Abstract
Innate immune mechanisms play a deterministic role in the rate of disease progression during acute infection in HIV infected humans and SIV infection of nonhuman primates. The role NK cells play in mediating such an effect has thus gained importance. One of the major sets of molecules that regulate NK cell function are the killer cell immunoglobulin like molecules (KIR's). Our laboratory has previously shown an association of KIR3DL alleles 13–14 with high plasma viral loads in a cohort of SIV infected rhesus macaques. To gain a more detailed understanding of the role of KIR polymorphisms, our laboratory herein conducted studies of 3 additional KIR loci and show that select KIR3DH alleles appear to be more strongly associated with high plasma viral loads than KIR3DL alleles 13–14. In addition, we herein document the existence of additional new alleles for the KIR1D, KIR2DL4, and the KIR3DH loci.
Keywords: Innate immunity, nonhuman primates, KIR's, MHC, NK cells, polymorphisms, SIV, rhesus macaques, genetics, viral loads
INTRODUCTION
It is becoming increasingly clear that the quality and quantity of innate immune responses that are induced by the host in response to lentivirus infections in humans and nonhuman primates plays a critical role in establishing the course of infection and the dynamics of disease [1; 2; 3; 4]. Among the cell lineages that play a prominent role in mediating such innate immune responses are cells of the NK cell lineage. While previously NK cells were considered as non-specific killer cells that target non-MHC expressing tumor or virus infected cells, it is now clear that NK cells appear to have a more sophisticated role than previously appreciated. Thus, NK cells undergo a process of negative/positive selection termed “licensing” and “arming”; they have been shown to undergo “education” in terms of recognizing self v/s non-self; execute regulatory function giving rise to the term “NKregs” and an “editing” function targeted on immature dendritic cells [5; 6; 7; 8; 9; 10]. Of interest is also the finding that they appear to have “memory” [11; 12] which clearly has an impact on vaccine design for a number of infectious agents including HIV and the simian immunodeficiency virus (SIV). Our laboratory has previously described the expression of a variety of cell surface markers and receptors expressed by NK cells from nonhuman primates which were utilized to define their heterogeneity [13]. Among these are the diverse arrays of receptors which can mediate either activating or inhibitory signals. These NK cells therefore utilize such receptors to continuously survey other cells for the presence or absence of cognate ligands for these receptors [14]. The ligation of such receptors on NK cells can either lead to killing of the target cells, inhibition of the killing activity of the NK cells or synthesis of cytokines [15; 16; 17]. The receptors that mediate such function can be broadly categorized into the family of killer cell immunoglobulin-like receptors (KIR's), the killer cell lectin-like receptor family such as the hetero-dimeric CD94/NKG2a molecules (KLR's) and the Killer cell non-immunoglobulin like receptors (NKCR) such as NKp46, p30 and p44 [18]. While the ligands for KIR's and KLR's are, to a large extent, the MHC class I molecules [14], the ligands for NKCR remain undefined. The KIR's and their cognate MHC class I ligands are encoded by unlinked polymorphic gene families which serve to distinguish all but the most related individuals. A large body of data has recently accumulated that documents the biological effects that have been associated with the inheritance of select MHC class I and KIR polymorphisms. These include the association of KIR's and their cognate MHC class I alleles with resistance to infection, susceptibility to autoimmune diseases, complications of pregnancies and the outcome of hematopoietic stem-cell transplantation [reviewed in 19]. Pertinent to the present communication, the role of MHC class I and KIR polymorphisms has been highlighted as one of the first host genes that appear to be associated with disease resistance in human HIV-1 infection [20; 21; 22].
Our laboratory has been studying the mechanisms of pathogenesis induced by the experimental infection of rhesus macaques with SIV, a model that shows a pattern of disease remarkably similar to that noted for human HIV-1 infection [23]. The findings using this model by several laboratories including ours has been that a certain fraction of a group of rhesus macaques experimentally infected with aliquots of the same pool and dose of SIV, infected via the same route, appear to show peak plasma viremia similar to the rest of the animals in the cohort but spontaneously control the level of viremia, in some cases below the level of detection. Although a role for MHC genes in contributing to such a state has been recognized [24; 25; 26], the precise mechanisms by which such “elite” and long term non-progressors (LTNP) achieve such a clinical status is far from clear and unraveling such mechanisms is considered clearly a very important research target. The published findings of an association between KIR polymorphisms and MHC class I genes in disease resistance of human HIV-1 infection [20; 21; 22] prompted us to initiate studies on defining the extent of KIR polymorphisms in the rhesus macaque with the ultimate goal of defining whether a similar association could define such “elite” LTNP rhesus macaques. More specifically, our objective has been to define the precise mechanisms by which the outcome of such interactions between KIR's and MHC class I alleles is induced and maintained. There have been limited studies so far on defining KIR polymorphisms in nonhuman primates including rhesus macaques. The initial studies were carried out by Hershberger et al [27] who defined 5 different families of KIR molecules. These studies have been followed by those from the laboratory of O'Connor et al [28] that have provided a foundation for the study of KIR polymorphisms in nonhuman primates. The data to date indicate that there are at least 8 different KIR loci with the likelihood of the existence of perhaps up to 12–16 loci and a couple of pseudogenes. The 8 loci identified include 1 locus for KIR1D, two for KIR2DL, one locus for KIRDH and four loci for KIR3DL [28]. While the degrees of polymorphisms for each of the loci remain to be fully documented, the finding of 22 alleles for the relatively conserved KIR2DL4 locus [29] highlights the degree of polymorphisms that exist for the KIR molecules. Our laboratory has previously documented the association of certain Mamu-KIR3DL alleles expressed by enriched population of NKG2a+ cells with influencing the level of plasma viremia in a cohort of SIV infected rhesus macaques [30] which also correlated with the level of NK cell function. In the present study we have extended these observations to 3 additional Mamu-KIR loci with the goal to further increase our knowledge of KIR polymorphisms and its potential role in influencing plasma viral loads and SIV pathogenesis. Results of these studies constitute the basis of this report.
MATERIALS AND METHODS
Blood samples
The heparinized peripheral blood samples utilized for the studies reported herein were obtained from a group of outbred rhesus macaques (Macaca mulatto; n = 38) bred in our colonies of this species at the Yerkes National Primate Center of Emory University. The monkeys were randomly assigned by the veterinary staff to the studies described herein. PBMC samples from each monkey was analyzed for MHC-class I Mamu-A01, Mamu-B01, Mamu-B08 and Mamu-B17 and the results of such typing in addition to clinical history and other salient data on these monkeys is illustrated in Table 1. Each monkey was infected intravenously with 1000 TCID50 of either SIVmac239 or SIVmac251 in a volume of 1 ml of sterile saline. Plasma samples from each of the 38 monkeys were routinely monitored for viral loads weekly for the first 4 weeks, every other week up to 12 weeks and monthly thereafter. Based on the average viral loads of each monkey following reaching viral load “set point”, the samples were divided into two groups: those that represented monkeys with high viral load (HVL > 106 viral copies/ml of plasma, n = 20, viral load range 1.2 × 106 to 4 × 106 copies/ml of plasma) and those with low viral load (LVL < 105 viral copies/ml of plasma, n = 18, viral load range 3 × 102 to 7 × 104 copies/ml of plasma). The clinical history, VL at set point and the frequency of CD4+ T cells at baseline and post infection during the chronic infection period when the samples for the studies reported herein were utilized is also detailed in Table 1. All monkeys were housed at the Yerkes Regional Primate Research Center of Emory University and were maintained according to the guidelines of the Committee on the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council and the Health and Human Services guidelines Guide for the Care and Use of Laboratory Animals.
Table 1.
