recognition in vitro and in vivo
UGGT is a unique protein that combines the specificity of a classic chaperone with the activity of a glycosyltransferase. Early observations demonstrated that UGGT was highly active towards misfolded glycoproteins [15]. UGGT can also recognize high mannose glycopeptides, provided that they are hydrophobic and long enough (at least approximately 12 residues) [16]. A problem when studying the specificity of UGGT is that high mannose glycoprotein folding intermediates (UGGT natural substrates) are transient species within the ER, thus making extremely difficult the preparation of sufficient amounts of substrates for in vitro studies. In addition, since those species are partially misfolded, their high tendency to aggregate hinders a correct interpretation of kinetic data. By using well characterized neoglycoproteins derived from truncated versions of chymotrpsin inhibitor 2 (CI2) as folding intermediates, it was shown that UGGT recognition mirrors the anilinonaphtalene sulfonic acid (ANS) binding capacity of the substrates [17, 18]. ANS is a hydrophobic probe that binds to collapsed folding intermediates provided that they expose hydrophobic patches, while native proteins or highly disordered conformations do not bind the drug. These experiments showed that UGGT recognizes the degree of exposition of the hydrophobic core in folding intermediates, focusing its biological activity on advanced folding stages rather than on early non collapsed folding intermediates. Similar results have been reported with slightly destabilized mutants of RNAseB and β-glucanase [19, 20]. The preference of UGGT for advanced folding intermediates was also observed in vivo in T. cruzi [21] and CHO cells [22]. Presumably other chaperones such as BiP are better suited to deal with early folding intermediates. In this sense, the ER seems to be endowed with a battery of chaperones able to assist the complete folding process, starting from highly unstructured polypeptides after they enter the ER to more advanced molten globule-like intermediates before completing the folding process. Interestingly, the recognition of hydrophobic moieties goes beyond proteins, since UGGT is able to glucosylate high mannose oligosacharides bound to diverse synthetic aglycones [23, 24], and recognition improves as the hydrophobicity of the modifying non proteinaceous aglycone increases.
The activity of UGGT also depends on the structure of the acceptor oligosaccharide, as the rat liver enzyme showed the highest rate toward Man9GlcNAc2 and diminished to 50 % and 15 % with Man8GlaNAc2 and Man7GlcNAc2, respectively [25]. This selectivity apparently depends on the organism, since T. brucei UGGT recognizes a wide variety of N-glycans, ranging from Man9GlcNAc2 to Man5GlcNAc2 [26]. This last glycan is also recognized by the UGGT from Entamoeba histolytica and Trichomonas vaginalis [27]. An early study found that misfolded glycoprotein proteins inhibit UGGT activity provided that they have attached the innermost GlcNAc unit (but not the rest of the glycan), suggesting that this residue is involved in recognition. For instance rat liver UGGT activity is inhibited by denatured RNAseB treated with endo-β-N-acetylglucosaminidase H, an enzyme that leaves the innermost GlcNAc residue but not by denatured RNAseA that, although having the same amino acid sequence as RNAseB, lacks covalently bound sugars [28]. Although this observation was challenged by a latter report that found inhibitory activity by a scrambled version of RNAseA [19], it was recently shown that an oligosaccharide lacking the innermost GlcNAc was not recognized by UGGT [24]. In addition, this last report also found that UGGT has a strong affinity for the core pentasaccharide Man3GlcNAc2. Recognition of the inner GlcNAc residue by UGGT may be an additional element supporting the high preference of the enzyme for glycoproteins not displaying their native structures, as that residue, contrary to what happens with the other constituent monosaccharides, highly interacts with neighboring amino acids exclusively in native conformers.
QC not only supervises the completion of tertiary structures, but also ensures the proper assembly of oligomeric proteins. For instance, UGGT-mediated reglucosylation of the subunits of the T-cell receptor persists until the final assembly of the oligomer [29]. Recent experiments carried out by Dan Hebert and co-workers using influenza HA nascent chains expressed in CHO MI8-5 cells showed that UGGT preferentially targets the slow folding membrane-proximal stem domain [22]. These cells transfer oligosaccharides devoid of glucose residues and, similarly to T. cruzi, the presence of monoglucosylated glycans is solely due to the activity of UGGT. In this system substrate recognition takes place preferentially after their dissociation from the ribosome, presumably because in this particular protein the motifs recognized by UGGT are hindered in the vicinity of the translocon. On the other hand, UGGT recognition was abolished upon complete reduction of HA, in line with previous observations suggesting that highly misfolded proteins are poor substrates of UGGT. Regarding HA quaternary structure, it is likely that UGGT recognizes a hydrophobic patch that becomes hindered upon HA trimer assembly. This observation agrees with in vitro studies showing that UGGT recognizes incompletely assembled complexes formed by properly folded subunits of soybean agglutinin (normally a homotetramer) [30]. Here, the sensing mechanism is similar to that observed with the CI2-derived family, since monomeric soybean agglutinin exposes a hydrophobic surface that binds ANS and becomes occluded upon oligomer assembly. In such scenario, UGGT would be impaired to sense the oligomerization state of complexes linked through hydrophilic interfaces.
There have been conflicting reports regarding the minimum distance between the glucose acceptor site and the position of the hydrophobic surface that triggers UGGT recognition. Dimers made by combining different forms of RNAseA and RNAseB are recognized only when the glycan is bound to a misfolded subunit, suggesting that UGGT recognition is local [19]. By contrast, studies performed with a slightly destabilized mutant β-glucanase showed that UGGT can recognize glycans located distal from the structural perturbation [20]. On the other hand, studies using BODIPY-derivatized oligosaccharides found that UGGT recognition diminishes as the polyethyleneglycol linker length increases [23]. At first sight this result suggests that the distance between the glucose acceptor site and the hydrophobic patch must be short, but it is possible that substrates displaying long linkers have a higher entropic cost to accommodate into the substrate binding site, thus explaining the decreased UGGT activity.
Although UGGT is predicted to recognize in vivo a wide range of substrates [31], studies performed so far have been restricted to a narrow set of glycoproteins, most of then expressed exogenously. The first endogenous UGGT substrate described was cruzipain (CZ), an abundant lysosomal protease from T. cruzi that displays three potential N-glycosylation sites, at least two of which are occupied [32]. Interestingly, the transit of CZ to the lysosome was delayed in the presence of GII inhibitors. In most organisms, GII inhibition precludes the association of glycoproteins with CNX and CRT as glycan processing is stopped at the diglucosylated stage, and in the absence of other retention mechanism they leave the ER faster. On the contrary, since T. cruzi transfers unglucosylated oligosaccharides from the lipid derivative, the only pathway to generate monoglucosylated glycans is through UGGT activity. As expected, in this protozoan GII inhibition leads to a more lasting association of glycoproteins with CRT [21]. Interestingly, by using non-reducing gels it was determined that CZ association with CRT takes places at advanced stages during its folding pathway, when many of its disulfide bridges have been formed [21]. This observation agrees with the in vitro experiments previously described, in which a higher UGGT recognition was observed for collapsed folding intermediates rather than for extended, random coil conformations. The ability of UGGT to recognize minor folding defects was nicely illustrated in Arabidopsis thaliana. The UGGT gene was isolated using a complementation genetic screen aiming to restore the normal growth of a plant expressing a mutant brassinosteroid receptor (bri1-9) [33]. The phenotype due to the bri1-9 mutation arises from receptor retention in the ER by CNX. Interestingly, upon disruption of the UGGT encoding gene the mutant receptor reached the plasma membrane in a functional form, thus showing that the structural perturbation that triggered UGGT recognition did not involve the receptor activity. This observation suggests that some diseases resulting in the ER retention of defective glycoproteins could be ameliorated by inhibitors of UGGT activity.
