Abstract
Central nervous system (CNS) development, homeostasis, stress responses, and plasticity are all mediated by epigenetic mechanisms that modulate gene expression and promote selective deployment of functional gene networks in response to complex profiles of interoceptive and environmental signals. Thus, not surprisingly, disruptions of these epigenetic processes are implicated in the pathogenesis of a spectrum of neurological and psychiatric diseases. Epigenetic mechanisms involve chromatin remodeling by relatively generic complexes that catalyze DNA methylation and various types of histone modifications. There is increasing evidence that these complexes are directed to their sites of action by long non-protein-coding RNAs (lncRNAs), of which there are tens if not hundreds of thousands specified in the genome. LncRNAs are transcribed in complex intergenic, overlapping and antisense patterns relative to adjacent protein-coding genes, suggesting that many lncRNAs regulate the expression of these genes. LncRNAs also participate in a wide array of subcellular processes, including the formation and function of cellular organelles. Most lncRNAs are transcribed in a developmentally regulated and cell-type specific manner, particularly in the CNS, wherein over half of all lncRNAs are expressed. While the numerous biological functions of lncRNAs are yet to be characterized fully, a number of recent studies suggest that lnRNAs are important for mediating cell identity. This function seems to be especially important for generating the enormous array of regional neuronal and glial cell subtypes that are present in the CNS. Further studies have also begun to elucidate additional roles played by lncRNAs in CNS processes, including homeostasis, stress responses and plasticity. Herein, we review emerging evidence that highlights the expression and function of lncRNAs in the CNS and suggests that lncRNA deregulation is an important factor in various CNS pathologies including neurodevelopmental, neurodegenerative and neuroimmunological disorders, primary brain tumors, and psychiatric diseases.
Keywords: CoREST, epigenetic, long non-coding RNA (lncRNA), neural stem cell (NSC), neuron, non-coding RNA (ncRNA), oligodendrocyte, repressor element-1 silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF)
Introduction
The human central nervous system (CNS) is the most highly evolved and sophisticated biological system. It is comprised of an enormous array of distinct regional neuronal and glial cell subtypes that are organized into dynamic neural networks, which are, in turn, responsible for mediating the functional repertoire of the CNS including its ability to perform higher order cognitive and behavioral functions (Graff and Mansuy, 2008). One of the central aims of modern neurobiology is to understand the molecular mechanisms that underpin the elaboration of these neural cells and neural networks, and recent advances in epigenetic sciences have uncovered novel insights into these processes (MacDonald and Roskams, 2009; Mehler and Mattick, 2007; Mehler, 2008). Developmental stage- and cell type-specific epigenetic mechanisms are now thought to be responsible for producing, maintaining, and refining neural cell identity and function, by regulating the selective deployment of gene networks throughout life in response to interoceptive and environmental stimuli. Therefore, epigenetic processes are also implicated in mediating CNS homeostasis, stress responses, plasticity and disease (MacDonald and Roskams, 2009; Robertson, 2005; Tsankova et al., 2007). These epigenetic regulatory mechanisms involve chromatin remodeling via DNA methylation and histone code modifications at a plethora of sites around the genome and are mediated by an extraordinary array of generic enzymes/complexes/molecular scaffolds that include Polycomb- and Trithorax-group proteins, which are essential for most if not all developmental processes and programs (Kouzarides, 2007; Ringrose and Paro, 2007; Schwartz and Pirrotta, 2007). What determines the locus-selectivity of these enzymes is uncertain, but recent evidence suggests that they are recruited to their sites of action by non-protein-coding RNAs (ncRNAs) (Dinger et al., 2008; Khalil et al., 2009; Mattick et al., 2009).
The precise temporal and spatial expression of ncRNAs appears to be exceptionally important for mediating CNS form and function. Genomic organization is extremely intricate and encompasses multiple layers of regulatory and functional elements, including many interleaved, overlapping and antisense protein-coding mRNA and ncRNA transcripts (Carninci et al., 2005; Cheng et al., 2005; Kapranov et al., 2007a; Kapranov et al., 2007b; Katayama et al., 2005; Mattick and Makunin, 2006). Indeed, the genome is transcribed into a spectrum of ncRNAs that are implicated in a wide range of structural, regulatory, and catalytic processes. Some of these classes of ncRNAs are well known, such as ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), whose roles in mediating the pathogenesis of CNS disorders (e.g., mitochondrial enchephalopathies) have been characterized, at least in part (Sproule and Kaufmann, 2008). Additional classes of ncRNAs that have more recently been identified include a wide range of long ncRNAs (lncRNAs) and various types of short ncRNAs (microRNAs [miRNAs], piwiRNAs [piRNAs], small nucleolar RNAs [snoRNAs], promoter-associated small RNAs [PASRs] and transcription initiation RNAs [tiRNAs], among others) (Taft et al., 2010). This ncRNA circuitry appears to be selectively and dynamically deployed in each neural cell, and the developing and adult nervous systems exhibit specific regional, cellular and subcellular localization profiles of ncRNAs (Fineberg et al., 2009; Mehler and Mattick, 2007; Mehler, 2008; Mercer et al., 2008b; Mercer et al., 2010; Ponjavic et al., 2009; Royo and Cavaille, 2008). These environmentally sensitive ncRNA networks are thought to efficiently couple bioenergetic properties with information storage and processing capacity and to be responsible for orchestrating a wide array of biological processes (Mattick, 2003; St Laurent and Wahlestedt, 2007). In the CNS, these factors are implicated in mediating critical functions including brain patterning, neural stem cell (NSC) maintenance, neurogenesis and gliogenesis, stress responses, homeostasis, and synaptic and neural network connectivity and plasticity (Mehler and Mattick, 2007; Mehler, 2008). Therefore, not surprisingly, perturbations in the expression and function of these ncRNAs are increasingly being linked to the molecular pathophysiology of CNS disorders (Mehler and Mattick, 2007; Mehler, 2008; Taft et al., 2010).
Although lncRNAs are one of the most abundant classes of ncRNAs encoded within the genome and are highly expressed in brain (Mercer et al., 2008b; Ponjavic et al., 2009; Ravasi et al., 2006); see below), they remain poorly characterized, and their roles in the CNS have not been studied in detail. This class of ncRNA generally encompasses transcripts longer than 200 nt, of which there are tens if not hundreds of thousands expressed from mammalian genomes (Birney et al., 2007; Carninci et al., 2005; Cheng et al., 2005; Kapranov et al., 2007a; Katayama et al., 2005). Many lncRNAs are transcribed from genomic loci exhibiting chromatin signatures that indicate their transcription is dynamically regulated in a cell type-specific manner (Guttman et al., 2009). In addition, many lncRNAs are 5′ capped, polyadenylated, and spliced, like mRNAs (Carninci et al., 2005; Kapranov et al., 2007a; Okazaki et al., 2002; Ponjavic et al., 2007), although others are not (Cheng et al., 2005). Biophysical analyses of lncRNAs suggest that they can form a myriad of functional secondary structures (Pedersen et al., 2006; Torarinsson et al., 2008; Washietl et al., 2005). Some lncRNAs also serve as precursors for shorter regulatory ncRNAs (e.g., snoRNAs and miRNAs) (Mattick and Makunin, 2005). The functional properties of lncRNAs seem to be associated, in part, with their genomic architecture. Some lncRNAs are found in intergenic regions while others are organized in antisense, bi-directional, or intronic configurations with key protein-coding genes. These pairs of lncRNAs and protein-coding mRNAs exhibit expression profiles that are often highly complex, including concordant and discordant patterns (Dinger et al., 2008; Guttman et al., 2009; Khalil et al., 2009; Mercer et al., 2008b; Mercer et al., 2009; Mercer et al., 2010; Pang et al., 2009; Ponjavic et al., 2009), suggesting that lncRNAs play diverse roles in regulating the expression of associated protein-coding genes.
A major function of lncRNAs appears to be to modulate the epigenetic status of proximal and distal protein-coding genes through cis- and trans-acting mechanisms that include the recruitment of chromatin remodeling complexes to specific genomic loci thereby regulating chromatin structure over a single gene promoter, a gene cluster, or an entire chromosome (Dinger et al., 2008; Khalil et al., 2009; Mattick et al., 2009; Ng et al., 2007; Redrup et al., 2009). For example, HOTAIR is a lncRNA transcribed from the HOXC locus that recruits the Polycomb group (PcG) chromatin remodeling complex, PRC2, to the HOXD locus where it creates a repressive chromatin environment across 40 kb of the locus (Rinn et al., 2007). Moreover, a recent study examining a subset of human intergenic lncRNAs showed that a significant proportion is bound by PRC2, either alone or in combination with other chromatin remodeling complexes, such as those formed by CoREST and SMCX (Khalil et al., 2009).