Clinical history of the 38 SIV-infected RMs in the cohort.
| Sample ID | MHC Class I typing (Mamu)* | SIV strains | viral loads (copies/ml) | NK cell activity (lytic units) | % CD4+T cell count | ||
|---|---|---|---|---|---|---|---|
| Pre-infection | Post-infection | Pre-infection | Post-infection | ||||
| RM-HVL cohort | |||||||
| mm 1 | SIVmac239 | 6,000,000 | 776 | 101 | 33.4 | 15.5 | |
| mm 3 | SIVmac239 | 28,841,600 | 673 | 824 | 30.6 | 25.3 | |
| mm 5 | SIVmac239 | 8,763,600 | 926 | 148 | 28.9 | 27.1 | |
| mm 6 | SIVmac239 | 6,912,600 | 887 | 88 | 32.1 | 15.4 | |
| mm 7 | B.01 | SIVmac239 | 1,200,000 | 776 | 172 | 24.4 | 9.5 |
| mm 8 | B.01 | SIVmac239 | 31,426,100 | 721 | 122 | 35.1 | 34.4 |
| mm 9 | B.01 | SIVmac239 | 27,000,000 | 709 | 154 | 25.1 | 30.5 |
| mm 10 | SIVmac239 | 6,200,000 | 921 | 210 | 35.9 | 10.5 | |
| mm 16 | SIVmac239 | 18,000,000 | 578 | 122 | 26.4 | 21.6 | |
| mm 17 | SIVmac239 | 36,831,700 | 822 | 321 | 23.3 | 8.9 | |
| mm 18 | SIVmac239 | 3,434,500 | 876 | 212 | 27.6 | 15.7 | |
| mm 19 | A.01 | SIVmac251 | 2,950,000 | 984 | 98 | 29.1 | 11.8 |
| mm 22 | SIVmac239 | 2,000,000 | 498 | 98 | 32.2 | 3.2 | |
| mm 23 | A.01 | SIVmac251 | 6,000,000 | 574 | 108 | 31.9 | 22.9 |
| mm 24 | A.01,B.01 | SIVmac239 | 3,300,000 | 582 | 232 | 21.1 | 10.8 |
| mm 25 | SIVmac251 | 4,000,000 | 621 | 164 | 31.4 | 5.2 | |
| mm 26 | SIVmac239 | 5,041,100 | 602 | 173 | 30.8 | 13.9 | |
| mm 27 | SIVmac251 | 2,000,000 | 548 | 182 | 41.1 | 11.8 | |
| mm 29 | SIVmac239 | 1,500,000 | 733 | 167 | 22.2 | 12.6 | |
| mm 33 | SIVmac239 | 3,000,000 | 678 | 186 | 31.8 | 10.7 | |
|
| |||||||
| RM-LVL cohort | |||||||
| mm 2 | SIVmac239 | 11,000 | 645 | 764 | 34.5 | 35.8 | |
| mm 4 | SIVmac239 | 70,184 | 833 | 722 | 31.1 | 37.2 | |
| mm 11 | A.01 | SIVmac239 | 800 | 887 | 776 | 19.3 | 15.7 |
| mm 12 | SIVmac239 | 2,000 | 567 | 698 | 23.2 | 18.1 | |
| mm 13 | A.01,B.01,B.17 | SIVmac239 | 1,000 | 587 | 859 | 28.8 | 17.2 |
| mm 14 | A.01 | SIVmac239 | 10,000 | 902 | 770 | 31.2 | 29.6 |
| mm 15 | A.01 | SIVmac239 | 20,000 | 768 | 623 | 33.8 | 25.9 |
| mm 20 | A.01 | SIVmac239 | 15,000 | 772 | 632 | 32.7 | 31.4 |
| mm 21 | SIVmac239 | 4,000 | 664 | 662 | 25.8 | 20.7 | |
| mm 28 | SIVmac239 | 2,770 | 776 | 589 | 31.1 | 27.2 | |
| mm 30 | SIVmac239 | 1,100 | 784 | 634 | 28.3 | 38.1 | |
| mm 31 | SIVmac239 | 300 | 698 | 702 | 38.2 | 33.5 | |
| mm 32 | SIVmac239 | 700 | 802 | 874 | 25.5 | 15.4 | |
| mm 34 | A.01 | SIVmac251 | 4,000 | 712 | 668 | 32.6 | 14.8 |
| mm 35 | SIVmac239 | 300 | 823 | 668 | 41.9 | 34.1 | |
| mm 36 | B.08 | SIVmac239 | 9,100 | 776 | 721 | 35.9 | 26.8 |
| mm 37 | B.17 | SIVmac239 | 20,000 | 715 | 693 | 29.1 | 16.3 |
| mm 38 | A.01 | SIVmac251 | 1,500 | 756 | 922 | 36.7 | 18.1 |
NK cell functional activity
An aliquot of the PBMC samples prior to infection (baseline) and post infection during the chronic infection stage were analyzed for NK cell functional activity as described elsewhere [30] using the K562 cell line as the target cells. Results of the functional activity were expressed as lytic units per a million PBMC's, with one lytic unit being the number of effector PBMC's that could kill 1/3 of maximal lysis. Data obtained are described under Table 1.
NK cell isolation
Peripheral blood mononuclear cells (PBMC's) were isolated from heparinized blood samples using the standard Ficoll-Hypaque gradient centrifugation technique (Sigma, St. Louis, MO, USA). The CD3+ cells were depleted using mouse anti-rhesus CD3 antibody (1 ug/1 × 106 cells, clone FN-18; BioSource) followed by incubation with anti-mouse Ig conjugated magnetic beads (Cellection kit; Invitrogen, Carlsbad, CA, USA). Bead-bound T cells were discarded and the remaining CD3 depleted cells incubated with mouse anti-rhesus NKG2A antibody (1 ug/1 × 106 cells, clone Z199; Beckman Coulter, Fullerton, CA, USA) and the NKG2a+ cells positively selected using anti-mouse Ig conjugated magnetic beads (Cellection kit; Invitrogen, Carlsbad, CA, USA). An aliquot of the cells was analyzed for purity and found to be > 90% enriched.
RNA isolation and cDNA synthesis
Total RNA was isolated from the highly enriched population of NKG2A+ NK cells by using the RNeasy kit (Qiagen, Valencia, CA, USA) and cDNA synthesized using Protoscript® First Strand cDNA Synthesis Kit (New England Biolabs, Ipswich, MA, USA). The resulting cDNA products were stored at −20°C until use.
KIR primer pairs
The primer pairs utilized for amplifying Mamu-KIRID, KIR2DL4, KIR2DL5 and KIR3DH are described in Table 2. The primer pairs were designed using PrimerSelect software (DNASTAR, Madison, WI) based on the published Mamu-KIR sequences (AY728181, AY728182, AF334646, and AY728190), respectively. The PCR cycling conditions included initial denaturation at 94°C for 5 min followed by 40 cycles of denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec; extension at 72°C for 90 sec, and a final extension at 72°C for 10 min. The amplified PCR products were visualized on a 1% agarose-ethidium bromide gel following electrophoresis and only bands giving the expected molecular weight were excised, purified and ligated into the pGEM-T vector (Promega, Madison, WI, USA). A minimum of five KIR sequence-containing colonies from each cloning procedure/transformation were selected and sequenced using T7 and SP6 primers (Promega) and sequences were analyzed using the DNASTAR analysis package (DNASTAR, Inc., Madison, MA, USA).
TABLE 2.