UGGT biological relevance
In unicellular organisms UGGT is not essential for viability under physiological conditions, but in general growth is impaired under ER stress conditions. In some cases it has been observed that UGGT deletion triggers the unfolded protein response, thus compensating for the lack of the folding sensor. UGGT minus S. pombe mutants grow normally but cells are approximately 30 % shorter than wild type ones [6]. However cell growth at 37 °C is inhibited when the alg6 gene, in addition to that coding for UGGT, is also eliminated (the alg6 gene codes for the enzyme that transfers glucose from Glc-P-dolichol to Man9GlcNAc2–P-P-dolichol). The OST of cells normally transferring the glycan depicted in Fig. 1 requires the presence of the full complement of glucose units to catalyze an efficient N-glycosylation reaction. As disruption of the alg6 gene results in the transfer of Man9GlcNAc2, the presence of UGGT is apparently required for overcoming the excessive ER stress caused by high temperature and protein underglycosylation [34]. UGGT knock out in a protozoon as T. cruzi results in a reduced infectivity and in an increased BiP level [11]. UGGT knock out In A. thaliana does not lead to an obvious phenotype, but several ER chaperones and folding assisting enzymes such as BiP, PDI, CNX and CRT are upregulated [33]. By contrast, UGGT knock out is embryonically lethal in mouse but MEF cells derived from this mouse are viable [35].
Chemical crosslinking experiments revealed that UGGT can associate with some ER chaperone and folding assisting enzymes such as BiP, Grp94 and PDI [36]. Interestingly, neither CRT nor CNX were found in those complexes. This observation suggests that UGGT may connect the folding systems based on classical chaperones to that based on glycans. Interestingly, it has been found that Sep15, a thioredoxin-like selenoprotein, forms a tight complex with UGGT through its N-terminal cysteine rich domain [37]. Sep15 lacks an ER retention signal, and its cellular localization is achieved through its association with UGGT. Sep15 is transcriptionally upregulated by treatments that trigger an adaptative stress response, such as tunicamycin and brefeldin A, while it is degraded by the proteasome upon sharper treatments such as DTT or thapsigargin [38]. Contrary to what happens upon elimination of UGGT, Sep15 knock down does not trigger the UPR, suggesting that its activity may be focused on a restricted set of glycoproteins. These observations open the possibility that UGGT activity could be regulated by associated proteins, similarly to the situation found with other chaperones and lectins of the ER such as BiP and CRT/CNX.
New findings concerning UGGT and open issues
As mentioned above, although UGGT knock out is lethal in mice, MEF cells derived from their embryos are viable. When studying the association of glycoproteins to CNX in this cell line the following intriguing findings were observed [39]. According to the classical view, monoglucosylated proteins can be formed by two alternative pathways: partial deglucosylation of the transferred glycan and/or UGGT activity. In the case of glycoproteins associated to the lectins by monoglucosylated formed exclusively by the former pathway, UGGT deletion is not expected to modify the kinetics of glycoprotein association with CNX, whereas in those associated additionally through UGGT activity, a faster dissociation is predicted. Indeed examples for both behaviors were observed. Proteins whose association time with CNX was unaffected by UGGT knock out (type I, as VSV G protein) are assumed to complete their folding process in a single round of association with CNX without involvement of UGGT. On the other hand, a second subset of substrates (type II as BACE501), dissociate faster from CNX in UGGT knock out cells. Binding of type II glycoproteins with CNX is mediated at least in part by UGGT. Highly striking was the detection of some glycoproteins (type III) whose association with CNX was prolonged upon UGGT deletion. One example of this type of proteins is influenza HA. One possible explanation for this counterintuitive observation would be that UGGT could play an active role during the folding of these proteins, going beyond its function as folding sensor. The folding maturation of type III proteins would be facilitated directly by UGGT or, alternatively, by an UGGT-associated protein. For instance Sep15, which may display protein disulfide isomerase activity, might facilitate the disulfide formation of certain substrates associated with UGGT, similarly as ERp57 does with glycoprotein-CNX/CRT complexes. In proteins analyzed the rate of reglucosylation seems to be rather slow. This parameter could vary depending on the particular substrate, since for example the T-cell receptor can undergo several cycles of deglucosylation-reglucosylation in a relatively short period of time [40]. Clearly more examples are needed to define this issue. Finally, even though it is clear that UGGT preferentially recognizes hydrophobic patches exposed in collapsed folding intermediates, the mechanism behind this exceptional specificity is so far unknown.
The heterodimeric nature of GII
As mentioned above, GII is the opposite UGGT partner. At first assumed to be a simple glycosidase, recent work has revealed it to be endowed with unsuspected features. GII is a soluble ER resident heterodimer composed of two tightly but noncovalently bound chains (GIIα and GIIβ) as first described by A. Helenius and co-workers upon purification of the rat liver enzyme [41]. A 100-110 KDa polypeptide chain (GIIα), reported to be the catalytic subunit, could not be separated from a smaller polypeptide (GIIβ) using conditions short of denaturation. The heterodimeric nature of GII was confirmed using a genetic approach and the fission yeast S. pombe as model system (this microorganism displays a quality control mechanism similar to that occurring in mammalian cells) [42]. Microsomes prepared not only from mutant cells lacking GIIα but also from those devoid of GIIβ were completely devoid of GII activity when assayed using Glc1Man9GlcNAc2 (G1M9) as substrate. Slightly different results were obtained in vivo: although cells lacking GIIα only produced Glc2Man9GlcNAc2 (G2M9), low but detectable amounts of G1M9 were formed in those lacking GIIβ, thus confirming the catalytic role of GIIα. Moreover, mutants devoid of either one of the subunits displayed the so called unfolded protein response, as shown by the induction of BiP-encoding mRNA, thus suggesting the ER accumulation of misfolded glycoproteins in both subunit knock out strains.