LncRNAs also fulfill a range of other functions in cell and developmental biology, including interaction with promoter elements and transcription factors to modulate transcriptional activity. For example, the lincRNA Evf2 is transcribed from an ultraconserved distal enhancer that recruits positive (i.e., DLX) and negative (i.e., MECP2) transcription factors to the enhancer to modulate the expression of adjacent protein-coding genes (Bond et al., 2009). Furthermore, through sequence specific interactions, lncRNAs can regulate mRNA post-transcriptional processing and translation (Beltran et al., 2008). LncRNAs can also participate in forming structural compartments of the cell. For example, the lncRNA Neat1 (also known as MEN ε/β) is an essential architectural and functional component of paraspeckles, a nuclear subdomain implicated in the regulation of mRNA nuclear export (Bond and Fox, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009), which is specifically induced upon cell differentiation (Chen and Carmichael, 2009), including neuronal differentiation (M.B. Clark and J.S. Mattick, unpublished observations). Similarly, the lncRNA Gomafu is expressed in a subset of differentiating neural progenitor cells and post-mitotic neurons, and is localized in a novel nuclear microdomain (Sone et al., 2007).
Many lncRNAs are also dynamically expressed in the nervous system of other species, including insects (for review see (Amaral and Mattick, 2008). Some lncRNAs are relatively conserved across different species implying that they play important biological roles common to those species (Marques and Ponting, 2009; Ponting et al., 2009). On the other hand, some lncRNAs are not well conserved but are known to be functional, suggesting that they may have been subject to lineage-specific selection pressures and evolutionary innovations associated with phenotypic divergence (Pang et al., 2006; Pheasant and Mattick, 2007). It is also possible that many transcripts may have diverged in primary sequence but still retained elements of conserved secondary structure (Torarinsson et al., 2008; Washietl et al., 2005). The potential adaptive roles played by lncRNAs in the human CNS are highlighted by a recent study that reported non-coding sequences comprise 47 of 49 regions of the human genome that are highly conserved among mammalian species but show accelerated changes in the human lineage since divergence from our common primate ancestor (Pollard et al., 2006). These observations suggest that recent adaptive selection of these regions may have given rise to innovations in human brain form and function. In fact, the lncRNA HAR1F is transcribed from one of these regions and specifically co-expressed in Cajal-Retzius cells of the human neocortex with the critical neural factor RELN (Pollard et al., 2006), which mediates seminal neural developmental processes and is implicated in the pathophysiology of a broad range of neurological and psychiatric disorders (Botella-Lopez et al., 2009; D’Arcangelo, 2006; Muller et al., 2009; Pisante et al., 2009; Serajee et al., 2006; Shifman et al., 2008; Tamura et al., 2007; Won et al., 2006).
lncRNAs in the central nervous system
Highly environmentally sensitive epigenetic processes are responsible for integrating complex cell-intrinsic and local- and long-distance environmental signals that include specific temporal and spatial profiles of gradient morphogens, growth factors, additional cell signaling cues, combinatorial transcription factor codes and neuronal activity, which together orchestrate the selective deployment of genes and functional gene networks that establish, maintain, and refine neural cell identity and connectivity throughout life (Mehler, 2008). LncRNAs are often located proximal to genes encoding regulatory proteins suggesting that they play a key role in these processes. Indeed, a number of transcriptomic studies have begun to reveal dynamic profiles of lncRNA expression and function in developing and adult tissues, including embryonic stem (ES) cells (Dinger et al., 2008; Sheik Mohamed et al., 2009), the immune system (Pang et al., 2009), muscle (Sunwoo et al., 2009), the vascular system (Li et al., 2009), the retina (Blackshaw et al., 2004; Rapicavoli and Blackshaw, 2009; Young et al., 2005), neural cell subtypes (Mercer et al., 2010), and the brain (Mercer et al., 2008b; Ponjavic et al., 2009).
lncRNA expression in brain and neural differentiation
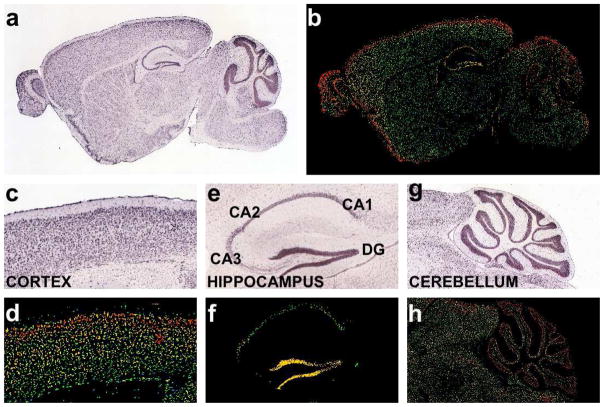
A study utilizing data from the Allen Brain Atlas found that, of 1,328 lncRNAs examined, 849 are expressed within the adult mouse brain, with almost half (623) exhibiting selective profiles for specific regions, cell types, and subcellular compartments (Mercer et al., 2008b). It should be noted that the majority are easily detectable by in situ hybridization in particular cells in, for example, the olfactory bulb, hippocampus, cortex or cerebellum (Fig. 1), which might be expected if these RNAs are involved in regulating specific processes, and which also explains their low abundance in whole brain transcriptomic profiling analyses. Similarly the Caenorhabditis elegans miRNA lsy-6, which was discovered by genetic studies to control left-right asymmetry in taste-receptor neurons, is expressed in a very limited subset of neurons and was initially difficult to verify biochemically (Johnston and Hobert, 2003). Thus, it will be important to undertake deeper and more focused transcriptomic studies on specific regions and specific cells in order to reveal the full repertoire of lncRNAs in the brain.
Figure 1. Illustrative examples of regionally enriched long non-coding RNA expression profiles in adult mouse brain.
(a) In situ hybridization and (b) expression views displaying expression levels (red - high, yellow - medium, green - low) for AK019375, which is broadly but preferentially expressed in the olfactory bulb, hippocampus, and cerebellar cortex. (c) In situ hybridization and (d) expression views for AK017599, which is enriched in multiple layers of the cerebral cortex. (e) In situ hybridization and (f) expression views for AK157548, which is enriched in hippocampal CA1-3 subregions and the dentate gyrus (DG). (g) In situ hybridization and (h) expression views for AK050124, which is enriched in the granule cell and Purkinje cell layers of the cerebellum (Images courtesy of the Allen Brain Atlas - Allen Institute for Brain Science, Seattle).
The first round of three-dimensional studies in the adult mouse brain (Mercer et al., 2008b) provides clear evidence that the majority of all lncRNAs, of which there are at least 30,000 (Carninci et al., 2005) and possibly an order of magnitude more, are expressed in the nervous system. Many of these lncRNAs are derived from complex genomic loci including those that are imprinted and those that encompass key neural protein-coding genes, in cis-antisense, intronic, or bidirectional configurations. Further, many of these pairs exhibited conservation of their genomic organization in other species, implying that the relationships are meaningful. A complementary study examining a more restricted subset of intergenic lncRNAs enriched for evolutionarily constrained sequences showed that over 200 of these lncRNAs are expressed in the developing and adult mouse brain (Ponjavic et al., 2009). Intriguingly, these lncRNAs are largely derived from genomic loci located proximal to protein-coding genes with similar expression profiles in the brain. Moreover, the majority of these protein-coding genes are transcriptional regulators and other factors implicated nervous system development. Intriguingly, a significant proportion of lncRNAs expressed in the mouse brain are transcribed from genomic loci adjacent to protein-coding genes expressed in the vomeronasal organ and olfactory bulb supporting the emerging view that the expression of the odorant receptor repertoire is coordinated by epigenetic processes and specifically implicating lncRNAs as a key part of this regulatory mechanism (Kambere and Lane, 2007).
An additional study identified more than 1,000 evolutionarily conserved intergenic lncRNAs in mouse by analyzing chromatin signatures from four mouse cell types, including neural precursor cells (NPCs) (Guttman et al., 2009). A functional analysis of the expression of these lncRNAs revealed the presence of a “brain cluster” of lncRNAs that is associated with biological processes including hippocampal development, oligodendrocyte (OL) myelination, brain aging, CREB and PGC1-alpha transcriptional regulation, and GABAergic neuronal (GABAN), G protein coupled receptor and calcineurin signaling pathways. Another recent study demonstrated that 169 lncRNAs are differentially expressed during the sequential processes of mouse ventral forebrain-derived NSC mediated lineage restriction, GABAN and OL lineage specification, progressive OL lineage maturation, and terminal differentiation including myelination (Mercer et al., 2010). These dynamically regulated lncRNAs are also associated with protein-coding genes that play roles in diverse neural developmental processes including, for example, AK053922, a lncRNA transcribed from the Gli3 locus, and Sox8OT, a lncRNA transcribed from the Sox8 locus. The dynamic and context-selective Gli transcriptional response code mediates gradient morphogen signaling by Sonic hedgehog (SHH), a master regulator of brain development (Yu et al., 2009). Similarly, Sox8 is a SRY-box transcription factor that mediates progressive stages of OL maturation (Stolt and Wegner, 2009). In addition, this study found that lncRNAs not associated with neural genes are also differentially expressed during developmental transitions. For example, the BIC (B-cell integration cluster) lncRNA was upregulated during neurogenesis but downregulated during oligodendrogliogenesis suggesting that it may play a role in neuronal-glial fate transitions.
lncRNA regulation of cell fate decisions, cellular differentiation, synaptic plasticity and behavior
Detailed analyses of specific lncRNAs that are dynamically expressed in the CNS reveal potential roles in mediating neural cell fate decisions. For example, Sox2 is a key transcription factor that is required for neural induction and maintenance of neural stem and progenitor cells, and a recent study demonstrated that the Sox2OT lncRNA, which contains the Sox2 gene within one of its introns and is transcribed in the same direction (Fantes et al., 2003), is expressed in regions of constitutive adult neurogenesis (Mercer et al., 2008b). Another recent study demonstrated that Sox2OT is dynamically regulated in CNS structures during development, where it may be responsible for modulating Sox2 expression (Amaral et al., 2009). Similarly, Nkx2.2, a transcription factor that is critical for OL lineage specification, is also subject to regulation by a lncRNA, Nkx2.2AS, which is transcribed antisense to the Nkx2.2 gene. A recent study reported that forced expression of Nkx2.2AS in NSCs in vitro enhances their differentiation along the OL lineage, in part, by inducing an increase in Nkx2.2 mRNA levels (Tochitani and Hayashizaki, 2008). This observation implies not only that Nkx2.2AS has a regulatory effect on the transcription of Nkx2.2 in cis but also influences other factors responsible for OL lineage specification in trans. Together, these observations suggest that, in concert with cell–intrinsic and environmental signals, a range of lncRNA-mediated epigenetic mechanisms participate in orchestrating neural cell identity.