List of primer pairs utilized
| KIR gene | Primer name | Direction | Sequence 5' to 3' | length (bp) |
|---|---|---|---|---|
| KIR1D: | ||||
| 1 KIR1DFL | Forward | TCATGGTCGTTAGCGTGGCGTGTG | 24 | |
| 2 KIR1DR | Reverse | CCTGCTGCTGGTACATTGGAACTG | 24 | |
| KIR2DL4 extracellular domain: | ||||
| 1 2DL4PSF7 | Forward | CTGGCCTGTCTTGGGTTCTTCT | 22 | |
| 2 2DL4PSR12 | Reverse | GGTCCATTACAGCAGCATTCTT | 22 | |
| KIR2DL4 intracellular domain: | ||||
| 1 KIR2D4FL | Forward | CAGTTCCCGGCGCTCCTTTGAC | 22 | |
| 2 2DL4RL1 | Reverse | CTAAGCAAAGGAGTGCGTTTTC | 22 | |
| KIR2DL 5: | ||||
| 1 KIR2D5FL2 | Forward | ATGGCATGTGTTGGGTTCTTCTTG | 24 | |
| 2 KIR2D5RL1 | Reverse | GCAGAGTCGCGCCTTCAGATTCCT | 24 | |
| KIR3DH: | ||||
| 1 KIR3HFL | Forward | TGTGTTGGGTTCTTCTTGGTCCAG | 24 | |
| 2 KIR3HFL1 | Forward | CACACGGGTGGTCAGGACAAGAC | 23 | |
| 3 KIR3HRL | Reverse | TCTGAGAAGGGCGAGTGATTTTTC | 24 | |
Data Analysis
Our strategy for assignment of alleles for each of the Mamu-KIR loci was similar to that described previously [30]. Briefly, the translated nucleotide sequences were first aligned. Those sequences, where a point mutation, deletion or insertion resulted in the early termination of the protein resulting in a truncated protein, were not included either for allele assignment and/or for further data analyses. Sequences that included the extracellular domain, the transmembrane domain and tail domains were included for allele assignments and for further data analysis. Splice variants were also included and assigned as variants to a given allele based on homology to the allele from which it was spliced. Accession numbers for new alleles identified are listed in Supplement Table 1. Based on utilizing such a strategy for allele assignment, the data obtained on alleles for each of the 3 loci described herein in addition to the Mamu-KIR3DL allele data published previously on these same monkeys [30] were subjected to statistical analysis. Statistical analysis was performed using the SPSS version 10.0 (Chicago, USA) unless otherwise noted. The Odds Ratio (OR) with 95 % confidence intervals (CI) for the KIR genotypes of each animal were calculated; additionally p-values were determined by Pearson's Chi-square (Cz) test. The KIR genotypes that showed significant difference (p<0.05) associated with either high or low viral load by using OR were determined.
RESULTS
Mamu-KIR2DL4 polymorphisms
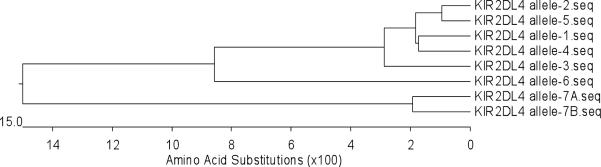
The primer pair utilized for amplifying the Mamu-KIR2DL4 locus (Table 2) covers the signal sequence, the D0 and D2 domain, the stem, the transmembrane domain, and part of the cytoplasmic tail. This primer pair successfully amplified products from 27 (15 with HVL and 12 with LVL) of the 38 macaques analyzed providing a minimum of 5 clones from each of these 27 monkeys (for a total of 135 clones) that were sequenced. There were 12 of these 135 sequences that showed an “early” (a stop codon that does not result in a protein with extracellular domains) stop codon which were thus not utilized for further analysis. The grouping and identification of sample sequences were determined as previously described [30]. Briefly, all individual sequences with “silent” SNPs that did not translate into a change in the amino acid sequence were disregarded. The protein sequences were subsequently grouped based on the phylogenetic distance in a stepwise fashion. Once the protein analysis was complete, a reverse analysis was performed using the corresponding DNA sequences. Such a strategy for analysis led to the assignment of the sequences to a total of 7 Mamu-KIR2DL4 alleles which included two previously identified alleles (GenBank acc. no. EU702486 and AY728182), five new alleles, and one new variant (GenBank acc. no. GU564176 to GU564181). Fig. 1 illustrates the phylogenetic tree of the 7 alleles encoded by the Mamu-KIR2DL4 locus.
FIGURE 1.
Phylogenetic tree of the Mamu-KIR2DL4 extracellular domain at the protein level
Alleles 1 to 4 of the Mamu-KIR2DL4 locus contain an AGG sequence in their transmembrane domain which codes for a positively charged arginine (R). The potential of the positively charged arginine residue to facilitate binding to the corresponding negatively charged transmembrane residues of the ITAM encoding adaptor proteins, such as DAP-12, which could contribute to the activation of the KIR2DL4 molecule has been previously documented [31]. The extracellular domain of Mamu-KIR2DL4 alleles-1 and 2 were found to be the two most dominant alleles in this group of rhesus macaques with each of the 27 expressing either allele 1, allele 2 or both in addition to the other alleles identified. Thus, 17 of the 27 monkeys (12 HVL and 5 LVL) expressed the KIR2DL4 allele-1 sequence and 13 of the 27 monkeys (5 HVL and 8 LVL) were found to express the KIR2DL4 allele-2. Three or more clones of each monkey expressed allele-1 in 15 of the 17 monkeys (11 HVL and 4 LVL) and allele-2 in 10 of the 13 monkeys (3 HVL and 7 LVL). All 5 clones from six monkeys (3 HVL and 3 LVL) expressed only allele-1 (Table 3). Seven different monkeys expressed only allele-2 (Table 3) in all 5 clones tested, however 6 of the 7 monkeys were from the LVL cohort and only 1 monkey was from the HVL cohort. Interestingly, the specific motif localized to the region between the stem and the transmembrane domain appeared to be a signature motif for the Mamu-KIR2DL4 allele-1 or allele-2 as previously described [27]. Thus, whereas allele-1 showed a `VT-X3-A-X18-H-X5-D' motif, the allele-2 showed either a `TR-X3- I-X18-L-X5-D/N' or `VR-X3-A-X18-L-X5-D' motifs at the same position. The other Mamu-KIR2DL4 alleles which were identified in this study (allele 3–7), were expressed at a much lower frequency. Thus, they were found in only one or two clones. Mamu-KIR2DL4 allele-3 (803 bp or 252 aa) and allele-4 (802 bp or 252 aa) coded for a full length extracellular region. Alleles-3 and 4 (GenBank acc. no. GU564176 and GU564177, respectively) coded for amino acids that were different from alleles-1 and 2 in the D0 domain. Additionally, the amino acids within the D2 domain in allele-4 were distinct from those expressed by alleles lto 3. Sequences representing allele-5 were found to be closely related to allele-2 with 98.1% and 99.3% homology at the amino acid and DNA level, respectively (Fig. 1). Allele-5 was 802 bp long and had a nonsense mutation causing the adenine to be replaced with thymine at position 673 yielding TAA (A673AA → T673AA) in the cDNA sequence (supplement Fig. 1A) and since TAA is a stop codon, such a stop codon would result in a truncated protein product (supplement Fig. IB). Allele-6 (803 bp or 236 aa) had one adenine base insertion within the cDNA sequence at position 676 (A676CT → A676ACT) that led to a frameshift translation (a shift in the reading frame) resulting in a truncated protein. Allele-7 was identified from two clones which appear to encode for closely related proteins. However, these clones had different deleted bases. Therefore, these clones were designated as two variants, allele-7 variant A (allele-7A) which lacks a guanine at position 591 and variant B (allele-7B) which lacks a cytosine at position 608. These proteins, although truncated, were included in this analysis and assigned to alleles since all three code for the full length extracellular region of the Mamu-KIR2DL4 locus. It was reasoned that if these proteins were expressed on the cell surface, they could still bind their ligands resulting in an inhibitory signal to the NK cells. There have been several reports that have identified HLA-G as the ligand for human KIR2DL4 [29; 32] and since HLA-G is relatively conserved, it is reasonable to assume that it is also the ligand for Mamu- KIR-2DL4. However, the precise sequence that is involved in recognition of the HLA-G molecule by human KIR2DL4 remains unknown at present. It is possible that the Ig-like domains of KIR2DL4 might function in ligand-receptor interactions similar to the interaction previously described for the HLA Bw4-KIR3DL binding [33; 34; 35]. Interestingly, four monkeys in the HVL cohort expressed the low frequency alleles-5, 6 and 7, while only one monkey in the LVL cohort expressed allele-7.