GIIα is a 95-110 kDa protein conserved in yeast, mammals, parasites and plants that contains the consensus sequence (G/F)(L/I/V/M)WXDMNE) of the active site of family 31 glycosylhydrolases [41-47]. This subunit lacks a consensus ER retention/retrieval sequence at its C-terminus in all species studied so far. Two GIIα cDNAs are expressed in humans and mice as splicing variants differing in a 66 bp stretch, rendering catalytically active isoforms [48-50].
GII substrate specificity
GII activity has been mainly assayed using either the small artificial substrate analogue p-nitro phenyl-α-D-glucopyranoside (pNPG) in which the absorbance of p-nitrophenol in alkaline medium is measured or N-glycans (G2M9 or G1M9 or derivatives) in which either the liberated glucose units or the glycans found after the biochemical reaction are quantified using different procedures [41, 51-53]. Although similar results were obtained with both methods when dealing with the dimeric enzyme, those yielded when assaying the isolated GIIα subunit were strikingly different. As will be discussed below, it was this discrepancy that prompted the study of the role of GIIβ in N-glycan deglucosylation by GIIα [54, 55].
GII has an almost neutral optimum pH value, no cation requirement and is inhibited by 1-deoxynojirimycin, castanospermine and bromoconduritol [56]. It has been proposed that GII first- and second-mediated cleavages have different kinetics, being faster for the formation of G1M9 than for that of Man9GlcNAc (M9) [56-58]. It has been speculated that the differential hydrolysis rates allow recognition of the monoglucosylated glycoprotein folding intermediates by CNX and CRT. More recent work suggests, however, that the differential trimming rates of both Glc units may be not be operative at the high protein concentrations occurring within the ER lumen [59].
It was initially determined that the activity of GII toward high mannose glycans decreased dramatically as mannose units were removed from the B or C arms (Fig. 1) [60]. A more recent work using metotrexate (MTX)-conjugated N-glycans showed that the deglucosylation rate by rat liver GII of G1M8B-MTX (lacking the terminal mannose of arm B, residue i in Fig. 1) was almost identical to that of G1M9-MTX whereas the activity toward G1M8C-MTX (lacking the terminal mannose of arm C, residue k in Fig. 1) was markedly lower, being almost identical to that of Glc1Man7GlcNAc (G1M7)-MTX (G1M7 stands for the glycan lacking residues i, k, m and n, Fig.1). This result suggested that the outermost mannose of the C arm (residue k in Fig. 1) is involved in substrate recognition [53]. In addition, it was shown that GII is inhibited by its end products [53, 61]. Similar inhibitory capacities by M8B and M9 toward G2M9-MTX trimming were observed but they were significantly higher than that of M8C. Surprisingly, Man7GlcNAc (M7, lacking residues i, k and l-n, Fig. 1) behaved as a much better inhibitor than M8C although G1M7 is a slightly poorer GII substrate than G1M8C. This result lead to the speculation that the accumulation of glycans with a low mannose content may regulate the entry of glycoproteins to CRT/CNX cycles by preventing the formation of monoglucosylated glycoproteins.
GIIβ involvement in GIIα maturation and stability
GIIβ is a 50-60 kDa polypeptide chain that bears the signal peptide required to deliver proteins into the ER and the canonical ER retention/retrieval sequence XDEL at its C-terminus [41, 42]. It also contains one or two (depending on the species) EF hand Ca2+ binding domains, a glutamic acid rich motif and a domain homologous to the Man 6-P receptor (MRH) responsible for delivering lysosomal enzymes to their final destination [41, 62, 63]. The roles of GIIβ have been object of growing interest in the last years, as autosomal dominant polycystic liver disease may develop in individuals carrying mutations in the GIIβ gene [64-66]. GIIβ (but not GIIα) is induced in differentiating rat neural progenitor cells in response to the glial cell derived neurotrophic factor (GDNF) [67]. Concerning its involvement in QC, GIIβ has been suggested to be responsible for GIIα maturation and stability, for ER localization and for enhancing N-glycan processing rates [42-49, 54, 55, 68-70]. These roles are discussed in this and following sections.
Co-expression of the human GIIα and GIIβ subunits in COS-1 cells resulted in more than a fourfold increase of GII activity toward G1M9. However, no activity increase was observed upon transfection with only the GIIα subunit encoding gene, indicating that both subunits were necessary for either GIIα folding, solubilization, and/or stability [48, 49]. Later work by Trombetta et al. [71] showed that rat liver GIIα and GIIβ formed a defined complex displaying a highly non-globular shape, from which GIIβ subunit could been specifically proteolyzed under restricted conditions. The resulting GIIα obtained was active toward pNPG, indicating that GIIβ was not required for the catalytic activity once the heterodimer had been formed.
However, the presence of GIIβ for GIIα folding, solubilization and/or stability does not appear to be a universal feature as we have recently shown that microsomes derived from S. pombe cells lacking GIIβ displayed a sizable activity toward pNPG, although they were almost completely inactive when N-glycans were used as substrates. In agreement with this last result, trimming of G2M9 and G1M9 was severely delayed in vivo in those mutants [54]. A rat liver GII purified preparation from which GIIβ had been removed by chymotrypsin treatment retained activity toward pNPG, as had been demonstrated by Trombetta et al. [71] but was inactive toward N-glycans, thus confirming that GIIβ subunit was required for physiological substrate trimming by GIIα but not for that of pNPG [54]. An approach similar to that described above for S. pombe but using microsomes derived from Aspergillus oryzae lead Watanabe et al. [55] to the same conclusions. In addition, Wilkinson et al. [69] showed that S. cerevisiae GIIα subunit was similarly active and stable when expressed in the presence or absence of GIIβ. It may be speculated that perhaps in previous work mentioned above expression of an active GIIα alone had apparently failed not because its folding or stability required the presence of GIIβ but because this last subunit is required for N-glycan hydrolysis but not for that of pNPG. However, in a recent work, a 2-fold overexpression of human GIIα in 293T cells did not significantly increase hydrolysis of pNPG indicating that the expression of an active mammalian GIIα may require the presence of GIIβ [72].
In summary, recent evidence leads to the conclusion that at least in fungi GIIβ is not involved in GIIα folding, maturation, stability or activity toward pNPG but that it is certainly involved in N-glycan trimming. This last feature is also shared by mammalian GIIβ.