LncRNAs are also implicated in processes responsible for modulating synaptic plasticity and promoting long-term changes in synaptic strength. For example, the rodent-specific BC1 and primate-specific BC200 lncRNAs, which are derived from transposable elements and transcribed by RNA polymerase III, are selectively targeted to postsynaptic dendritic compartments, where they modulate local protein synthesis by repressing the initiation of translation through an eIF4A-dependent mechanism (Brosius, 1999; Kondrashov et al., 2005; Lin et al., 2008; Martignetti and Brosius, 1993). Knockout of BC1 in mice produces no overt phenotype in the cage, but behavioral phenotypes including reduced exploration and increased anxiety in less constrained environments, leading to reduction in survival rates (Lewejohann et al., 2004); see also below). Similarly, NTAB is a lncRNA that is expressed in developing and adult rat brain, where it is also found in neuronal processes (French et al., 2001).
LncRNAs are also involved in retinal development. In mouse, the lncRNA TUG1 was identified as being up-regulated by taurine, a cysteine derivative required for proper neural development. TUG1 is expressed during retinal development and its inactivation causes loss or malformation of the outer segments of photoreceptors and affects the expression of other genes involved in eye development (Young et al., 2005). TUG1 is also upregulated in T-cell differentiation (Pang et al., 2009), adding to the list of intriguing similarities and functional parallels between the brain and the immune system (Habibi et al., 2009; Mattick and Mehler, 2008). TUG1 is highly conserved in mammals but not found in other vertebrates, as is also the case for the lncRNA Gomafu discussed earlier.
Regulation of lncRNA expression in the nervous system
The factors influencing the expression of lncRNAs, in general, and within the CNS in particular, are not well characterized. However, the expression of lncRNAs and protein-coding genes is mediated by some common regulatory mechanisms, including morphogens and transcription factors (Cawley et al., 2004; Dinger et al., 2008; Guttman et al., 2009; Mercer et al., 2010; Zhang et al., 2009). For example, the transcription factor, Pax2, plays a role in patterning of the mouse embryonic midbrain and hindbrain, and Ncrms, a lncRNA, is specifically regulated by Pax2 in this region (Bouchard et al., 2005). Intriguingly, Ncrms serves as a host gene for miR-135a (Rodriguez et al., 2004), an oncogenic miRNA that is dysregulated in medulloblastoma (Ferretti et al., 2009). These observations illustrate the complex bidirectional relationships that exist between the genetic and epigenetic networks mediating oncogenesis.
In addition, recent evidence suggests not only that lncRNAs regulate epigenetic processes (Mattick et al., 2009) but also that perturbations of these processes can alter the expression of lncRNAs. For example, a study performed utilizing an in vitro OL developmental paradigm showed dynamic changes in long ncRNA expression profiles in response to treatment with trichostatin A (TSA), a histone deacetylase inhibitor that prevents maturation of OL progenitors by suppressing OL-specific gene expression (Mercer et al., 2010). These findings indicate that lncRNAs are regulated by the same transcriptional and epigenetic mechanisms as protein-coding genes.
Furthermore, a recent study found that the master epigenetic regulator, repressor element-1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF), plays a role in modulating the expression of lncRNAs (Johnson et al., 2009). This study determined that a significant proportion of repressor element 1/neuron restrictive silencer element (RE1/NRSE) REST binding motifs are within 10 kb of lncRNAs and subsequently verified that a number of these lncRNAs are targets of REST regulation, in both mouse and human (Johnson et al., 2009). This study raises a number of interesting possibilities because the roles of REST in the CNS have been the focus of intense examination: Firstly, RE1-associated genes can be modulated by the independent or combinatorial actions of REST and CoREST, which both serve as dynamic modular platforms for the recruitment of a diverse array of factors that participate in genomic locus-specific and more widespread epigenetic remodeling (Qureshi and Mehler, 2009). Thus, CoREST is also likely to have a role in regulating the expression of a subset of lncRNAs. Secondly, distinct but overlapping cell type- and developmental stage-specific REST and CoREST transcriptional networks are implicated in modulating seminal neural developmental and homeostatic processes (Qureshi and Mehler, 2009). Therefore, lncRNAs that are regulated by REST and CoREST may play context-specific roles in NSC maintenance, neuronal and glial cell specification, progressive maturation, terminal differentiation, and activity dependent plasticity. Thirdly, many lncRNAs are bound to chromatin remodeling complexes containing CoREST or other factors, implying that chromatin remodeling complexes containing REST are similarly associated, directly or indirectly, with the function of lncRNAs (Khalil et al., 2009). In addition, deregulation of REST and CoREST functions is linked to a range of CNS pathologies that include cancer (i.e., glioblastoma, medulloblastoma, and neuroblastoma), neurodegenerative disease (i.e., Huntington’s disease), neurodevelopmental disorders (i.e., Down syndrome and X-linked mental retardation [XLMR]), epilepsy, and ischemia (Qureshi and Mehler, 2009).
lncRNAs in diseases of the central nervous system
Neurodevelopmental disorders
LncRNAs are implicated in the pathophysiology of neurodevelopmental disorders associated with genomic imprinting, such as Prader-Willi syndrome (PWS) and Angelman syndrome (AS) (Koerner et al., 2009). In fact, a number of paternally expressed lncRNAs are derived from this imprinted cluster, although their specific roles are not well characterized. Some of these lncRNAs are responsible for epigenetic gene regulation within the imprinted cluster indirectly by serving as hosts for snoRNAs. Other lncRNAs may be directly involved in modulating gene expression within the imprinted cluster. For example, Ube3a-as is a lncRNA transcribed antisense to the maternally expressed Ube3a gene, a candidate gene for AS, suggesting that Ube3a-as may be responsible for repressing paternal Ube3a expression. Indeed, some studies have shown that repression of Ube3a is dependent on Ube3a-as (Chamberlain and Brannan, 2001; Johnstone et al., 2006). However, other data has demonstrated that silencing of paternal Ube3a can occur in the absence of Ube3a-as and implies a more complex regulatory relationship underlying the imprinting of Ube3a (Le Meur et al., 2005). Intriguingly, a recent finding suggests that lncRNAs derived from the PWS-AS domain are nuclear-retained mediating the spatial organization of gene expression through dynamic modulation of nuclear architecture (Vitali et al., 2010).
LncRNAs may influence the pathogenesis of fragile X syndrome (FXS) and fragile X-associated tremor and ataxia syndrome (FXTAS), which are, respectively, caused by mutation and pre-mutation in the protein-coding FMR1 gene. FMR4 is a primate-specific lncRNA that appears to share a bidirectional promoter with the FMR1 gene (Khalil et al., 2008). A recent study showed that, like FMR1, FMR4 is also silenced in FXS patients because of a CGG expansion repeat in the 5′ untranslated region (UTR) of the FMR1 gene and up regulated in pre-mutation carriers (Khalil et al., 2008). Further, short interfering RNA (siRNA) mediated knockdown of FMR4 did not affect FMR1 expression, suggesting that FMR4 does not simply regulate FMR1 and also that its expression might independently contribute to the clinical presentation of FXS. In fact, siRNA knockdown of FMR4 resulted in alterations in cell cycle regulation and increased apoptotic cell death, whereas over-expression of FMR4 caused an increase in cell proliferation. Similarly, another lncRNA, ASFMR1, which is derived from the FMR1 locus, may also be important in mediating the complex clinical phenotypes associated with mutations at this genomic site. A recent study showed that ASFMR1 is a spliced and polyadenylated antisense transcript, which overlaps the 5′ UTR CGG repeat region of FMR1 (Ladd et al., 2007). Intriguingly, alternative splicing of ASFMR1 seems to exhibit premutation-specific profiles. Further, like FMR1 and FMR4, ASFMR1 is also silenced in FXS patients and up regulated in pre-mutation carriers suggesting that a common process is responsible for regulating the expression these transcripts. In fact, binding sites for the CTCF chromatin insulator protein flank the CGG repeat suggesting that CTCF establishes the local chromatin structure at the repeats. Indeed, this mechanism plays a role in regulation of ncRNA transcription and establishing local chromatin structure at other expansion repeat disease loci (e.g., DM1) (Cho et al., 2005; Filippova et al., 2001).