TABLE 3.
Association of the Mamu-KIR2DL4 alleles in SIV infected rhesus macaques according to plasma viral loads
| Allele | HVL (n) | LVL (n) | Total (n) |
|---|---|---|---|
| 1 | 12 | 5 | 17 |
| 2 | 5 | 8 | 13 |
| 3 | 1 | 0 | 1 |
| 4 | 1 | 0 | 1 |
| 5 | 2 | 0 | 2 |
| 6 | 2 | 0 | 2 |
| 7A | 0 | 1 | 1 |
| 7B | 1 | 0 | 1 |
| Predicted allele-5 | 4 | 2 | 6 |
| allele-1/allele-2 ratio | 2.40 | 0.63 | 1.31 |
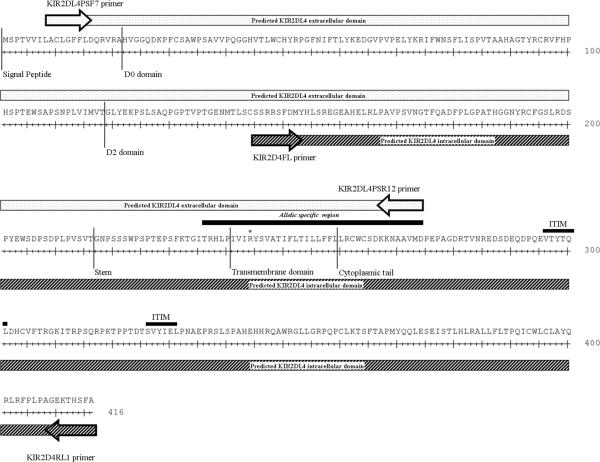
In addition to the truncated proteins of allele-5, samples from six monkeys, (four monkeys in the HVL cohort and two monkeys in the LVL cohort) resulted in clones which coded for an early stop codon within either the D0 or D2 domain. These clones had a high degree of sequence homology (>98%) at both the DNA and protein level (supplement Fig. 2) with allele-5. They were thus termed “predicted” allele-5 proteins in this study (Table 3). A preponderance of the HVL monkeys (9 of the 15) appeared to express truncated KIR2DL4 proteins if one takes into account all of the truncated KIR2DL4 proteins (which include alleles-5, 6, and 7 and the predicted allele-5). The stem, transmembrane domain and the cytoplasmic tail of the Mamu- KIR2DL4 sequences were also amplified from 9 of the 27 monkeys from our cohort, by using the KIR2D4FL-KIR2D4RL1 primer pair (Table 2). These sequences including the ITIMs were found to share >98% homology to previously published data and, surprisingly, each of these were associated with the extracellular domain of allele 2 (data not shown). Although use of a large series of primer pairs failed to amplify the full-length KIR2DL4 sequences, the use of separate pairs of primers that covered the two end of the KIR2DL4 sequence with a significant portion that included overlapping sections of the extracellular and intracellular domains yielded sequences that were aligned to ensure complete coverage of the KIR2DL4 cDNA (Figure 2).
FIGURE 2.
The predicted Mamu-KIR2DL4 intra- and extra-cellular domains
Mamu-KIR1D Polymorphisms
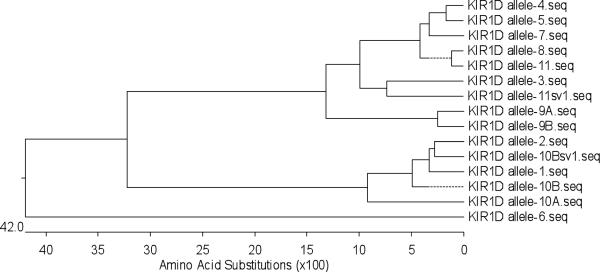
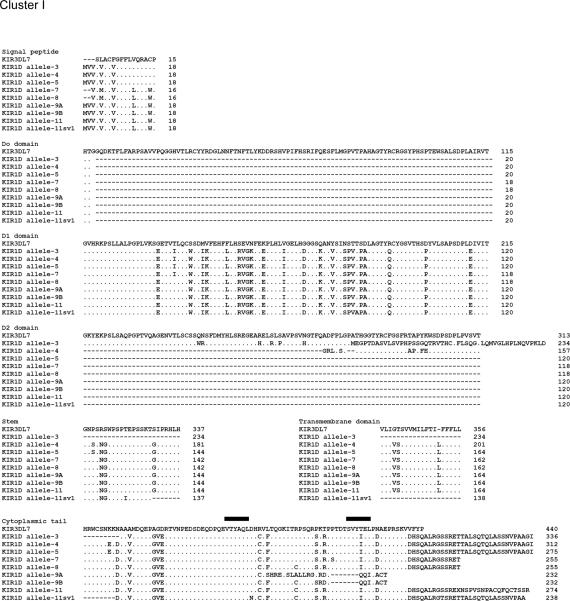
The Mamu-KIR1D sequences analyzed encompassed the full length sequence of KIR1D which thus included the signal peptide, the D1 domain, part of the D2 domain, the stem, the transmembrane domain, and the cytoplasmic tail with a predicted length of ~1.1 kb. The Mamu-KIR1D primer pair (Table 2) utilized resulted in amplified products from the cDNA prepared from highly enriched population of NKG2a+ cells from only 15 of the 38 monkeys (9 RM-HVL and 6 RM-LVL) tested. As with the Mamu-KIR2DL4 analysis, the close phylogenetic relationship/sequence identity (>98%) between sequences of clones from at least two different monkeys led to the assignment of the sequences to a total of 11 alleles (Fig. 3). Of these alleles, allele-1 to allele-8 have been previously published (GenBank acc. no. AY728181, AF334635, AF334636, AF334638, AF334640, AF334643, FJ217804, and FJ217805, respectively). In addition, there were 3 new alleles identified. Alleles 9 and 11 each had two variants and Allele 10 had 3 variants (GenBank acc. no. GU564169 to GU564175). The previously published Mamu-KIR3DL7 sequence (GenBank acc. no. AF334622) now designated to be part of the KIR1D locus, was utilized as a consensus sequence for the analysis of the Mamu-KIR1D alleles [26]. As seen in Fig. 4, the 11 Mamu-KIR1D alleles identified appeared to cluster into two groups, a KIR1D cluster I and a KIR1D cluster II, based on the presence or absence of an ITIM(s). Thus, while the members of Mamu-KIR1D cluster I had ITIM(s) expression in their cytoplasmic tail and included alleles-3, 4, 5, 7, 8, 9, and 11, those belonging to Cluster II showed no ITIM expression and consists of alleles-1, 2, 6, and 10. Interestingly, cluster I was found in ~1.7 fold more monkeys from the HVL cohort than the LVL cohort; thus, monkeys in the HVL cohort tended to express the inhibitory KIR molecules (ITIMs in the cytoplasmic region). There was no significant difference between HVL and LVL groups in macaques expressing Mamu-KIR1D cluster II alleles (no ITIMs) (Table 4). Additionally, none of the monkeys in our cohort expressed four of the previously described [27] Mamu-KIR1D splice variants, sv-3, 5, 7, and 8 (GenBank acc. no. AF334637, AF334639, AF334641 and AF334642). Mamu-KIR1D allele-9 showed close homology to allele-8 at the DNA level. However, a one base deletion was found upstream of the secondary ITIM, resulting in a frame-shift. This allele was identified as two variants (allele-9 variant A and variant B) based on different clones from the same HVL monkey. In allele-9 variant A (allele-9A) the thymine at position 623 was deleted and in allele-9 variant B (allele-9B) the adenine at position 662 was deleted (supplement Fig. 3). These deletions result in a reading frame mutation within the cytoplasmic tail which causes the loss of an ITIM. Furthermore, Mamu-KIR1D allele-11 splice variant 1 (allele-11sv1) was found which expressed an ITIM but lost part of the transmembrane motif, similar to allele-3. Whether these proteins are still functional remains unknown.