GIIβ participation in GIIα localization
GIIα has been shown to be present predominantly in the ER: the enzyme was concentrated in the rough and smooth ER but was not detectable in Golgi cisternae, although transitional elements of ER close to the Golgi were positive for the enzyme [73]. The localization correlated well with that of UGGT, CRT, and pre-Golgi intermediate markers in ultra thin cryosections of D. melanogaster salivary gland, rat pancreas and liver cell lines [10]. The localization mechanism of GIIα lacking any known retention/retrieval sequence for soluble ER-resident proteins remained intriguing until GIIβ was shown to be intimately linked to the catalytic subunit. As the former protein displayed indeed a retention/retrieval sequence it was proposed GIIβ to be responsible for the heterodimer ER localization [41, 42]. In agreement with this proposal, Pelletier et al. [49] showed that higher amounts of GIIα were secreted when transfection of COS7 cells with GIIα- and GIIβ- encoding genes was replaced by a similar transfection in which the GIIβ gene displayed the code for a (His)6 tag instead of the HDEL retention/retrieval sequence. Addition of S. pombe GIIβ retention/retrieval sequence (VDEL) to GIIα C-terminus expressed in mutant cells lacking both subunits improved the ER retention of the catalytic one to levels similar to those of wild type cells [54]. However, a normal ER localization of GIIβ occurred in cells in which the VDEL sequence was occluded by YFP [54]. Moreover, S. pombe cells lacking GIIβ had an ER GIIα content that varied between 20-50 % of that found in wild type cells, thus suggesting that the GIIα subunit itself may bear another yet unknown ER localization signal [54]. In addition, the S. cerevisiae GIIβ localized to the ER although not displaying any known retention/retrieval sequence, it physically interacted with GIIα as revealed by co-immunoprecipitation and, furthermore, disruption of the GIIβ encoding gene did not affect GIIα ER localization [69]. Moreover, it was shown that 50% of a 1338-2A-G mutated human GIIβ (named hepatocystin or protein kinase C substrate 80K-H) that produces a truncated protein lacking the HDEL retention signal in a patient with autosomal dominant polycystic liver disease was partially (50 %) retained in the ER of transfected HeLa cells [63]. The truncated protein also failed to assemble with the GIIα subunit, probably because it lacked the interacting sequence.
It may be concluded, therefore, that GIIβ and its ER retention/retrieval sequence at its C-terminus certainly play a role in GIIα ER localization, but not an absolute one as other retention mechanisms seem to be operative.
Involvement of GIIβ MRH domain in GIIα-mediated glycan trimming
A new role proposed for GIIβ subunit in the QC of glycoprotein folding in the ER is that of being a lectin enhancing GIIα-mediated N-glycan trimming. As mentioned above, GIIβ displays at its C-terminus a domain (MRH) homologous to that responsible for Man 6-P binding in the receptor driving lysosomal enzymes to their final destination. Several lines of evidence indicate that the interaction of the MRH domain with mannoses in B and/or C arms participates in the enhancement of hydrolysis rate by GIIα mentioned above. Work performed with S. pombe showed that mutations in amino acids conserved in several MRH domain-containing proteins and known to be involved in the interaction of the receptor with mannose units in lysosomal glycoprotein enzymes sharply decreased the GIIβ enhancing capability of G2M9 and G1M9 hydrolysis by the catalytic subunit both in vivo and in vitro [54, 74, 75]. In addition, removal of mannose units from B and C arms of the N-glycan drastically decreased N-glycan trimming rates by S. pombe GII in cell free assays. Using frontal affinity chromatography Hu et al. [72] demonstrated a direct binding of a phycoerythrin-labeled GIIβ MRH domain tetramer to synthetic high mannose type glycans. Binding was lost when MRH residues involved in mannose recognition were mutated. Comparison of sugar–binding activity of GIIβ-MRH to a set of mono and unglucosylated glycans showed that the affinity of G1M9 or M9 diminished when the terminal mannose residues on either the B or C arms (residues i or k, Fig. 1) were trimmed but residue k showed a higher relevance for binding. GIIβ MRH domain-glycan binding appeared to be cation independent. However, as GIIβ EF hand Ca2+ binding domain was absent from the MRH domain tetramer a possible cation influence on sugar binding affinity cannot be discarded.
The interaction between GIIα and GIIβ subunits was not affected by mutations in the MRH domain as GIIα co-immunoprecipitated with MRH-mutated GIIβ in S. cerevisiae cells [70]. In addition, GIIα in microsomes, as measured by pNPG hydrolysis, increased to wild type cell levels when MRH-mutated GIIβ was expressed in GIIβ null cells [54]. It should be mentioned that the surface mediating the interaction between both subunits maps to GIIβ N-terminus [76]. Surprisingly, a point mutation in a conserved N-terminal domain of S. cerevisiae GIIβ resulted in a reduced G2M9 trimming, suggesting that this region may be also important for glucose trimming [70].
At least two mechanisms may be envisaged for the MRH-mediated enhancement of N-glycan deglucosylation rate (Fig. 3). In the first one the MRH domain, upon binding mannose units in the B and/or C arms of the glycan, presents bonds to be cleaved to the catalytic site in GIIα. This possibility, however, is at odds with the known 3-D structure of Glc2Man9GlcNAc2. As determined by NMR, the bond to be cleaved first (Glcα1,3Glc epitope) is exposed to the external face of the A arm (residues d, f and g, Fig. 1), whereas the second bond (Glcα1,3Man epitope) is on the internal side (that is, facing the B and C arms, residues e and h-k in Fig. 1) [77]. The need to reorient the substrate makes the static mechanism suggested above highly improbable as both bonds to be cleaved lie far apart in space, and cannot be conceivably be reached by the single GIIα catalytic site without such reorientation. However, this mechanism cannot be ruled out altogether as the known flexibility of mannoses in arm A (Fig. 1) may allow the successive cleavage of both glucoses in the same glycan [78]. The second mechanism was initially proposed by A. Helenius and co-workers to explain the apparent need of two glycans in the same glycoprotein to efficiently generate monoglucosylated glycans [68]. It was reported several years ago that in most cases only glycoproteins having more than one N-glycan interacted with CNR/CRT. It was then assumed that the lectins were either monomeric with two binding sites per monomer or, alternatively, at least homodimeric complexes with one binding site per monomer. Further characterization of the lectin structure and binding features proved this assumption to be wrong as both CNX and CRT behaved as monomers with a single binding site per monomer. The explanation of the observation, instead, lies in an interesting property of the mammalian cell GII: two glycans in the same glycoprotein molecule are apparently required for removing the middle glucose unit (residue m, Fig. 1). It was then proposed that GII has a basal glucosidase activity (that responsible for pNPG hydrolysis in the absence of GIIβ) but that interaction of the B and/or C arms of an N-glycan (glycan 1) with the MRH domain would induce a conformational change in the catalytic subunit, thus increasing the enzymatic activity and allowing the first cleavage to proceed in a neighboring N-glycan (glycan 2). On the other hand, two glycans would not be required for the second cleavage to proceed because interaction of the B/C arms of glycan 2 with the MRH site would allow the second cleavage to occur as those arms and the Glcα1,3Man epitope lie on the same face. Nevertheless, there are exceptions and cases in which glycoproteins bearing a single N-glycan interact with CNX/CRT are known. These may be due to transactivation of the first cleavage by an N-glycan in different glycoprotein molecules in the crowded ER environment or, for glycoproteins that have a rather long folding process, to GII basal activity. The mechanism proposed allows glycoproteins to enter the CNX/CRT cycle before total deglucosylation, that is, independently from UGGT activity.