In addition, lncRNAs may play a role in the development of brain malformations. Genetic defects of Sox2 cause various syndromes of micropthalmia and of optic nerve hypoplasia associated with a number of CNS developmental abnormalities. The Sox2OT gene encompasses the entire Sox2 gene, is dynamically regulated in the CNS during development, and is implicated in modulating Sox2 expression (Amaral et al., 2009). Therefore, a direct role for Sox2OT or an indirect role through effects on Sox2 in mediating the clinical features of these and related syndromes cannot be excluded. Similarly, velocardiofacial syndrome (VCFS) or DiGeorge Syndrome is a clinically heterogeneous disorder characterized by developmental brain malformations, cognitive and behavioral abnormalities, and an increased risk of psychiatric disorders (i.e., schizophrenia and bipolar disorder). Intriguingly, VCFS is caused by deletions of the 2q11.2 chromosomal region that includes DGCR5, a REST regulated lncRNA, which suggests a potential role for this lncRNA in mediating neural developmental processes and the phenotype of this disorder (Johnson et al., 2009).
Further, lncRNAs may be involved in the pathobiology of Down’s syndrome (DS). NRON is a lncRNA that mediates the cytoplasmic to nuclear shuttling of the NFAT transcription factor (Willingham et al., 2005). In animal models, deregulation of the DSCR1 and DYRK1A genes act synergistically to prevent nuclear occupancy of NFATc transcription factors leading to reduced NFATc activity and to many features of DS, suggesting a potential link between NRON activity and DS pathophysiology (Arron et al., 2006).
Neurodegenerative disorders
A lncRNA may influence the pathogenesis of Alzheimer’s disease (AD). BACE1 (β-site amyloid precursor protein-cleaving enzyme 1) is an enzyme that cleaves amyloid precursor protein (APP) and generates amyloid β (Aβ) peptides, which form amyloid plaques in the brains of patients with AD. The mechanisms that regulate the expression and function of BACE1 in AD are complex and not completely understood, but they include a conserved antisense transcript, BACE1-AS, which modulates BACE1 gene expression. BACE1-AS levels are increased in tissues from AD patients and in an APP transgenic mouse model of AD (Faghihi et al., 2008). In addition, neuronal cells exposed to diverse cell stressors (e.g., reactive oxygen species, chronic hypoxia, and Aβ1–42) exhibited increased expression and nuclear to cytoplasmic translocation of BACE1-AS transcripts, where BACE1-AS promotes the stabilization of BACE1 mRNA and up-regulation of BACE1 protein, which, in turn, leads to production of Aβ peptide. These findings imply that BACE1-AS is deregulated in AD, which induces feed-forward regulation of BACE1, increases Aβ levels, and thus may promote the pathogenesis of AD. These observations are particularly interesting because modulating amyloidogenic APP metabolism represents an important candidate strategy for treating AD, and targeting BACE1-AS may circumvent some of the challenges posed by current approaches aimed at inhibiting β-secretase activity. In fact, identification of potent and selective β-secretase inhibitors has been difficult, with only a single β-secretase inhibitor drug candidate, CTS-21166, having advanced into Phase I clinical trials because of medicinal chemistry issues, such as the properties of the β-secretase active site (Frisardi et al., 2009).
Another study utilized human AD brain tissue to link alterations in levels of a lncRNA, BC200, with AD pathogenesis (Mus et al., 2007). Increased levels of BC200 were found in brain regions that are preferentially affected in AD, such as Brodmann’s area 9 and the hippocampus, which correlated with disease severity measured by Clinical Dementia Rating scores. Further, in advanced stages of AD, BC200 was mis-localized and clustered in the perikaryon. These observations suggest that deregulation of these synaptic lncRNAs is involved in the synaptic and neural network dysfunction that is found in both early and later stages of AD.
In addition, a lncRNA also plays a key role in the pathogenesis of spinocerebellar ataxia type 8 (SCA8), an autosomal dominant disorder caused by an expansion repeat. SCA8 is characterized by bidirectional transcription of this expansion repeat from opposite strands forming both a protein-coding transcript encoding a polyglutamine expansion, ATXN8, and a lncRNA transcript containing a CUG expansion, ATXN8OS (Moseley et al., 2006). Both of these are implicated in the molecular pathophysiology of the disease implying that SCA8 may be caused by toxic protein and RNA functions (Daughters et al., 2009; Koob et al., 1999; Moseley et al., 2006). Indeed, a recent study found that the expanded ATXN8OS transcript accumulates in ribonuclear inclusions in the cerebellar cortex (i.e., Purkinje cells, Bergmann glia, and molecular layer interneurons) of SCA8 patients and in the cerebellar cortex and the deep cerebellar nuclei of transgenic mice expressing the SCA8 expansion (Daughters et al., 2009). These inclusions co-localized with splicing factor, MBNL1. In addition, evaluation of the GABA-A transporter 4 (GAT4) revealed dysregulation of CUGBP1-MBNL1-mediated alternative splicing and loss of GABAN-mediated inhibition within the granular cell layer, which is a hallmark of the disease. Together, these observations suggest that the mutant ATXN8OS transcript contributes to SCA8 pathogenesis by altering the activity of MBNL/CELF alternative splicing proteins. This mechanism is similar to the pathogenic role played by the mutant DMPK transcript that causes myotonic dystrophy type 1 (DM1), which is encoded by a protein-coding gene containing a CUG expansion repeat in its 3′-untranslated region (Lee and Cooper, 2009). Intriguingly, these examples suggest a common pathogenic mechanism for both lncRNAs and protein-coding RNAs.
Furthermore, a lncRNA may be implicated in amyotrophic lateral sclerosis (ALS). Mutations in the FUS/TLS gene cause a subset of ALS cases and, intriguingly, FUS/TLS acts as an RNA-binding protein (RBP) that can be recruited by a lncRNA to the genomic locus encoding cyclin D1, where it represses cyclin D1 transcription (Wang et al., 2008). Because members of the cyclin-dependent kinase (Cdk) family are implicated in mediating apoptotic death of neurons, these observations may link aberrant FUS/TLS to neurodegeneration in ALS through abnormal lncRNA-mediated cyclin D1 transcriptional regulation. This mechanism may also influence the pathogenesis of other neurodegenerative diseases because FUS/TLS is also implicated in the neuropathology of spinal cerebellar ataxia types 1–3, dentatorubral-pallidoluysian atrophy, and Huntington’s disease (HD) (Doi et al., 2008; Doi et al., 2009).
HD is particularly interesting because it is caused by an expansion repeat mutation in the Htt gene, which encodes a mutant protein with a polyglutamine stretch that seems to be at the nexus of PRC2-, REST-, and ncRNA-associated transcriptional dysregulation, one of the hallmarks of HD (Benn et al., 2008; Johnson et al., 2008; Marullo et al., 2008; Packer et al., 2008; Qureshi and Mehler, 2009; Seong et al., 2010; Zuccato et al., 2007). Indeed, mutant Htt promotes aberrant nuclear-cytoplasmic trafficking of REST and leads to the deregulation of REST target gene expression in tissues from animal models of HD and human HD. These genes include both protein-coding genes as well as ncRNAs, such as miRNAs. Because REST also regulates the expression of lncRNAs, it is therefore likely that HD tissues are also characterized by dysregulation of lncRNA expression. Furthermore, if a subset of lncRNAs binds to REST chromatin-remodeling complexes as it does to CoREST macromolecular complexes, then the potential disruption of REST-regulated lncRNA expression in HD may lead to additional disturbances in lncRNA-mediated chromatin and transcriptional regulatory processes through a feed-forward mechanism. In fact, a recent study demonstrated that Htt acts as a molecular facilitator of the PRC2 complex, which is bound by a subset of lncRNAs, providing further evidence for a lncRNA-mediated final common pathway for transcriptional dysregulation and, thus, neurodegeneration in HD (Seong et al., 2010).