FIGURE 3.
Phylogenetic tree of the Mamu-KIR1D locus at the protein level
FIGURE 4.
Cluster I and II alleles at the protein level comprising the Mamu-KIR1D alleles (the solid black lines reflect the ITIM's)
TABLE 4.
Association of the Mamu-KIR1D alleles in SIV infected rhesus macaques according to plasma viral loads
| Cluster | Allele | HVL (n) | LVL (n) | Total (n) |
|---|---|---|---|---|
| I | 3 | 3 | 1 | 4 |
| 4 | 0 | 1 | 1 | |
| 5 | 1 | 2 | 3 | |
| 7 | 2 | 1 | 3 | |
| 8 | 2 | 1 | 3 | |
| 9A | 1 | 0 | 1 | |
| 9B | 1 | 0 | 1 | |
| 11 | 2 | 0 | 2 | |
| 11sv1 | 0 | 1 | 1 | |
| Total (n) | 12 (60%)* | 7 (39%)* | 19 (50%)** | |
| II | 1 | 4 | 5 | 9 |
| 2 | 2 | 1 | 3 | |
| 6 | 1 | 1 | 2 | |
| 10A | 0 | 1 | 1 | |
| 10B | 1 | 0 | 1 | |
| 10Bsv1 | 1 | 0 | 1 | |
| Total (n) | 9 (45%) | 8 (44%) | 17 (45%) |
% of total monkey in group. HVL total = 20, LVL total = 18
% of total of all 38 monkeys
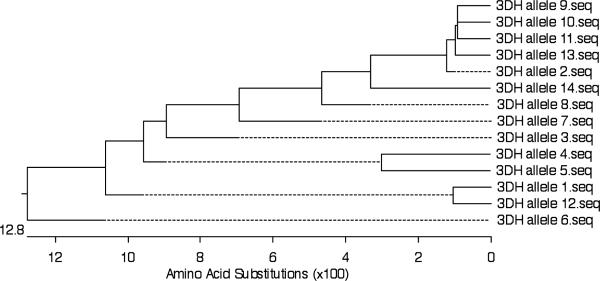
Mamu-KIR3DH Polymorphisms
KIR3DH as noted by its name is a hybrid KIR between Mamu-KIR3DL and Mamu-KIR2DL4 allele-2 which has a truncated cytoplasmic tail. Thus, while the D0, D2 domains and the stem sequence appear to be derived from KIR3DL, the transmembrane domain and the cytoplasmic tail appear to be derived from the KIR2DL4 allele-2. The products of this locus were amplified using the primer pairs listed in Table 2 resulting in 65 full length Mamu-KIR3DH sequences from cDNA derived from highly enriched population of NKG2a+ cells from the PBMC's of a total of 21 of the 38 monkeys (13 RM-HVL and 8 RMLVL). The cDNA from the other 17 monkeys in our cohort did not yield Mamu- KIR3DH sequences using these primer pairs. Again based on 98% homology, the 65 sequences were assigned to 14 Mamu-KIR3DH alleles and a phylogenetic tree developed (see Fig. 5). Mamu-KIR3DH allele-11 and allele-13 were each defined by two independent clones from one monkey. Most of the other alleles were, in fact, defined by four or more clones from at least two different monkeys (data not shown). According to the alignment, Mamu-KIR3DH had either one or two stop codons around amino acid 370, depending on the clone, similar to previously published sequences for Mamu-KIR3DH. All of these sequences contain an arginine in their transmembrane domain as found in the Mamu-KIR2DL4 locus product. These Mamu-KIR3DH alleles appeared to segregate into three groups based on VL of the monkeys from whom they were derived. They were thus identified as HVL-alleles, LVL-alleles, and other alleles. The Mamu-KIR3DH-HVL alleles contained 7 alleles that included alleles-2, 3, 6, 10, 11, 13, and 14. In contrast, Mamu-KIR3DH-LVL included 3 alleles that were alleles-4, 5 and 12. The remaining alleles were identified as other Mamu-KIR3DH alleles (Table 5).
FIGURE 5.
Phylogenetic tree of the Mamu-KIR3DH extracellular domain at the protein level
*Mamu-KIR3DH allele-1 and 2 have been described previously
TABLE 5.
Association of the Mamu-KIR3DH alleles in SIV infected rhesus macaques according to plasma viral loads
alleles 2,3,6,10,11,13,14
alleles 4,5,12
alleles 1,7,8,9
Univariate and Multivariate analysis of an association between Mamu-KIR alleles and viral loads
Statistical analysis of the data aimed at identifying Mamu-KIR genotype association with high as compared with low viral load revealed an Odds Ratio (OR) with statistical significance (p < 0.05) for 3 pairs (6 genotypes) of Mamu-KIR loci and alleles that were associated with levels of viral loads. The first pair of Mamu-KIR alleles showing statistical significance associated with viral loads are the monkeys that express the KIR3DH high viral load alleles (alleles 2, 3, 6, 10, 11, 13 and 14) as compared with monkeys that express the Mamu-KIR3DH low viral load alleles (4, 5 and 12) giving a p-value of 0.015 (Table 6). The second pair of Mamu-KIR alleles that were associated with VL and showing statistical significance were the monkeys that were homozygous for the Mamu-KIR3DH high viral load alleles as compared with monkeys that were homozygous for the Mamu-KIR3DH low viral load alleles with a p-value of 0.047. As a matter of fact, using multiple logistic regression analysis (using the Forward-Likelihood ratio), monkeys expressing the Mamu-KIR3DH high viral load alleles as compared with any of the other Mamu- KIR loci showed a highly significant (p < 0.01) association with high viral load. Lastly, when association between multiple Mamu-KIR loci and viral loads were analyzed, there was a statistically significant association between monkeys that expressed the Mamu-KIR3DH high viral load alleles along with Mamu-KIR3DL alleles 13 or 14 and high viral loads as compared with monkeys that expressed any of the other 5 Mamu-KIR genotypes (p = 0.034). These data provide support to the view that select Mamu-KIR loci appear to be associated with influencing plasma viral loads in SIV infected rhesus macaques.
TABLE 6.