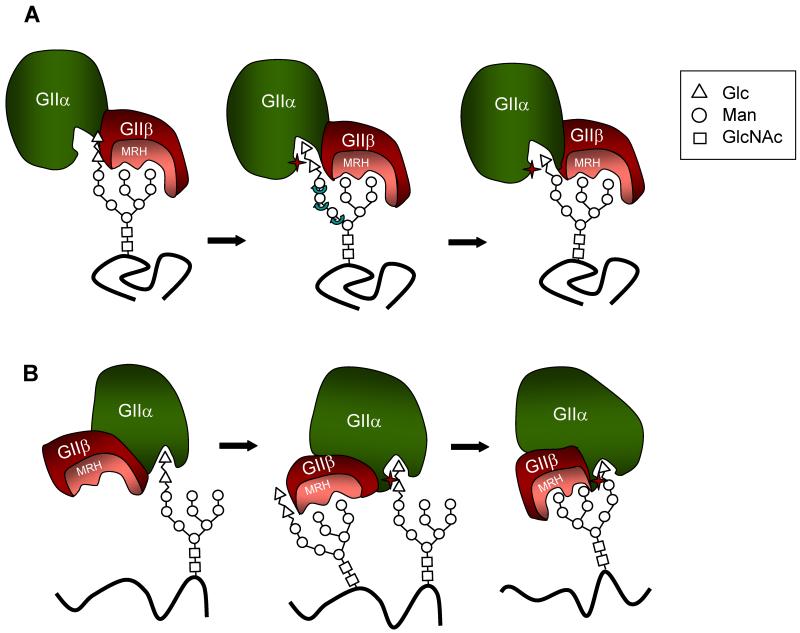
Fig. 3.
Models proposed for GIIβ-mediated enhancement of N-glycan deglucosylation. (A) Upon binding mannose units in the B and/or C arms of the glycan, the GIIβ MRH domain presents bonds to be cleaved to the GIIα catalytic site (star). Please note that in this model a rotation of A arm is required for both cleavages to proceed. (B) Binding of a glycan to the GIIβ MRH domain would result in a conformational change in their GIIβ subunit, which in turn would modify GIIα structure thus activating the catalytic site and allowing hydrolysis of the middle Glc residue in another glycan. Cleavage of the inner Glc in this last glycan would be enhanced by the interaction of Man units in its own B and/or C arms with the MRH domain as the bond to be cleaved and the arms lie on the same face.
GIIβ is involved in yet poorly understood cellular processes
Although most of the efforts have been directed to understanding the role of GIIβ in GIIα maturation and stabilization, intracellular localization and N-glycan recognition, there are still mechanisms involving GIIβ that remain to be fully understood. For instance, why is GIIβ induced in rat progenitor cell lines in response to the expression of the GDNF gene and why GIIβ expression regulation does not correlate with that of GIIα [67] The finding that GIIβ is able to interact with the 3′ UTR of the R1-subunit mRNA of the NMDA receptor in mouse fetal cortical neurons and is significantly up-regulated in the presence of ethanol [79] indicates that this protein may act by influencing not only the glycosylation process within the ER but also as a regulator of distinct developmental processes.
Acknowledgements
This review is dedicated to the memory of Rodolfo A. Ugalde, a brilliant scientist and a dear friend. It was Rodolfo who provided the first evidence that two different specific glucosidases were involved in N-glycan processing (ref. 51). Financial support of the Howard Hughes Medical Institute and of the National Institutes of Health (Grant GM044500) for work performed by the authors is gratefully acknowledged.
Abbreviations
- ANS
anilinonaphtalene sulfonic acid
- BiP
binding protein
- CNX
calnexin
- CRT
calreticulin
- CZ
cruzipain
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERGIC
ER-Golgi intermediate compartment
- GI
glucosidase I
- GII
glucosidase II
- HA
hemagglutinin
- PDI
protein disulfide isomerase
- pNPG
p-nitro phenyl-α-D-glucopyranoside
- QC
quality control
- UGGT
UDP-Glc:glycoprotein glucosyltransferase
- VSV
vesicular stomatitis virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Parodi AJ, Cazzulo JJ. Protein glycosylation in Trypanosoma cruzi. II. Partial characterization of protein-bound oligosaccharides labeled “in vivo”. J Biol Chem. 1982;257:7641–5. [PubMed] [Google Scholar]
- [2].Trombetta SE, Parodi AJ. Purification to apparent homogeneity and partial characterization of rat liver UDP-glucose:glycoprotein glucosyltransferase. J Biol Chem. 1992;267:9236–40. [PubMed] [Google Scholar]
- [3].Ware FE, Vassilakos A, Peterson PA, Jackson MR, Lehrman MA, Williams DB. The molecular chaperone calnexin binds Glc1Man9GlcNAc2 oligosaccharide as an initial step in recognizing unfolded glycoproteins. J Biol Chem. 1995;270:4697–704. doi: 10.1074/jbc.270.9.4697. [DOI] [PubMed] [Google Scholar]
- [4].Hammond C, Braakman I, Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci USA. 1994;91:913–7. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Babour A, Beckerich JM, Gaillardin C. Identification of an UDP-Glc:glycoprotein glucosyltransferase in the yeast Yarrowia lipolytica. Yeast. 2004;21:11–24. doi: 10.1002/yea.1051. [DOI] [PubMed] [Google Scholar]
- [6].Fernandez F, Jannatipour M, Hellman U, Rokeach LA, Parodi AJ. A new stress protein: synthesis of Schizosaccharomyces pombe UDP-GIc:glycoprotein glucosyltransferase mRNA is induced by stress conditions but the enzyme is not essential for cell viability. EMBO J. 1996;15:705–13. [PMC free article] [PubMed] [Google Scholar]
- [7].Parker CG, Fessler LI, Nelson RE, Fessler JH. Drosophila UDP-glucose:glycoprotein glucosyltransferase: sequence and characterization of an enzyme that distinguishes between denatured and native proteins. EMBO J. 1995;14:1294–303. doi: 10.1002/j.1460-2075.1995.tb07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tessier DC, Dignard D, Zapun A, Radominska-Pandya A, Parodi AJ, Bergeron JJ, Thomas DY. Cloning and characterization of mammalian UDP-Glc:glycoprotein glucosyltransferase and the development of a specific substrate for this enzyme. Glycobiology. 2000;10:403–12. doi: 10.1093/glycob/10.4.403. [DOI] [PubMed] [Google Scholar]
- [9].Arnold SM, Fessler LI, Fessler JH, Kaufman RJ. Two homologues encoding human UDP-glucose:glycoprotein glucosyltransferase differ in mRNA expression and enzymatic activity. Biochemistry. 2000;39:2149–63. doi: 10.1021/bi9916473. [DOI] [PubMed] [Google Scholar]
- [10].Zuber C, Fan J, Guhl B, Parodi A, Fessler JH, Parker C, Roth J. Immunolocalization of UDP-glucose:glycoprotein glucosyltransferase indicates involvement of pre-Golgi intermediates in protein quality control. Proc Natl Acad Sci USA. 2001;98:10710–5. doi: 10.1073/pnas.191359198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Conte I, Labriola C, Cazzulo JJ, Docampo R, Parodi AJ. The interplay between folding-facilitating mechanisms in Trypanosoma cruzi endoplasmic reticulum. Mol Biol Cell. 2003;14:3529–40. doi: 10.1091/mbc.E03-04-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Banerjee S, Vishwanath P, Cui J, Kelleher DJ, Gilmore R, Robbins PW, Samuelson J. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci USA. 2007;104:11676–81. doi: 10.1073/pnas.0704862104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guerin M, Parodi AJ. The UDP-glucose:glycoprotein glucosyltransferase is organized in at least two tightly bound domains from yeast to mammals. J Biol Chem. 2003;278:20540–6. doi: 10.1074/jbc.M300891200. [DOI] [PubMed] [Google Scholar]