Neuroimmunological disorders
Multiple sclerosis (MS) is a complex autoimmune disease, and recent immunopathological studies implicate abnormal CD8+ T cell activity in the pathophysiology of MS (Friese and Fugger, 2009). Because lncRNAs are involved in CD8+ T cell differentiation and activation, lncRNAs may also be important in the development and progression of MS. In fact, lncRNA transcripts derived from the mouse T early α (TEA) promoter are responsible, in part, for regulating downstream promoter usage and, thus, for generating the diversity of the T cell receptor repertoire (Abarrategui and Krangel, 2007). Further, a recent study utilizing mouse CD8+ T cells identified hundreds of lncRNAs that are dynamically expressed during T cell differentiation and activation, including many transcribed from genomic loci encompassing protein-coding genes important for immune system functions and potentially for MS pathogenesis (Pang et al., 2009). For example, the mouse IL2RA locus encodes a number of lncRNAs that are nested within individual introns of the IL2RA gene, and the expression of one of these lncRNAs, M21981, is strongly up regulated with T cell activation. Homologous lncRNAs are present within the human IL2RA gene locus, and intriguingly, the human IL2RA locus has been identified by genome-wide association studies (GWAS) with susceptibility to MS (Hafler et al., 2007). In addition, Tmevpg1 is another lncRNA that may be involved in MS. It is found in human and mouse immune cells and is transcribed from a cluster of cytokine genes, which includes γ-interferon (Vigneau et al., 2003). In mouse, Tmevpg1 is believed to play a role in controlling the persistence of Theiler’s murine encephalomyelitis virus (TMEV) infection (Vigneau et al., 2003). Notably, TMEV infection serves as an experimental model for MS because it is characterized, in part, by chronic inflammatory demyelination with oligodendrocyte apoptosis and axonal degeneration (Tsunoda and Fujinami, 2009). These observations suggest that lncRNAs are responsible, at least in part, for mediating immune responses in the CNS.
Neuro-oncological disorders
LncRNAs are implicated in promoting the acquisition and maintenance of cell identity, which are perturbed in cancer, suggesting that some lncRNA species may be important in the process of cellular transformation. In fact, lncRNAs are important for mediating a range of processes that are aberrant in cancer, such as X chromosome inactivation (XCI), genomic imprinting and transcriptional regulation, and a number of systemic cancer phenotypes, including leukemia, colon cancer, prostate cancer, breast cancer and hepatocellular carcinoma, exhbit dysregulation of lncRNAs as a primary feature (Calin et al., 2007; Fu et al., 2006; Guffanti et al., 2009; Lin et al., 2007; Pibouin et al., 2002). Perturbations in lncRNA expression are also associated with CNS tumors.
H19 is an imprinted lncRNA expressed from the maternal allele that is located in a gene cluster, which also includes IGF2. H19 is expressed during embryogenesis, and subsequent deregulation of H19 and genes in the imprinted cluster is linked directly to cellular transformation and also associated indirectly with the development of a number of other tumors, including medulloblastomas, meningiomas and gliomas. For example, a study of medulloblastomas and medulloblastoma cell lines showed partial loss of imprinting (LOI) and biallelic expression of H19 (Albrecht et al., 1996). An examination of meningiomas (World Health Organization grades I-III) demonstrated more robustly that the imprinting status of H19 is perturbed with LOI in a significant number of these tumors (Muller et al., 2000). Another study performed in CD133+ and CD133− glioblastoma derived primary cell lines revealed levels of H19 expression that were relatively high and low, respectively (Beier et al., 2007). Mechanistically, H19 is a target of the GLI1 transcription factor, which mediates SHH signaling and is amplified more than 50-fold in human gliomas (Kinzler et al., 1987; Yoon et al., 2002). H19 has also been linked to the tumor suppressor, p53, which negatively regulates its expression (Dugimont et al., 1998) and the oncogene, c-myc, which positively regulates its expression in diverse cell types including T98G human glioblastoma cells (Barsyte-Lovejoy et al., 2006). In addition, H19 transcription is positively regulated by the cell cycle regulatory factor, E2F1, during the S-phase of growth-stimulated cells (Berteaux et al., 2005). Intriguingly, a lncRNA has recently been described that is transcribed antisense to H19 but not imprinted, implicated in regulating IGF2, and overexpressed in human cancer cells (Berteaux et al., 2008). The presence of this antisense lncRNA and its deregulation in cancer further highlights the complexity of epigenetic regulation within this gene cluster and may be relevant for CNS tumors.
Similarly, anti-NOS2A is a lncRNA that is expressed in meningiomas and glioblastomas from a genomic locus that evolved by duplication of the NOS2A gene followed by internal DNA inversion (Korneev et al., 2008). This intronless, non-polyadenylated lncRNA is implicated in negatively regulating the expression of NOS2A, which plays a role in neuronal differentiation of ES cells (Korneev et al., 2008). Further, NOS2A is induced in human brain tumors including glioblastoma and in glioma cell lines, where it can differentially influence the efficacy of chemotherapeutic agents (Broholm et al., 2003). This example may be particularly important because many other genes that are deregulated in CNS tumors, including oncogenes and tumor suppressor genes, have antisense lncRNAs encoded in the genome (Grinchuk et al., 2010).
Furthermore, abnormalities in pathways related to lncRNA regulation and function are associated with CNS tumors, supporting the conclusion that lncRNAs are important in modulating cellular transformation. For example, a recent study found a subset of p53 cis-regulatory element-associated lncRNAs that are specifically induced in response to DNA damage in p53 wild type cells but not in p53−/− cells (Guttman et al., 2009). These and other lncRNAs may be involved in the p53-mediated induction of cell-cycle arrest, DNA repair and apoptosis that protect neural cells from DNA damage and transformation (Tedeschi and Di Giovanni, 2009). REST also regulates lncRNA expression, and abnormalities of REST expression and function are associated with the development of medulloblastomas, neuroblastomas and glioblastomas (Qureshi and Mehler, 2009). These observations, coupled with the potential roles of lncRNAs in establishing and maintaining neural cell identity, suggest that REST-mediated lncRNA dysregulation may contribute to the development of these tumors. Disruption of chromatin regulation may be one the potential mechanisms by which lncRNAs play roles in oncogenesis. In fact, many lncRNAs participate in chromatin modulation through interactions with PRC2, and recent evidence suggests that genes epigenetically deregulated in glioblastoma are highly enriched for targets of PRC2 (Martinez et al., 2009). Further, subunits of the PRC2 complex, such as EZH2, are implicated in the development of glioblastomas and maintenance of cancer initiating stem cells (Abdouh et al., 2009). Similarly, many lncRNAs participate in chromatin modulation through interactions with CoREST, which is thought to modulate genes in key pathways responsible for CNS tumors (Abrajano et al., 2009a; Abrajano et al., 2009b).
Other neurological and psychiatric disorders
LncRNAs, such as BC1/BC200 and Evf2, may be involved in mediating the process of epileptogenesis because they modulate neural network plasticity and excitability (Mattick and Mehler, 2008; Mehler, 2008; Mercer et al., 2008a). For example, a recent study showed that BC1−/− animals exhibit neuronal hyperexcitability, significantly elevated gamma frequency oscillations on cortical electroencephalogram (EEG) recordings, and epileptogenesis (Zhong et al., 2009). Similarly, Evf2 mouse mutants display abnormal development of GABAN circuitry in the hippocampus and dentate gyrus leading to a reduction in inhibitory synaptic activity suggesting a predisposition to neuronal hyperexcitability (Bond et al., 2009).
LncRNAs may also influence the pathogenesis of Restless Legs Syndrome (RLS), a sensorimotor disorder that is associated with abnormal cerebellar activity (Bucher et al., 1997). The leading genetic risk factor for this disorder is variation at the Meis1 gene locus, which encodes a homeobox protein with roles in development and oncogenesis (Winkelmann et al., 2007). A recent study suggested that the predisposition to RLS results from reduced expression of Meis1 mediated by intronic cis-regulatory elements (Xiong et al., 2009). Intriguingly, in the developing mouse brain, Meis1 is co-expressed in the developing cerebellar granule cell layer along with a genomically-associated lncRNA AK042766 (Ponjavic et al., 2009). These observations raise the interesting possibility that lncRNA mediated mechanisms may regulate the expression of the Meis1 gene during development and adult life, thereby modulating the pathogenesis of this complex disorder.
In addition to neurological diseases, a number of psychiatric disorders have also been associated with lncRNAs. Specifically, the disruption of the DISC genomic locus, which encodes both the DISC1 protein-coding gene and the DISC2 lncRNA, has been linked in a number of genetic analyses to the risk of developing schizophrenia, schizoaffective disorder, bipolar disorder, major depression, and autistic spectrum disorders (Chubb et al., 2008; Millar et al., 2000; Williams et al., 2009). DISC2 overlaps DISC1 and is transcribed in the opposite direction. Like other antisense transcripts, DISC2 is implicated in regulating the expression of its partner, DISC1, which modulates multiple aspects CNS structure and function including embryonic and adult neurogenesis (Brandon et al., 2009). However, DISC2 may also represent an important candidate gene for psychiatric disease separately from its effects on DISC1 (Chubb et al., 2008).
Perspectives
Recent studies have dramatically changed our understanding of the genome and the transcriptome, and encouraged us to focus on examining the roles not only of protein-coding genes but also of diverse classes of interleaved ncRNAs to understand biological systems in general, and the sophistication of the CNS in particular. Indeed, RNA molecules have properties that make them uniquely suited to perform a spectrum of regulatory, structural, and catalytic functions. For example, RNAs can efficiently serve as highly sensitive biosensors for interoceptive and environmental signals and can dynamically store, process and integrate information through sequence-specific, digital, and conformational, analog, features (St Laurent and Wahlestedt, 2007). Because of these properties, ncRNAs can specifically regulate the temporal and spatial deployment of genes and functional gene networks by modulating the transcription, post-transcriptional processing, and translation of mRNAs. These functions are critical in the CNS, where ncRNAs also play additional roles in mediating bidirectional axodendritic transport and activity-dependent plasticity. Short ncRNAs, such as miRNAs, have been studied in detail, but in this review we surveyed data that indicates lncRNAs, the most abundant and least studied class of ncRNAs, are similarly important for mediating nervous system development, homeostasis, stress responses, plasticity and the pathophysiology of a spectrum of CNS pathologies including neurodevelopmental, neurodegenerative and neuroimmunological disorders, primary brain tumors, and psychiatric diseases.