Uni- and multivariate analysis of the association between Mamu-KIR1D, KIR2DL4, KIR3DL and KIR3DH loci with plasma viral loads in SIV infected rhesus macaques
| Genotypes | HVL (n) | LVL (n) | OR | 95%CI | p-value | |
|---|---|---|---|---|---|---|
| lower | upper | |||||
| 1D*ITIM/*ITIM | 4 | 1 | 1.33 | 0.06 | 31.12 | 0.858 |
| 1D*null/*null | 3 | 1 | ||||
| 2DL4*1/*1 | 3 | 3 | 6.00 | 0.42 | 85.25 | 0.164 |
| 2DL4*2/*2 | 1 | 6 | ||||
| 3DH*h/*h | 7 | 1 | 21.67 | 0.64 | 730.08 | 0.047 |
| 3DH*l/*l | 0 | 3 | ||||
| 3DH*h/*z | 8 | 1 | 35.00 | 1.12 | 1094.79 | 0.015 |
| 3DH*l/*z | 0 | 4 | ||||
| 3DL*h/*h | 4 | 1 | 7.33 | 0.66 | 81.37 | 0.078 |
| 3DL*o/*o | 6 | 11 | ||||
| 1D*ITIM/*x+2DL4*1/*y | 6 | 3 | 7.86 | 0.28 | 217.12 | 0.182 |
| 1D*null/*x+2DL4*2/*y | 0 | 3 | ||||
| 1D*ITIM/*x+3DL*h/*a | 2 | 1 | 1.33 | 0.07 | 26.62 | 0.850 |
| 1D*null/*x+3DL*o/*a | 3 | 2 | ||||
| 2DL4*1/*y+3DH*h/*z | 6 | 0 | 11.00 | 0.08 | 1438.12 | 0.270 |
| 2DL4*2/*y+3DH*l/*z | 0 | 1 | ||||
| 2DL4*1/*y+3DL*h/*a | 5 | 0 | 14.14 | 0.57 | 352.02 | 0.064 |
| 2DL4*2/*y+3DL*o/*a | 3 | 6 | ||||
| 3DH*h/*z+3DL*h/*a | 5 | 1 | 21.00 | 0.64 | 690.03 | 0.053 |
| 3DH*l/*z+3DL*o/*a | 0 | 4 | ||||
| 3DH*h/*h+3DL*o/*o | 3 | 0 | 15.00 | 0.18 | 1236.28 | 0.192 |
| 3DH*l/*l+3DL*o/*o | 0 | 2 | ||||
| 3DH*h/*z+3DL*o/*a | 6 | 1 | 25.67 | 0.80 | 824.78 | 0.034 |
| 3DH*l/*z+3DL*o/*a | 0 | 4 | ||||
| 1D*ITIM/*x+2DL4*1/*y+3DL*h/*a | 2 | 0 | 3.00 | 0.02 | 473.1 | 0.665 |
| 1D*null/*x+2DL4*2/y+3DL*o/*a | 0 | 1 | ||||
| 2DL4*1/*y+3DH*h/*z+3DL*h/*a | 3 | 0 | 5.00 | 0.04 | 711.87 | 0.505 |
| 2DL4*2/*y+3DH*l/*z+3DL*o/*a | 0 | 1 | ||||
Abbreviation: 1D*ITIM is Mamu-KIR1D allele-3, 4, 5, 7, 8, 9 and 11; 1D*null is Mamu-KIR1D allele-1, 2, 6 and 10; 1D*x is Mamu-KIR1D allele-1 to allele-11; 2DL4*1 is Mamu-KIR2DL4 allele-1; 2DL4*2 is Mamu-KIR2DL4 allele-2; 2DL4*y is Mamu-KIR2DL4 allele-1 to allele-7; 3DH*h is Mamu-KIR3DH allele-2, 3, 6, 10, 11, 13, and 14; 3DH*l is Mamu-KIR3DH allele-4, 5, and 12; 3DH*z is Mamu-KIR3DH allele-1 to allele-14; 3DL*h is Mamu-KIR3DL allele-13 and 14; 3DL*o is Mamu-KIR3DL allele-1 to allele-12 and variant A; 3DL*a is Mamu-KIR3DL allele-1 to allele-14 and variant A; OR is the odds ratio; and CI is confidence interval
DISCUSSION
Studies from a number of laboratories indicate that the kinetics of viral replication in both HIV-1 infected humans and SIV infected rhesus macaques shows a marked and rapid decline following reaching peak levels of plasma viremia within a 2–3 week period post infection. The fact that the adaptive immune responses have not fully matured within this time period strongly indicates that cell lineages of the innate immune system most likely play a dominant if not a contributing role in this decline of viremia during this initial acute infection period [2]. In addition, the fact that the degree of this initial decline in viremia during the acute infection sets a footprint for the pace at which disease progression ensues, suggests that a study of the detailed mechanisms by which such differences in the level and rate of decline in viremia occurs during the acute infection period is highly warranted. Recognizing this issue has prompted our laboratory to focus on the potential role of NK cells as perhaps one of the major effector mechanisms that potentially mediate such an effect. As stated above, NK cells are in fact phenotypically and functionally heterogenous [2] and mediate their effector function by the net interactions of a large number of activating and inhibiting receptors decorating their cell surface. Among these receptors are the KIR molecules. Our studies have therefore focused on identifying the various KIR gene molecules expressed by rhesus macaques with the goal of identifying those KIR loci and alleles that are associated with influencing the initial decline of viremia described above. It is important to note that we utilized highly enriched population of NKG2a+ cells as the source of the cDNA for the studies presented herein. This may become an important issue since the KIR gene products have also been known to be expressed by other than the NK cell lineage which has not heretofore been taken into consideration in the analysis of KIR/MHC association data on cohorts of HIV-1 infected humans. In addition, it is not clear at present whether all the KIR genes are expressed by all hematopoietic cell lineages and whether they are expressed at the same levels. Further studies are clearly needed to sort out these issues. We also chose to assign Mamu-KIR alleles based on sequences of a single locus that shared 98% homology at the nucleotide level as belonging to a single allele. The rationale for using this strategy is that if we were to use the classical definition of an allele as being sequences that would differ by a single amino acid, it would lead to the assignments of hundreds of alleles for a single locus which would require a study of a very large number of SIV infected rhesus macaques and make it difficult if not impossible to examine the association between KIR alleles and SIV viral loads. In addition, our focus has been to scrutinize sequences that are associated with function such as those that are primarily localized to extracellular domains that have been previously shown to participate in binding to MHC molecules and/or are involved in select cases in coding for intracellular sequences involved in signaling such as ITIMs and ITAMs. Such a strategy clearly we submit has led to some interesting data as described herein.