- [14].Arnold SM, Kaufman RJ. The noncatalytic portion of human UDP-glucose:glycoprotein glucosyltransferase I confers UDP-glucose binding and transferase function to the catalytic domain. J Biol Chem. 2003;278:43320–8. doi: 10.1074/jbc.M305800200. [DOI] [PubMed] [Google Scholar]
- [15].Trombetta SE, Bosch M, Parodi AJ. Glucosylation of glycoproteins by mammalian, plant, fungal, and trypanosomatid protozoa microsomal membranes. Biochemistry. 1989;28:8108–16. doi: 10.1021/bi00446a022. [DOI] [PubMed] [Google Scholar]
- [16].Taylor SC, Thibault P, Tessier DC, Bergeron JJM, Thomas DY. Glycopeptide specificity of the secretory protein folding sensor UDP-glucose glycoprotein:glucosyltransferase. EMBO Rep. 2003;4:405–11. doi: 10.1038/sj.embor.embor797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Caramelo JJ, Castro OA, Alonso LG, de Prat-Gay G, Parodi AJ. UDP-Glc:glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates. Proc Natl Acad Sci USA. 2003;100:86–91. doi: 10.1073/pnas.262661199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Caramelo JJ, Castro OA, de Prat-Gay G, Parodi AJ. The endoplasmic reticulum glucosyltransferase recognizes nearly native glycoprotein folding intermediates. J Biol Chem. 2004;279:46280–5. doi: 10.1074/jbc.M408404200. [DOI] [PubMed] [Google Scholar]
- [19].Ritter C, Quirin K, Kowarik M, Helenius A. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 2005;24:1730–8. doi: 10.1038/sj.emboj.7600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Taylor SC, Ferguson AD, Bergeron JJM, Thomas DY. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat Struct Mol Biol. 2004;11:128–34. doi: 10.1038/nsmb715. [DOI] [PubMed] [Google Scholar]
- [21].Labriola C, Cazzulo JJ, Parodi AJ. Trypanosoma cruzi calreticulin is a lectin that binds monoglucosylated oligosaccharides but not protein moieties of glycoproteins. Mol Biol Cell. 1999;10:1381–94. doi: 10.1091/mbc.10.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pearse BR, Gabriel L, Wang N, Hebert DH. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J Cell Biol. 2008;181:309–20. doi: 10.1083/jcb.200712068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Totani K, Ihara Y, Tsujimoto T, Matsuo I, Ito Y. The recognition motif of the glycoprotein-folding sensor enzyme UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 2009;48:2933–40. doi: 10.1021/bi8020586. [DOI] [PubMed] [Google Scholar]
- [24].Totani K, Ihara Y, Matsuo I, Koshino H, Ito Y. Synthetic substrates for an endoplasmic reticulum protein-folding sensor, UDP-glucose:glycoprotein glucosyltransferase. Angew Chem Int Ed. 2005;44:7950–4. doi: 10.1002/anie.200502723. [DOI] [PubMed] [Google Scholar]
- [25].Sousa MC, Ferrero-Garcia MA, Parodi AJ. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- [26].Izquierdo L, Atrih A, Rodrigues JA, Jones DC, Ferguson MAJ. Trypanosoma brucei UDP-glucose:glycoprotein glucosyltransferase has unusual substrate specificity and protects the parasite from stress. Eukar Cell. 2009;8:230–40. doi: 10.1128/EC.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Magnelli P, Cipollo JF, Ratner DM, Cui J, Kelleher D, Gilmore R, Costello CE, Robbins PW, Samuelson J. Unique Asn-linked oligosaccharides of the human pathogen Entamoeba histolytica. J Biol Chem. 2008;283:18355–64. doi: 10.1074/jbc.M800725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sousa M, Parodi AJ. The molecular basis for the recognition of misfolded glycoproteins by the UDP-GIc:glycoprotein glucosyltransferase. EMBO J. 1885;14:4196–203. doi: 10.1002/j.1460-2075.1995.tb00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gardner TG, Kearse KP. Modification of the T cell antigen receptor (TCR) complex by UDP-glucose:glycoprotein glucosyltransferase. TCR folding is finalized covergent with formation of αβδεγε complexes. J Biol Chem. 1999;274:14094–9. doi: 10.1074/jbc.274.20.14094. [DOI] [PubMed] [Google Scholar]
- [30].Keith N, Parodi AJ, Caramelo JJ. Glycoprotein tertiary and quaternary structures are monitored by the same quality control mechanism. J Biol Chem. 2005;280:18138–41. doi: 10.1074/jbc.M501710200. [DOI] [PubMed] [Google Scholar]
- [31].Gañán S, Cazzulo JJ, Parodi AJ. A major proportion of N-glycoproteins are transiently glucosylated in the endoplasmic reticulum. Biochemistry. 1991;30:3098–104. doi: 10.1021/bi00226a017. [DOI] [PubMed] [Google Scholar]
- [32].Labriola C, Cazzulo JJ, Parodi AJ. Retention of glucose units added by the UDP-Glc:glycoprotein glucosyltransferase delays exit of glycoproteins from the endoplasmic reticulum. J Cell Biol. 1995;130:771–9. doi: 10.1083/jcb.130.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26:821–30. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fanchiotti S, Fernandez F, D’Alessio C, Parodi AJ. The UDP-Glc:glycoprotein glucosyltransferase is essential for Schizosaccharomyces pombe viability under conditions of extreme endoplasmic reticulum stress. J Cell Biol. 1998;143:625–35. doi: 10.1083/jcb.143.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Molinari M, Galli C, Vanoni O, Arnold SM, Kaufman RJ. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Mol Cell. 2005;20:503–12. doi: 10.1016/j.molcel.2005.09.027. [DOI] [PubMed] [Google Scholar]