It also appears increasingly likely that ncRNAs may be the primary substrate for environment-epigenome interactions mediated by RNA editing, especially in the brain. There are two classes of RNA editing/DNA recoding enzymes in animals, which function by deamination to catalyze adenosine-to-inosine (A-I) and cytidine-to-uridine (C-U) conversions (adenosine deaminases acting on RNAs [ADARs] and apolipoprotein B mRNA editing enzymes, catalytic polypeptide-like [APOBECs], respectively) (Bass, 2002; Navaratnam and Sarwar, 2006; Valente and Nishikura, 2005). ADARs are highly expressed in brain and ADAR3 is both vertebrate- and brain-specific. There is extensive A-I editing of RNAs in the brain, which is far more intensive in humans than mouse and mostly occurs in noncoding transposon-derived transcribed sequences (Athanasiadis et al., 2004; Blow et al., 2004; Kim et al., 2004; Levanon et al., 2004; Levanon et al., 2005; Neeman et al., 2006). The APOBEC C-U editing enzymes are vertebrate-specific and have expanded greatly in mammals and especially in the primates, where the APOBEC3 subfamily shows strong signatures of positive selection (Sawyer et al., 2004; Zhang and Webb, 2004). The functions of the various orthologs of the APOBEC C-U editing enzymes are not well understood, but they are, in part at least, variously involved in the class switch recombination and somatic hypermutation of immunoglobulins (see (Navaratnam and Sarwar, 2006)) and the control of the movement of retroviral sequences and transposable elements such as LINEs (Aguiar and Peterlin, 2008; Schumann, 2007). These sequences are differentially expressed during development (Faulkner et al., 2009), including in the brain where they have been suggested to contribute to somatic neuronal diversity (Coufal et al., 2009). Given that the locus-specificity of epigenetic marks appears to be regulated by lncRNAs (Mattick et al., 2009), this raises the prospect that the editing of these transcripts modulates the epigenetic trajectories that underpin brain development and function (Mattick, 2009b; Mattick et al., 2009) and that the expansion of RNA editing, concomitant with the selection of responsive cassettes spread by retrotransposition, was central to the evolution of higher order brain function and cognition (Mattick and Mehler, 2008). If so, variation in, and/or dysregulation, of this system and its target repertoire of protein-coding and regulatory RNAs may play a significant role in psychological and cognitive variation, as well as various disorders. Indeed there is evidence that aberrant RNA editing is associated with an increased risk of neurodevelopmental, neurodegenerative and neuropsychiatric diseases (Maas et al., 2006; Valente and Nishikura, 2005) as well as primary brain tumors (Cenci et al., 2008).
A raft of transcriptomic and functional studies are now needed to poll the full range of lncRNAs expressed in different parts of the CNS and to further elucidate the epigenetic and other functional contributions made by lncRNAs and RNA editing to the complexity of CNS structure and function, as well as to characterize their roles in disease processes. Indeed, GWAS have been performed for many CNS disorders and revealed a number of susceptibility loci but only a paucity of disease-causing protein-coding genes (Simon-Sanchez and Singleton, 2008). Because a significant percentage of disease association signals map to non-protein-coding regions of the genome and because of the abundance of antisense and other lncRNAs encoded by the genome, it is important to consider whether these previously identified disease association signals are linked to lncRNAs, although it is a challenging problem to determine the causative variation in such sequences, in contrast to identifying nonsense or missense mutations in protein-coding exons (Mattick, 2009a). In addition, therapeutic strategies that target endogenous mRNA molecules, such as those employing RNA interference (RNAi) and other customized oligonucleotide approaches with the capacity to reprogram disease-associated mRNAs, are now being developed (Wood et al., 2007). These approaches may readily be adapted to target lncRNAs whose sequence or expression may be aberrant in CNS disorders. Together, these observations suggest that lncRNAs represent a versatile class of factors that are centrally important to the modulation of diverse CNS processes and may represent the major layer underlying the genetic programming of brain development and its ability to learn, which may potentially be utilized for developing novel diagnostic and therapeutic strategies to combat CNS disorders.
Acknowledgments
M.F.M. is supported by grants from the National Institutes of Health (NS38902, MH66290), as well as by the Roslyn and Leslie Goldstein, the Mildred and Bernard H. Kayden, the F. M. Kirby and the Alpern Family Foundations. J.S.M is supported by an Australian Research Council Federation Fellowship (FF0561986), the University of Queensland, and the Queensland State Government.
Abbreviations
- Aβ
amyloid β
- AD
Alzheimer’s disease
- ADAR
adenosine deaminases acting on RNAs
- ALS
amyotrophic lateral sclerosis
- APOBEC
apolipoprotein B mRNA editing enzymes, catalytic polypeptide-like
- APP
amyloid precursor protein
- AS
Angelman syndrome
- BIC
B-cell integration cluster
- CNS
central nervous system
- DM1
myotonic dystrophy
- DS
Down syndrome
- EEG
electroencephalogram
- ES
embryonic stem
- FXS
fragile X syndrome
- FXTAS
fragile X-associated tremor and ataxia syndrome
- GABAN
GABAergic neuron
- GAT4
GABA-A transporter 4
- GWAS
genome wide association study
- HD
Huntington’s disease
- lncRNA
long non-coding RNA
- LOI
loss of imprinting
- miRNA
microRNA
- MS
multiple sclerosis
- NPC
neural precursor cell
- NSC
neural stem cell
- ncRNA
non-coding RNA
- OL
oligodendrocyte
- PASR
promoter-associated small RNA
- PcG
Polycomb group
- piRNA
piwiRNA
- PWS
Prader-Willi syndrome
- RBP
RNA binding protein
- RE1/NRSE
repressor element 1/neuron restrictive silencer element
- REST/NRSF
repressor element-1 silencing transcription factor/neuron-restrictive silencer factor
- RNAi
RNA interference
- RLS
restless legs syndrome
- rRNA
ribosomal RNA
- SCA8
spinocerebellar ataxia 8
- SHH
Sonic hedgehog
- siRNA
short interfering RNA
- snoRNA
small nucleolar RNA
- TEA
T early α
- tiRNA
transcription initiation RNA
- TMEV
Theiler’s murine encephalomyelitis virus
- tRNA
transfer RNA
- TSA
trichostatin A
- UTR
untranslated region
- VCFS
velocardiofacial syndrome
- XCI
X chromosome inactivation
- XLMR
X-linked mental retardation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abarrategui I, Krangel MS. Noncoding transcription controls downstream promoters to regulate T-cell receptor alpha recombination. EMBO J. 2007;26:4380–90. doi: 10.1038/sj.emboj.7601866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci. 2009;29:8884–96. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. REST and CoREST modulate neuronal subtype specification, maturation and maintenance. PLoS One. 2009a;4:e7936. doi: 10.1371/journal.pone.0007936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS One. 2009b;4:e7665. doi: 10.1371/journal.pone.0007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar RS, Peterlin BM. APOBEC3 proteins and reverse transcription. Virus Res. 2008;134:74–85. doi: 10.1016/j.virusres.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Albrecht S, Waha A, Koch A, Kraus JA, Goodyer CG, Pietsch T. Variable imprinting of H19 and IGF2 in fetal cerebellum and medulloblastoma. J Neuropathol Exp Neurol. 1996;55:1270–6. doi: 10.1097/00005072-199612000-00011. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–92. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, Mattick JS. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15:2013–27. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arron JR, Winslow MM, Polleri A, Chang CP, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, Yamasaki N, Miyakawa T, Francke U, Graef IA, Crabtree GR. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–7. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–46. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–5. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–69. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn CL, Sun T, Sadri-Vakili G, McFarland KN, DiRocco DP, Yohrling GJ, Clark TW, Bouzou B, Cha JH. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J Neurosci. 2008;28:10720–33. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280:29625–36. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- Berteaux N, Aptel N, Cathala G, Genton C, Coll J, Daccache A, Spruyt N, Hondermarck H, Dugimont T, Curgy JJ, Forne T, Adriaenssens E. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol. 2008;28:6731–45. doi: 10.1128/MCB.02103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Xu M, Haidar JN, Yu Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–87. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–7. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–44. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella-Lopez A, Cuchillo-Ibanez I, Cotrufo T, Mok SS, Li QX, Barquero MS, Dierssen M, Soriano E, Saez-Valero J. beta-amyloid controls altered Reelin expression and processing in Alzheimer’s disease. Neurobiol Dis. 2009 doi: 10.1016/j.nbd.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Bouchard M, Grote D, Craven SE, Sun Q, Steinlein P, Busslinger M. Identification of Pax2-regulated genes by expression profiling of the mid-hindbrain organizer region. Development. 2005;132:2633–43. doi: 10.1242/dev.01833. [DOI] [PubMed] [Google Scholar]
- Brandon NJ, Millar JK, Korth C, Sive H, Singh KK, Sawa A. Understanding the role of DISC1 in psychiatric disease and during normal development. J Neurosci. 2009;29:12768–75. doi: 10.1523/JNEUROSCI.3355-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm H, Rubin I, Kruse A, Braendstrup O, Schmidt K, Skriver EB, Lauritzen M. Nitric oxide synthase expression and enzymatic activity in human brain tumors. Clin Neuropathol. 2003;22:273–81. [PubMed] [Google Scholar]
- Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–34. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- Bucher SF, Seelos KC, Oertel WH, Reiser M, Trenkwalder C. Cerebral generators involved in the pathogenesis of the restless legs syndrome. Ann Neurol. 1997;41:639–45. doi: 10.1002/ana.410410513. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–29. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O’Connell MA, Gallo A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem. 2008;283:7251–60. doi: 10.1074/jbc.M708316200. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–22. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–78. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–9. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcangelo G. Reelin mouse mutants as models of cortical development disorders. Epilepsy Behav. 2006;8:81–90. doi: 10.1016/j.yebeh.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H, Okamura K, Bauer PO, Furukawa Y, Shimizu H, Kurosawa M, Machida Y, Miyazaki H, Mitsui K, Kuroiwa Y, Nukina N. RNA-binding protein TLS is a major nuclear aggregate-interacting protein in huntingtin exon 1 with expanded polyglutamine-expressing cells. J Biol Chem. 2008;283:6489–500. doi: 10.1074/jbc.M705306200. [DOI] [PubMed] [Google Scholar]
- Doi H, Koyano S, Suzuki Y, Nukina N, Kuroiwa Y. The RNA-binding protein FUS/TLS is a common aggregate-interacting protein in polyglutamine diseases. Neurosci Res. 2009 doi: 10.1016/j.neures.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Dugimont T, Montpellier C, Adriaenssens E, Lottin S, Dumont L, Iotsova V, Lagrou C, Stehelin D, Coll J, Curgy JJ. The H19 TATA-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene. 1998;16:2395–401. doi: 10.1038/sj.onc.1201742. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–3. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, Hornig N, Arakawa T, Takahashi H, Kawai J, Forrest AR, Suzuki H, Hayashizaki Y, Hume DA, Orlando V, Grimmond SM, Carninci P. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Po A, Di Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R, Giangaspero F, Farcomeni A, Nofroni I, Laneve P, Gioia U, Caffarelli E, Bozzoni I, Screpanti I, Gulino A. MicroRNA profiling in human medulloblastoma. Int J Cancer. 2009;124:568–77. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet. 2001;28:335–43. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–9. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- French PJ, Bliss TV, O’Connor V. Ntab, a novel non-coding RNA abundantly expressed in rat brain. Neuroscience. 2001;108:207–15. doi: 10.1016/s0306-4522(01)00408-0. [DOI] [PubMed] [Google Scholar]
- Friese MA, Fugger L. Pathogenic CD8(+) T cells in multiple sclerosis. Ann Neurol. 2009;66:132–41. doi: 10.1002/ana.21744. [DOI] [PubMed] [Google Scholar]
- Frisardi V, Solfrizzi V, Imbimbo BP, Capurso C, D’Introno A, Colacicco AM, Vendemiale G, Seripa D, Pilotto A, Capurso A, Panza F. Towards Disease-Modifying Treatment of Alzheimer’s Disease: Drugs Targeting beta-Amyloid. Curr Alzheimer Res. 2009 doi: 10.2174/156720510790274400. [DOI] [PubMed] [Google Scholar]
- Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–41. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- Graff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav Brain Res. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Grinchuk OV, Jenjaroenpun P, Orlov YL, Zhou J, Kuznetsov VA. Integrative analysis of the human cis-antisense gene pairs, miRNAs and their transcription regulation patterns. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkp954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti A, Iacono M, Pelucchi P, Kim N, Solda G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ, Callari M, Mignone F, Pesole G, Bertalot G, Bernardi LR, Albertini A, Lee C, Mattick JS, Zucchi I, De Bellis G. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi L, Ebtekar M, Jameie SB. Immune and nervous systems share molecular and functional similarities: memory storage mechanism. Scand J Immunol. 2009;69:291–301. doi: 10.1111/j.1365-3083.2008.02215.x. [DOI] [PubMed] [Google Scholar]
- Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–62. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis. 2008;29:438–45. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Jia H, Vanisri RR, Pandey T, Lu ZH, Buckley NJ, Stanton LW, Lipovich L. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA. 2009;15:85–96. doi: 10.1261/rna.1127009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–9. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Johnstone KA, DuBose AJ, Futtner CR, Elmore MD, Brannan CI, Resnick JL. A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Hum Mol Genet. 2006;15:393–404. doi: 10.1093/hmg/ddi456. [DOI] [PubMed] [Google Scholar]
- Kambere MB, Lane RP. Co-regulation of a large and rapidly evolving repertoire of odorant receptor genes. BMC Neurosci. 2007;8(Suppl 3):S2. doi: 10.1186/1471-2202-8-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007a;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007b;8:413–23. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–25. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O’Brien SJ, Wong AJ, Vogelstein B. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–3. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136:1771–83. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AV, Kiefmann M, Ebnet K, Khanam T, Muddashetty RS, Brosius J. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP) J Mol Biol. 2005;353:88–103. doi: 10.1016/j.jmb.2005.07.049. [DOI] [PubMed] [Google Scholar]
- Koob MD, Moseley ML, Schut LJ, Benzow KA, Bird TD, Day JW, Ranum LP. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat Genet. 1999;21:379–84. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- Korneev SA, Korneeva EI, Lagarkova MA, Kiselev SL, Critchley G, O’Shea M. Novel noncoding antisense RNA transcribed from human anti-NOS2A locus is differentially regulated during neuronal differentiation of embryonic stem cells. RNA. 2008;14:2030–7. doi: 10.1261/rna.1084308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–87. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- Le Meur E, Watrin F, Landers M, Sturny R, Lalande M, Muscatelli F. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Dev Biol. 2005;286:587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–6. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, Olshansky M, Rechavi G, Jantsch MF. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–5. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Levanon K, Eisenberg E, Rechavi G, Levanon EY. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome. EMBO Rep. 2005;6:831–5. doi: 10.1038/sj.embor.7400507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewejohann L, Skryabin BV, Sachser N, Prehn C, Heiduschka P, Thanos S, Jordan U, Dell’Omo G, Vyssotski AL, Pleskacheva MG, Lipp HP, Tiedge H, Brosius J, Prior H. Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav Brain Res. 2004;154:273–89. doi: 10.1016/j.bbr.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, Chun CZ, Samant GV, Zhao B, Garnaas MK, Horswill MA, Stanhope SA, North PE, Miao RQ, Wilkinson GA, Affolter M, Ramchandran R. A non-coding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2009 doi: 10.1182/blood-2009-09-242180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Pestova TV, Hellen CU, Tiedge H. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol. 2008;28:3008–19. doi: 10.1128/MCB.01800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–8. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JL, Roskams AJ. Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog Neurobiol. 2009;88:170–83. doi: 10.1016/j.pneurobio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J. BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc Natl Acad Sci U S A. 1993;90:11563–7. doi: 10.1073/pnas.90.24.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4:255–64. doi: 10.4161/epi.9130. [DOI] [PubMed] [Google Scholar]
- Marullo M, Valenza M, Mariotti C, Di Donato S, Cattaneo E, Zuccato C. Analysis of the Repressor Element-1 Silencing Transcription Factor/Neuron-Restrictive Silencer Factor Occupancy of Non-Neuronal Genes in Peripheral Lymphocytes from Patients with Huntington’s Disease. Brain Pathol. 2008 doi: 10.1111/j.1750-3639.2008.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–9. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14(Spec No 1):R121–32. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Mehler MF. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 2008;31:227–33. doi: 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009a;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Has evolution learnt how to learn? EMBO Rep. 2009b;10:665. doi: 10.1038/embor.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–9. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008;86:305–41. doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mariani J, Kosik KS, Mehler MF, Mattick JS. Noncoding RNAs in Long-Term Memory Formation. Neuroscientist. 2008a;14:434–45. doi: 10.1177/1073858408319187. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008b;105:716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, St Clair DM, Muir WJ, Blackwood DH, Porteous DJ. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–23. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, Day JW, Ranum LP. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–69. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- Muller MC, Osswald M, Tinnes S, Haussler U, Jacobi A, Forster E, Frotscher M, Haas CA. Exogenous reelin prevents granule cell dispersion in experimental epilepsy. Exp Neurol. 2009;216:390–7. doi: 10.1016/j.expneurol.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Muller S, Zirkel D, Westphal M, Zumkeller W. Genomic imprinting of IGF2 and H19 in human meningiomas. Eur J Cancer. 2000;36:651–5. doi: 10.1016/s0959-8049(99)00328-7. [DOI] [PubMed] [Google Scholar]
- Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2007;104:10679–84. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnam N, Sarwar R. An overview of cytidine deaminases. Int J Hematol. 2006;83:195–200. doi: 10.1532/IJH97.06032. [DOI] [PubMed] [Google Scholar]
- Neeman Y, Levanon EY, Jantsch MF, Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA. 2006;12:1802–9. doi: 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K, Pullirsch D, Leeb M, Wutz A. Xist and the order of silencing. EMBO Rep. 2007;8:34–9. doi: 10.1038/sj.embor.7400871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, Hill DP, Bult C, Hume DA, Quackenbush J, Schriml LM, Kanapin A, Matsuda H, Batalov S, Beisel KW, Blake JA, Bradt D, Brusic V, Chothia C, Corbani LE, Cousins S, Dalla E, Dragani TA, Fletcher CF, Forrest A, Frazer KS, Gaasterland T, Gariboldi M, Gissi C, Godzik A, Gough J, Grimmond S, Gustincich S, Hirokawa N, Jackson IJ, Jarvis ED, Kanai A, Kawaji H, Kawasawa Y, Kedzierski RM, King BL, Konagaya A, Kurochkin IV, Lee Y, Lenhard B, Lyons PA, Maglott DR, Maltais L, Marchionni L, McKenzie L, Miki H, Nagashima T, Numata K, Okido T, Pavan WJ, Pertea G, Pesole G, Petrovsky N, Pillai R, Pontius JU, Qi D, Ramachandran S, Ravasi T, Reed JC, Reed DJ, Reid J, Ring BZ, Ringwald M, Sandelin A, Schneider C, Semple CA, Setou M, Shimada K, Sultana R, Takenaka Y, Taylor MS, Teasdale RD, Tomita M, Verardo R, Wagner L, Wahlestedt C, Wang Y, Watanabe Y, Wells C, Wilming LG, Wynshaw-Boris A, Yanagisawa M, Yang I, Yang L, Yuan Z, Zavolan M, Zhu Y, Zimmer A, Carninci P, Hayatsu N, Hirozane-Kishikawa T, Konno H, Nakamura M, Sakazume N, Sato K, Shiraki T, Waki K, Kawai J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashizume W, Imotani K, Ishii Y, Itoh M, Kagawa I, Miyazaki A, Sakai K, Sasaki D, Shibata K, Shinagawa A, Yasunishi A, Yoshino M, Waterston R, Lander ES, Rogers J, Birney E, Hayashizaki Y. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–73. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–6. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009;182:7738–48. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, Kent J, Miller W, Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheasant M, Mattick JS. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–53. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- Pibouin L, Villaudy J, Ferbus D, Muleris M, Prosperi MT, Remvikos Y, Goubin G. Cloning of the mRNA of overexpression in colon carcinoma-1: a sequence overexpressed in a subset of colon carcinomas. Cancer Genet Cytogenet. 2002;133:55–60. doi: 10.1016/s0165-4608(01)00634-3. [DOI] [PubMed] [Google Scholar]
- Pisante A, Bronstein M, Yakir B, Darvasi A. A variant in the reelin gene increases the risk of schizophrenia and schizoaffective disorder but not bipolar disorder. Psychiatr Genet. 2009;19:212. doi: 10.1097/YPG.0b013e32832cebe6. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Jr, Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–72. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–65. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Regulation of non-coding RNA networks in the nervous system--what’s the REST of the story? Neurosci Lett. 2009;466:73–80. doi: 10.1016/j.neulet.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Blackshaw S. New meaning in the message: noncoding RNAs and their role in retinal development. Dev Dyn. 2009;238:2103–14. doi: 10.1002/dvdy.21844. [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, Grimmond SM, Hume DA, Hayashizaki Y, Mattick JS. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–9. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136:525–30. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–32. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol Cell. 2008;100:149–66. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–30. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35:637–42. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Seong IS, Woda JM, Song JJ, Lloret A, Abeyrathne PD, Woo CJ, Gregory G, Lee JM, Wheeler VC, Walz T, Kingston RE, Gusella JF, Conlon RA, Macdonald ME. Huntingtin facilitates polycomb repressive complex 2. Hum Mol Genet. 2010;19:573–83. doi: 10.1093/hmg/ddp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serajee FJ, Zhong H, Mahbubul Huq AH. Association of Reelin gene polymorphisms with autism. Genomics. 2006;87:75–83. doi: 10.1016/j.ygeno.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2009 doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Johannesson M, Bronstein M, Chen SX, Collier DA, Craddock NJ, Kendler KS, Li T, O’Donovan M, O’Neill FA, Owen MJ, Walsh D, Weinberger DR, Sun C, Flint J, Darvasi A. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Singleton A. Genome-wide association studies in neurological disorders. Lancet Neurol. 2008;7:1067–72. doi: 10.1016/S1474-4422(08)70241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- Sproule DM, Kaufmann P. Mitochondrial encephalopathy, lactic acidosis, and strokelike episodes: basic concepts, clinical phenotype, and therapeutic management of MELAS syndrome. Ann N Y Acad Sci. 2008;1142:133–58. doi: 10.1196/annals.1444.011. [DOI] [PubMed] [Google Scholar]
- St Laurent G, 3rd, Wahlestedt C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30:612–21. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Wegner M. SoxE function in vertebrate nervous system development. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010 doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Kunugi H, Ohashi J, Hohjoh H. Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol Psychiatry. 2007;12:519, 593–600. doi: 10.1038/sj.mp.4002014. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Di Giovanni S. The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 2009;10:576–83. doi: 10.1038/embor.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochitani S, Hayashizaki Y. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem Biophys Res Commun. 2008;372:691–6. doi: 10.1016/j.bbrc.2008.05.127. [DOI] [PubMed] [Google Scholar]
- Torarinsson E, Yao Z, Wiklund ED, Bramsen JB, Hansen C, Kjems J, Tommerup N, Ruzzo WL, Gorodkin J. Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions. Genome Res. 2008;18:242–51. doi: 10.1101/gr.6887408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tsunoda I, Fujinami RS. Neuropathogenesis of Theiler’s Murine Encephalomyelitis Virus Infection, An Animal Model for Multiple Sclerosis. J Neuroimmune Pharmacol. 2009 doi: 10.1007/s11481-009-9179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol. 2003;77:5632–8. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, Royo H, Marty V, Bortolin-Cavaille ML, Cavaille J. Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin organization at imprinted snoRNA gene arrays. J Cell Sci. 2010;123:70–83. doi: 10.1242/jcs.054957. [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol. 2005;23:1383–90. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- Williams JM, Beck TF, Pearson DM, Proud MB, Cheung SW, Scott DA. A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. Am J Med Genet A. 2009;149A:1758–62. doi: 10.1002/ajmg.a.32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–3. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Winkelmann J, Schormair B, Lichtner P, Ripke S, Xiong L, Jalilzadeh S, Fulda S, Putz B, Eckstein G, Hauk S, Trenkwalder C, Zimprich A, Stiasny-Kolster K, Oertel W, Bachmann CG, Paulus W, Peglau I, Eisensehr I, Montplaisir J, Turecki G, Rouleau G, Gieger C, Illig T, Wichmann HE, Holsboer F, Muller-Myhsok B, Meitinger T. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–6. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- Won SJ, Kim SH, Xie L, Wang Y, Mao XO, Jin K, Greenberg DA. Reelin-deficient mice show impaired neurogenesis and increased stroke size. Exp Neurol. 2006;198:250–9. doi: 10.1016/j.expneurol.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Wood M, Yin H, McClorey G. Modulating the expression of disease genes with RNA-based therapy. PLoS Genet. 2007;3:e109. doi: 10.1371/journal.pgen.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Catoire H, Dion P, Gaspar C, Lafreniere RG, Girard SL, Levchenko A, Riviere JB, Fiori L, St-Onge J, Bachand I, Thibodeau P, Allen R, Earley C, Turecki G, Montplaisir J, Rouleau GA. MEIS1 intronic risk haplotype associated with restless legs syndrome affects its mRNA and protein expression levels. Hum Mol Genet. 2009;18:1065–74. doi: 10.1093/hmg/ddn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JW, Kita Y, Frank DJ, Majewski RR, Konicek BA, Nobrega MA, Jacob H, Walterhouse D, Iannaccone P. Gene expression profiling leads to identification of GLI1-binding elements in target genes and a role for multiple downstream pathways in GLI1-induced cell transformation. J Biol Chem. 2002;277:5548–55. doi: 10.1074/jbc.M105708200. [DOI] [PubMed] [Google Scholar]
- Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501–12. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Yu W, Wang Y, McDonnell K, Stephen D, Bai CB. Patterning of ventral telencephalon requires positive function of Gli transcription factors. Dev Biol. 2009;334:264–75. doi: 10.1016/j.ydbio.2009.07.026. [DOI] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum Mol Genet. 2004;13:1785–91. doi: 10.1093/hmg/ddh183. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–34. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Chuang SC, Bianchi R, Zhao W, Lee H, Fenton AA, Wong RK, Tiedge H. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J Neurosci. 2009;29:9977–86. doi: 10.1523/JNEUROSCI.3893-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, MacDonald M, Fossale E, Zeitlin S, Buckley N, Cattaneo E. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J Neurosci. 2007;27:6972–83. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]