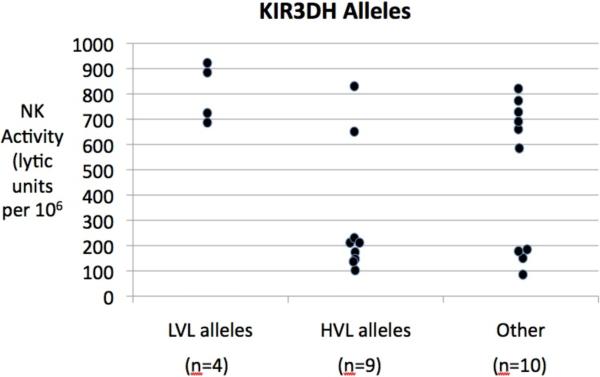
Data from the studies reported herein appear to show some interesting trends in terms of KIR polymorphisms and its association with viral loads. Thus, rhesus macaques that expressed KIR2DL4 allele-2 but not allele-1 appeared to fall into the LVL category. The fact that most of the monkeys that expressed KIR2DL4 either expressed allele-1 or 2 supports previous observations that indeed this locus is relatively conserved among Asian rhesus macaques. Other important associations between KIR loci and viral loads were the findings that the rhesus macaques that expressed the ITIM containing KIR1D alleles 3, 4, 5, 7, 8, 9, and 11 were associated with HVL. It was interesting to note that these sets of KIR1D alleles formed a Cluster that was distinct from the other non-ITIM containing alleles encoded by this locus. In addition, the hybrid KIR3DH locus appeared to code for certain alleles that segregated with high versus low viral loads. Thus, KIR3DH alleles 2, 3, 6, 10, 11, 13, and 14 were associated with HVL and the alleles 4, 5 and 12 with LVL. Statistical analysis of the data presented herein show a high degree of significance between plasma viral loads and the association with KIR3DH-HVL alleles and KIR3DH-HVL alleles combined with KIR3DL alleles as determined by calculating the odds ratio (O.R.) (Table 6). Clearly, the number of animals studied herein is a relatively small number to derive any major conclusions and larger groups of animals need to be studied. However, we submit that these findings are interesting and encourage the need for additional study. It is also important to note that these finding will take on added significance in terms of mechanisms when the MHC ligands for such Mamu-KIR loci are identified. In addition, when a more complete set of data on the other Mamu-KIR loci becomes available, it would be important to determine if the loci segregate into haplotypes much like in humans. Such assignments of KIR haplotypes for rhesus macaques has important practival implications. Thus, human KIR haplotypes have been shown to form 2 major groups. Haplotype A which is the most common haplotype contains two activating KIR genes (KIR2DL4 and KIR2DS4) and five inhibitory KIR genes (KIR2DL1, KIR2DL3, KIR3DL1, KIR3DL2, and KIR3DL3). On the other hand, haplotype B includes 6 activating KIR genes (KIR2DS1, S2, S3, S5.A, S5.B, and KIR3DS1), two inhibitory KIR genes (KIR2DL2 and KIR2DL5) and two KIR pseudogenes (KIR2DP1 and KIR3DP1) [36]. Several of the studies of HIV-1 infected human population have thus attempted to show association between rate of disease progression and inheritance of KIR haplotypes and thus such broader association studies may be more practical and useful for studies involving nonhuman primates once such haplotype assignments have been made. Linking specific KIR alleles with specific NK cell function such as killing of non-MHC expressing target cells as recently reported [37] would also provide an added dimension for understanding the biological role of such KIR and MHC association and HIV-1 infection. In this regard, as previously published there is clearly a major difference in NK cell function in the monkeys utilized for the studies reported herein and plasma viral loads (p<0.001). In addition, when data were analyzed to correlate the KIR3DH alleles with NK cell function, once again it appears that there are clear differences in the monkeys with KIR3DH HVL alleles as compared with KIR3DH LVL alleles (Fig. 6) although the number of animals in the latter group make it difficult to derive statistical power.
Figure 6.
It should also be noted that there are 2 other groups of molecules expressed by NK cells that could also potentially be involved in regulating their functional activity. These include the KLR's and the non-immunoglobulin like NK cell receptors [18]. Among the KLR's, both an activating (NKG2a) and an inhibitory form (NKG2C) have been identified in the rhesus macaque which interestingly have been shown to share 85% homology at the amino acid level with the human homologues [38]. The non-classical human MHC class I HLA-E molecule has been shown to be the ligand for these receptors and a rhesus homologue for HLA-E has been identified [39]. In this regard, it is important to note that the human HLA-E tetramer reagent binds to not only rhesus macaque NKG2a expressing NK cells but also CD3 expressing cells (data not shown) which is not surprising because HLA-E homologues have limited polymorphisms and are the among the most phylogenetically conserved MHC class I genes in primates [40]. Relevant to the present study, a role for NKG2C/C2 has been implicated in SIV infected rhesus macaques [41], and a role for NKG2a in human HIV-1 infection [42; 43]. It thus appears that the interaction between NK cells and its subsets with target cells following infection involve the interactions between multiple receptors and ligands. The nature of the net signals generating from such interaction results in either killing of the target cells or the generation of multiple cytokines and chemokines that influence the quantity and quality of the immune response. These issues with regards to a role for these other receptor/ligand interactions have to be to be taken into consideration in the interpretation of data such as those provided in the present communication.
It should also be noted that we attempted to amplify, clone and sequence alleles encoded by the Mamu-KIR2DL5 locus using the primer pairs listed in Table 2. Unfortunately, samples from only 3 of the 10 monkeys gave results with most of the sequences that have already been previously published for Mamu-KIR2DL5 (GenBank acc. no. AF334646 and AF 334647) or were identified as Mamu-KIR3DL20 (GenBank acc. no. NM 001104552). Further studies are currently in progress utilizing different sets of primer pairs in attempts to generate sufficient data for the KIR2DL5 locus that is amenable for analysis.
Clearly, one of the major issues concerning KIR locus and allele typing of unknown rhesus macaques requires a simplified, technically simple and reproducible assay system. In attempts to address this issue, our lab initiated studies to establish a TaqMan assay for the KIR3DL locus to begin with. Results of these initial studies showed that whereas we were quite successful in establishing a TaqMan assay for identifying KIR3DL allele 13/14 [30], establishing a TaqMan assay for the other KIR3DL alleles have so far not been successful most likely due to non-specific binding of reaction primers. In efforts to identify potential reasons for such failure, we subjected the entire KIR3DL locus to sequencing (25Kb) using a commercial vendor. Unfortunately, the sequence data obtained was not informative and was found to be difficult due to the occurrence of long stretches of repeat sequences within the exons. On the other hand, the availability of relatively inbred population of Mauritian cynomolgus macaques facilitated a more simplified study of the genetics of KIR gene inheritance performed by Bimber et al [28]. The results of these studies led to the construction of a model for KIR genomic organization and led the authors to define four KIR3DL loci, one KIR3DH locus, two KIR2DL loci and one KIR1D locus which they could apply to studies of rhesus macaques. This is clearly an approach that may provide some important clues as to the role of KIR haplotypes/MHC and its association with SIV VL and disease progression. However, to establish specific function association between KIR's and MHC molecules will require knowledge of the complete sequences of the entire KIR loci in efforts to establish TaqMan like procedures for the typing of KIR alleles for each of the loci. However, this is a large endeavor that will require a major resource.
In summary, results of these studies combined with previous studies [30] suggest that select KIR3DL and KIR3DH alleles appear to be associated with influencing high viral loads in SIV rhesus macaques in our colony at the Yerkes National Primate Research Center. It would be important to determine if a similar association is also seen in rhesus macaques of Indian origin at other Primate Centers by analysis of both previously archived samples and in prospective studies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are deeply grateful to the veterinary and support staff of the Yerkes National Primate Research Center of Emory University for maintaining and care of all the non-human primates utilized in the studies reported herein. The special efforts of Ms. Stephanie Ehnert are gratefully acknowledged.