- [36].Meunier L, Usherwood Y, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–69. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Labunskyy VM, Ferguson AD, Fomenko DE, Chelliah Y, Hatfield DL, Gladyshev VN. A novel cysteine-rich domain of Sep15 mediates the interaction with UDP-glucose:glycoprotein glucosyltransferase. J Biol Chem. 2005;280:37839–45. doi: 10.1074/jbc.M508685200. [DOI] [PubMed] [Google Scholar]
- [38].Labunskyy VM, Yoo M, Hatfield DL, Gladyshev VN. Sep15, a thioredoxin-like selenoprotein, is involved in the unfolded protein response and differentially regulated by adaptive and acute ER stresses. Biochemistry. 2009;48:8458–65. doi: 10.1021/bi900717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Solda T, Galli C, Kaufman RJ, Molinari M. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol Cell. 2007;27:238–49. doi: 10.1016/j.molcel.2007.05.032. [DOI] [PubMed] [Google Scholar]
- [40].Van Leeuwen JEM, Kearse KP. Reglucosylation of N-Linked glycans is critical for calnexin assembly with T cell receptor (TCR) α proteins but not TCRβ proteins. J Biol Chem. 1997;272:4179–86. doi: 10.1074/jbc.272.7.4179. [DOI] [PubMed] [Google Scholar]
- [41].Trombetta ES, Simons JF, Helenius A. Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J Biol Chem. 271:27509–16. doi: 10.1074/jbc.271.44.27509. 199. [DOI] [PubMed] [Google Scholar]
- [42].D’Alessio C, Fernández F, Trombetta ES, Parodi AJ. Genetic evidence for the heterodimeric structure of glucosidase II. The effect of disrupting the subunit-encoding genes on glycoprotein folding. J Biol Chem. 1999;274:25899–905. doi: 10.1074/jbc.274.36.25899. [DOI] [PubMed] [Google Scholar]
- [43].Arendt CW, Ostergaard HL. Identification of the CD45-associated 116-kDa and 80-kDa proteins as the alpha- and beta-subunits of alpha-glucosidase II. J Biol Chem. 1997;272:13117–25. doi: 10.1074/jbc.272.20.13117. [DOI] [PubMed] [Google Scholar]
- [44].Jones DC, Mehlert A, Güther ML, Ferguson MA. Deletion of the glucosidase II gene in Trypanosoma brucei reveals novel N-glycosylation mechanisms in the biosynthesis of variant surface glycoprotein. J Biol Chem. 2005;280:35929–42. doi: 10.1074/jbc.M509130200. [DOI] [PubMed] [Google Scholar]
- [45].Soussilane P, D’Alessio C, Paccalet T, Fitchette AC, Parodi AJ, Williamson R, Plasson C, Faye L, Gomord V. N-glycan trimming by glucosidase II is essential for Arabidopsis development. Glycoconj J. 2009;26:597–607. doi: 10.1007/s10719-008-9201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Frandsen TP, Svensson B. Plant alpha-glucosidases of the glycoside hydrolase family 31. Molecular properties, substrate specificity, reaction mechanism, and comparison with family members of different origin. Plant Mol Biol. 1998;37:1–13. doi: 10.1023/a:1005925819741. [DOI] [PubMed] [Google Scholar]
- [47].Feng J, Romaniouk AV, Samal SK, Vijay IK. Processing enzyme glucosidase II: proposed catalytic residues and developmental regulation during the ontogeny of the mouse mammary gland. Glycobiology. 2004;14:909–21. doi: 10.1093/glycob/cwh110. [DOI] [PubMed] [Google Scholar]
- [48].Treml K, Meimaroglou D, Hentges A, Bause E. The alpha- and beta-subunits are required for expression of catalytic activity in the hetero-dimeric glucosidase II complex from human liver. Glycobiology. 2000;10:493–502. doi: 10.1093/glycob/10.5.493. [DOI] [PubMed] [Google Scholar]
- [49].Pelletier MF, Marcil A, Sevigny G, Jakob CA, Tessier DC, Chevet E, Menard R, Bergeron JJ, Thomas DY. The heterodimeric structure of glucosidase II is required for its activity, solubility, and localization in vivo. Glycobiology. 2000;10:815–27. doi: 10.1093/glycob/10.8.815. [DOI] [PubMed] [Google Scholar]
- [50].Arendt CW, Dawicki W, Ostergaard HL. Alternative splicing of transcripts encoding the alpha- and beta-subunits of mouse glucosidase II in T lymphocytes. Glycobiology. 1999;9:277–83. doi: 10.1093/glycob/9.3.277. [DOI] [PubMed] [Google Scholar]
- [51].Ugalde RA, Staneloni RJ, Leloir LF. Microsomal glucosidases acting on the saccharide moiety of the glucose-containing dolichyl diphosphate oligosaccharide. Biochem Biophys Res Commun. 1979;91:1174–81. doi: 10.1016/0006-291x(79)92003-5. [DOI] [PubMed] [Google Scholar]
- [52].Herscovics A, Jelinek-Kelly S. A rapid method for assay of glycosidases involved in glycoprotein biosynthesis. Anal Biochem. 1987;166:85–9. doi: 10.1016/0003-2697(87)90550-1. [DOI] [PubMed] [Google Scholar]
- [53].Totani K, Ihara Y, Matsuo I, Ito Y. Substrate specificity analysis of endoplasmic reticulum glucosidase II using synthetic high mannose-type glycans. J Biol Chem. 2006;281:31502–8. doi: 10.1074/jbc.M605457200. [DOI] [PubMed] [Google Scholar]
- [54].Stigliano ID, Caramelo JJ, Labriola CA, Parodi AJ, D’Alessio C. Glucosidase II beta subunit modulates N-glycan trimming in fission yeast and mammals. Mol Biol Cell. 2009;20:3974–84. doi: 10.1091/mbc.E09-04-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Watanabe T, Totani K, Matsuo I, Maruyama J, Kitamoto K, Ito Y. Genetic analysis of glucosidase II beta-subunit in trimming of high-mannose-type glycans. Glycobiology. 2009;19:834–40. doi: 10.1093/glycob/cwp061. [DOI] [PubMed] [Google Scholar]
- [56].Alonso JM, Santa-Cecilia A, Calvo P. Effect of bromoconduritol on glucosidase II from rat liver. A new kinetic model for the binding and hydrolysis of the substrate. Eur J Biochem. 1993;215:37–42. doi: 10.1111/j.1432-1033.1993.tb18004.x. [DOI] [PubMed] [Google Scholar]
- [57].Alonso JM, Santa-Cecilia A, Calvo P. Glucosidase II from rat liver microsomes. Kinetic model for binding and hydrolysis. Biochem J. 1991;278:721–7. doi: 10.1042/bj2780721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Cumpstey I, Butters TD, Tennant-Eyles RJ, Fairbanks AJ, France RR, Wormald MR. Synthesis of fluorescence-labelled disaccharide substrates of glucosidase II. Carbohydr Res. 2003;338:1937–49. doi: 10.1016/s0008-6215(03)00255-6. [DOI] [PubMed] [Google Scholar]
- [59].Totani K, Ihara Y, Matsuo I, Ito Y. Effects of macromolecular crowding on glycoprotein processing enzymes. J Am Chem Soc. 2008;130:2101–7. doi: 10.1021/ja077570k. [DOI] [PubMed] [Google Scholar]
- [60].Grinna LS, Robbins PW. Substrate specificities of rat liver microsomal glucosidases which process glycoproteins. J Biol Chem. 1980;255:2255–8. [PubMed] [Google Scholar]
- [61].Bosis E, Nachliel E, Cohen T, Takeda Y, Ito Y, Bar-Nun S, Gutman M. Endoplasmic reticulum glucosidase II is inhibited by its end products. Biochemistry. 2008;47:10970–80. doi: 10.1021/bi801545d. [DOI] [PubMed] [Google Scholar]
- [62].Munro S. The MRH domain suggests a shared ancestry for the mannose 6-phosphate receptors and other N-glycan-recognising proteins. Curr Biol. 2001;11:499–01. doi: 10.1016/s0960-9822(01)00302-5. 2001. [DOI] [PubMed] [Google Scholar]
- [63].Drenth JP, Martina JA, Te Morsche RH, Jansen JB, Bonifacino JS. Molecular characterization of hepatocystin, the protein that is defective in autosomal dominant polycystic liver disease. Gastroenterology. 2004;126:1819–27. doi: 10.1053/j.gastro.2004.02.023. [DOI] [PubMed] [Google Scholar]
- [64].Li A, Davila S, Furu L, Qian Q, Tian X, Kamath PS, King BF, Torres VE, Somlo S. Mutations in PRKCSH cause isolated autosomal dominant polycystic liver disease. Am J Hum Genet. 2003;72:691–3. doi: 10.1086/368295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Drenth JP, Martina JA, van de Kerkhof R, Bonifacino JS, Jansen JB. Polycystic liver disease is a disorder of cotranslational protein processing. Trends Mol Med. 2005;11:37–42. doi: 10.1016/j.molmed.2004.11.004. [DOI] [PubMed] [Google Scholar]
- [66].Drenth JP, Morsche RH, Smink R, Bonifacino JS, Jansen JB. Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat Genet. 2003;33:345–7. doi: 10.1038/ng1104. 2003. [DOI] [PubMed] [Google Scholar]
- [67].Hoffrogge R, Beyer S, Hübner R, Mikkat S, Mix E, Scharf C, et al. 2-DE profiling of GDNF overexpression-related proteome changes in differentiating ST14A rat progenitor cells. Proteomics. 2007;7:33–46. doi: 10.1002/pmic.200600614. [DOI] [PubMed] [Google Scholar]
- [68].Deprez P, Gautschi M, Helenius A. More than one glycan is needed for ER glucosidase II to allow entry of glycoproteins into the calnexin/calreticulin cycle. Mol Cell. 2005;19:183–95. doi: 10.1016/j.molcel.2005.05.029. [DOI] [PubMed] [Google Scholar]
- [69].Wilkinson BM, Purswani J, Stirling CJ. Yeast GTB1 encodes a subunit of glucosidase II required for glycoprotein processing in the endoplasmic reticulum. J Biol Chem. 2006;10:6325–33. doi: 10.1074/jbc.M510455200. 2006. [DOI] [PubMed] [Google Scholar]
- [70].Quinn RP, Mahoney SJ, Wilkinson BM, Thornton DJ, Stirling CJ. A novel role for Gtb1p in glucose trimming of N-linked glycans. Glycobiology. 2009;19:1408–16. doi: 10.1093/glycob/cwp087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Trombetta ES, Fleming KG, Helenius A. Quaternary and domain structure of glycoprotein processing glucosidase II. Biochemistry. 2001;40:10717–22. doi: 10.1021/bi010629u. 2001. [DOI] [PubMed] [Google Scholar]
- [72].Hu D, Kamiya Y, Totani K, Kamiya D, Kawasaki N, Yamaguchi D, Matsuo I, Matsumoto N, Ito Y, Kato K, Yamamoto K. Sugar-binding activity of the MRH domain in the ER alpha-glucosidase II beta subunit is important for efficient glucose trimming. Glycobiology. 2009;19:1127–35. doi: 10.1093/glycob/cwp104. [DOI] [PubMed] [Google Scholar]
- [73].Lucocq JM, Brada D, Roth J. Immunolocalization of the oligosaccharide trimming enzyme glucosidase II. J Cell Biol. 1986;102:2137–46. doi: 10.1083/jcb.102.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Olson LJ, Hancock MK, Dix D, Kim JJ, Dahms NM. Mutational analysis of the binding site residues of the bovine cation-dependent mannose 6-phosphate receptor. J Biol Chem. 1999;274:36905–11. doi: 10.1074/jbc.274.52.36905. [DOI] [PubMed] [Google Scholar]
- [75].Sun G, Zhao H, Kalyanaraman B, Dahms NM. Identification of residues essential for carbohydrate recognition and cation dependence of the 46-kDa mannose 6-phosphate receptor. Glycobiology. 2005;15:1136–49. doi: 10.1093/glycob/cwi098. [DOI] [PubMed] [Google Scholar]
- [76].Arendt CW, Ostergaard HL. Two distinct domains of the beta-subunit of glucosidase II interact with the catalytic alpha-subunit. Glycobiology. 2000;10:487–92. doi: 10.1093/glycob/10.5.487. [DOI] [PubMed] [Google Scholar]
- [77].Petrescu AJ, Butters TD, Reinkensmeier G, Petrescu S, Platt FM, Dwek RA, Woremald MR. The solution NMR structure of glucosylated N-glycans involved in the early stages of glycoprotein biosynthesis and folding. EMBO J. 1997;16:4302–10. doi: 10.1093/emboj/16.14.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Woods RJ, Pathiaseril A, Wormald MR, Edge CJ, Dwek RA. The high degree of internal flexibility observed for an oligomannose oligosaccharide does not alter the overall topology of the molecule. Eur J Biochem. 1998;258:372–86. doi: 10.1046/j.1432-1327.1998.2580372.x. [DOI] [PubMed] [Google Scholar]
- [79].Anji A, Kumari M. A novel RNA binding protein that interacts with NMDA R1 mRNA: regulation by ethanol. Eur J Neurosci. 2006;23:2339–50. doi: 10.1111/j.1460-9568.2006.04776.x. [DOI] [PubMed] [Google Scholar]