Supported by NIH NIAID RO1 AI078773, NIH-NCRR-DRR base grant 000165 to the Yerkes National Primate Research Center, the Thailand Research Fund – Senior Research Scholar and Royal Golden Jubilee and the Grant Agency of the Czech Republic, grant no. P304-10-1161.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Alter G, Altfeld M. NK cells in HIV-1 infection: evidence for their role in the control of HIV-1 infection. J Intern Med. 2009;265:29–42. doi: 10.1111/j.1365-2796.2008.02045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pereira LE, Ansari AA. A case for innate immune effector mechanisms as contributors to disease resistance in SIV-infected sooty mangabeys. Curr HIV Res. 2009;7:12–22. doi: 10.2174/157016209787048465. [DOI] [PubMed] [Google Scholar]
- [3].Shattock RJ, Haynes BF, Pulendran B, Flores J, Esparza J. Improving defences at the portal of HIV entry: mucosal and innate immunity. PLoS Med. 2008;5:e81. doi: 10.1371/journal.pmed.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rhee EG, Barouch DH. Translational Mini-Review Series on Vaccines for HIV: Harnessing innate immunity for HIV vaccine development. Clin Exp Immunol. 2009;157:174–80. doi: 10.1111/j.1365-2249.2009.03928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jonsson AH, Yokoyama WM. Assessing licensing of NK cells. Methods Mol Biol. 612:39–49. doi: 10.1007/978-1-60761-362-6_4. [DOI] [PubMed] [Google Scholar]
- [6].Johansson S, Salmon-Divon M, Johansson MH, Pickman Y, Brodin P, Karre K, Mehr R, Hoglund P. Probing natural killer cell education by Ly49 receptor expression analysis and computational modelling in single MHC class I mice. PLoS One. 2009;4:e6046. doi: 10.1371/journal.pone.0006046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cooper MA, Colonna M, Yokoyama WM. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO Rep. 2009;10:1103–10. doi: 10.1038/embor.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Saito S, Nakashima A, Myojo-Higuma S, Shiozaki A. The balance between cytotoxic NK cells and regulatory NK cells in human pregnancy. J Reprod Immunol. 2008;77:14–22. doi: 10.1016/j.jri.2007.04.007. [DOI] [PubMed] [Google Scholar]
- [9].Lunemann A, Lunemann JD, Munz C. Regulatory NK-cell functions in inflammation and autoimmunity. Mol Med. 2009;15:352–8. doi: 10.2119/molmed.2009.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Moretta A, Marcenaro E, Parolini S, Ferlazzo G, Moretta L. NK cells at the interface between innate and adaptive immunity. Cell Death Differ. 2008;15:226–33. doi: 10.1038/sj.cdd.4402170. [DOI] [PubMed] [Google Scholar]
- [11].Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci U S A. 2009;106:1915–9. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sun JC, Lanier LL. Natural killer cells remember: an evolutionary bridge between innate and adaptive immunity? Eur J Immunol. 2009;39:2059–64. doi: 10.1002/eji.200939435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pereira LE, Johnson RP, Ansari AA. Sooty mangabeys and rhesus macaques exhibit significant divergent natural killer cell responses during both acute and chronic phases of SIV infection. Cell Immunol. 2008;254:10–9. doi: 10.1016/j.cellimm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- [14].Biassoni R. Human natural killer receptors, co-receptors, and their ligands. Curr Protoc Immunol Chapter. 2009;14 doi: 10.1002/0471142735.im1410s84. Unit 14 10. [DOI] [PubMed] [Google Scholar]
- [15].Jonsson AH, Yokoyama WM. Natural killer cell tolerance licensing and other mechanisms. Adv Immunol. 2009;101:27–79. doi: 10.1016/S0065-2776(08)01002-X. [DOI] [PubMed] [Google Scholar]
- [16].Brown MG, Scalzo AA. NK gene complex dynamics and selection for NK cell receptors. Semin Immunol. 2008;20:361–8. doi: 10.1016/j.smim.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Long EO. Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Averdam A, Petersen B, Rosner C, Neff J, Roos C, Eberle M, Aujard F, Munch C, Schempp W, Carrington M, Shiina T, Inoko H, Knaust F, Coggill P, Sehra H, Beck S, Abi-Rached L, Reinhardt R, Walter L. A novel system of polymorphic and diverse NK cell receptors in primates. PLoS Genet. 2009;5:e1000688. doi: 10.1371/journal.pgen.1000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–14. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- [20].Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C, Lifson JD, Allen TM, Carrington M, Altfeld M. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–36. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Carrington M, Martin MP, van Bergen J. KIR-HLA intercourse in HIV disease. Trends Microbiol. 2008;16:620–7. doi: 10.1016/j.tim.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].McClure HM, Anderson DC, Fultz PN, Ansari AA, Lockwood E, Brodie A. Spectrum of disease in macaque monkeys chronically infected with SIV/SMM. Vet Immunol Immunopathol. 1989;21:13–24. doi: 10.1016/0165-2427(89)90126-8. [DOI] [PubMed] [Google Scholar]
- [24].Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–30. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–32. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, Hawkins OE, Piaskowski S, Wilson NA, Hildebrand WH, Watkins DI, Sette A. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–75. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hershberger KL, Shyam R, Miura A, Letvin NL. Diversity of the killer cell Ig-like receptors of rhesus monkeys. J Immunol. 2001;166:4380–90. doi: 10.4049/jimmunol.166.7.4380. [DOI] [PubMed] [Google Scholar]
- [28].Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O'Connor DH. Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol. 2008;181:6301–8. doi: 10.4049/jimmunol.181.9.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. Evidence for balancing selection acting on KIR2DL4 genotypes in rhesus macaques of Indian origin. Immunogenetics. 2009;61:503–12. doi: 10.1007/s00251-009-0379-6. [DOI] [PubMed] [Google Scholar]
- [30].Bostik P, Kobkitjaroen J, Tang W, Villinger F, Pereira LE, Little DM, Stephenson ST, Bouzyk M, Ansari AA. Decreased NK cell frequency and function is associated with increased risk of KIR3DL allele polymorphism in simian immunodeficiency virus-infected rhesus macaques with high viral loads. J Immunol. 2009;182:3638–49. doi: 10.4049/jimmunol.0803580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol Rev. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- [32].Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093–100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].lannello A, Debbeche O, Samarani S, Ahmad A. Antiviral NK cell responses in HIV infection: I. NK cell receptor genes as determinants of HIV resistance and progression to AIDS. J Leukoc Biol. 2008;84:1–26. doi: 10.1189/jlb.0907650. [DOI] [PubMed] [Google Scholar]
- [34].Miah SM, Hughes TL, Campbell KS. KIR2DL4 differentially signals downstream functions in human NK cells through distinct structural modules. J Immunol. 2008;180:2922–32. doi: 10.4049/jimmunol.180.5.2922. [DOI] [PubMed] [Google Scholar]
- [35].Yu YR, Tian XH, Wang Y, Feng MF. Rapid production of human KIR2DL4 extracellular domain and verification of its interaction with HLA-G. Biochemistry (Mosc) 2006;71(Suppl 1):S60–4. 4–5. doi: 10.1134/s0006297906130104. [DOI] [PubMed] [Google Scholar]
- [36].Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- [37].Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, Tsoukas CM, Bernard NF. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J Immunol. 184:2057–64. doi: 10.4049/jimmunol.0902621. [DOI] [PubMed] [Google Scholar]
- [38].LaBonte ML, Levy DB, Letvin NL. Characterization of rhesus monkey CD94/NKG2 family members and identification of novel transmembrane-deleted forms of NKG2-A, B, C, and D. Immunogenetics. 2000;51:496–9. doi: 10.1007/s002510050650. [DOI] [PubMed] [Google Scholar]
- [39].Boyson JE, McAdam SN, Gallimore A, Golos TG, Liu X, Gotch FM, Hughes AL, Watkins DI. The MHC E locus in macaques is polymorphic and is conserved between macaques and humans. Immunogenetics. 1995;41:59–68. doi: 10.1007/BF00182314. [DOI] [PubMed] [Google Scholar]
- [40].Knapp LA, Cadavid LF, Watkins DI. The MHC-E locus is the most well conserved of all known primate class I histocompatibility genes. J Immunol. 1998;160:189–96. [PubMed] [Google Scholar]
- [41].LaBonte ML, McKay PF, Letvin NL. Evidence of NK cell dysfunction in SIV-infected rhesus monkeys: impairment of cytokine secretion and NKG2C/C2 expression. Eur J Immunol. 2006;36:2424–33. doi: 10.1002/eji.200635901. [DOI] [PubMed] [Google Scholar]
- [42].Zeddou M, Rahmouni S, Vandamme A, Jacobs N, Frippiat F, Leonard P, Schaaf-Lafontaine N, Vaira D, Boniver J, Moutschen M. Downregulation of CD94/NKG2A inhibitory receptors on CD8+ T cells in HIV infection is more pronounced in subjects with detected viral load than in their aviraemic counterparts. Retrovirology. 2007;4:72. doi: 10.1186/1742-4690-4-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yunis EJ, Romero V, Diaz-Giffero F, Zuniga J, Koka P. Natural Killer Cell Receptor NKG2A/HLA-E Interaction Dependent Differential Thymopoiesis of Hematopoietic Progenitor Cells Influences the Outcome of HIV Infection. J Stem Cells. 2007;2:237–248. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.