Abstract
BiP is the mammalian endoplasmic reticulum (ER) Hsp70 orthologue that plays a major role in all functions of this organelle including the seemingly opposing functions of aiding the maturation of unfolded nascent proteins and identifying and targeting chronically unfolded proteins for degradation. The recent identification of mammalian BiP co-factors combined with delineation of the ER degradation machinery and data suggesting that the ER is subdivided into unique regions helps explain how these different functions can occur in the same organelle and raises some unresolved issues.
1. Introduction
The ER is the site of synthesis of proteins destined for the cell surface or secretion. Although the concentration of nascent unfolded polypeptides in the ER is extremely high, >95% of nascent chains fold and assemble rapidly and correctly through the participation of chaperones and folding enzymes [1]. Proteins that do not meet the stringent requirements of the ER quality control system are retained in the ER, where the chaperones prevent them from aggregating and provide them with additional opportunities to achieve their correct conformation. Proteins that ultimately fail to mature properly are targeted for intracellular degradation [2,3]. Two major chaperone systems exist in the mammalian ER, the calnexin/calreticulin system and the BiP or Hsp70 system. Unlike calnexin, which relies on monitoring both N-linked glycans and unfolded regions on nascent polypeptide chains, BiP detects the latter and is therefore the major system used for nonglycosylated proteins [4] or glycoproteins in which the most N-terminal glycan occurs relatively late in the linear sequence [5]. In addition to its role in the folding and assembly of nascent proteins, BiP contributes to the maintenance of ER calcium stores, identifies defective proteins that must be targeted for proteasome-mediated degradation, and preserves the permeability barrier of the ER translocon during early stages of protein translocation [4]. Mechanisms exist to ensure that sufficient amounts of BiP are available to perform its multiple functions, and as necessary, can activate a signal transduction cascade known as the unfolded protein response (UPR), which controls levels of ER chaperones and folding enzymes [6]. Recent data suggest that the various BiP co-chaperones may play a critical role in regulating its various activities.
When protein folding ultimately fails either due to disruption of ER homeostasis or mutations in individual proteins, the defective proteins must be identified, extracted from the ER, and degraded by the 26S proteosome. Although first delineated in yeast [7], there have been major advances in the identification of mammalian components of the ER associated degradation (ERAD) machinery [8]. As many proteins synthesized in the ER are glycosylated, appropriately much of the initial work has focused on the disposal of glycoproteins [9]. Recently however, proteins involved in the turnover of nonglycosylated proteins have been identified, which reveal some differences in the handling of these two types of substrates [10]. Since the same chaperones that aid the folding of a particular substrate often participate in its destruction, a particularly perplexing question is how the ER quality control machinery can distinguish between nascent proteins that have not yet folded and those that cannot fold. Recent studies have begun to shed light on how these two types of BiP substrates are distinguished.
The ER is an oxidizing environment, and mammalian proteins largely enter the ER co-translationally. As nascent polypeptide chains begin to fold as they enter the ER and are often stabilized by the formation of disulfide bonds, if the misfolding event occurs relatively late in the biosynthesis of the protein, those portions of the polypeptide chain may have folded and disulfide bonds may have formed. Thus both reduction and unfolding of the protein may be required to pass the ERAD substrate through a protein channel in the ER membrane. How the ER can perform these two very different functions is an area of active research. Recent data suggesting that different regions of the ER are populated by distinct subsets of proteins might hold the clue to establishing unique environments to support opposing functions.
2. The BiP chaperone cycle
Like all Hsp70 family members BiP interacts with unfolded substrates through its C-terminal substrate-binding domain (SBD), which is tightly regulated by the highly conserved N-terminal nucleotide-binding domain (NBD). When ATP occupies the cleft of the NBD, the SBD is in an open configuration, which has both a high on and high off rate for unfolded proteins. The hydrolysis of ATP results in a closure of the lid on the SBD, thus stabilizing the interaction with bound protein [11]. Release of the unfolded protein occurs when ADP is exchanged for ATP. This reopens the lid on the SBD, allowing the bound substrate to be released and provides an opportunity for it to fold. Communication between these domains is controlled by a linker region that juxtaposes them and opens the SBD when ATP is bound [12]. This interaction is dependent on a surface exposed arginine on the NBD that is present in all Hsp70 proteins [12,13]. Since DnaJ proteins bind to the same residue on the NBD, it is likely that they stimulate tight binding of Hsp70s to substrates by associating with this residue and disrupting the interaction between the two domains. Recently, a number of BiP cofactors have been identified that control BiP’s ATPase cycle and allow it to participate in its various functions.
2.1 Nucleotide Exchange Factors
2.1.1 BAP
The release of substrates from BiP requires the exchange of ADP to ATP, which is catalyzed by nucleotide exchange factors (NEF). BAP/Sil1 was the first NEF to be identified in the mammalian ER [14]. BAP is highly expressed in secretory tissues showing the same overall distribution as BiP, and over-expression of BAP can inappropriately induce the release of BiP from substrates [15]. Unlike BiP, BAP is not induced by the UPR, which significantly changes the ER ratio of these two proteins during ER stress, although the effect of this imbalance on protein maturation has not been investigated. While there is an ever-growing number of conditions classified as protein folding diseases that arise due to a mutation in a secretory pathway protein, BAP was the first component of the chaperone machinery to be linked to a disease. Mutations in BAP cause an autosomal recessive form of ataxia and cerebellar atrophy known as Marinesco-Sjögren syndrome, which is characterized by multisystem defects [16,17]. In almost all cases the mutation would lead to loss of exons 6–10, which are predicted to form the BiP interacting site [18]. The BAP/Sil1p null or “woozy” mouse fairly closely phenocopies the human disease [19], with their Purkinje cells showing evidence of protein aggregation and ER stress, indicating that the symptoms in the MSS patients might be due to either the failure to release critical substrates from BiP or a resulting decrease in the available pool of BiP to chaperone new substrates.
2.1.2 Large Hsp70s
Recent studies in yeast [20,21], and mammals [22] suggest that the large Hsp70s may also serve as NEF for their conventional Hsp70 organellar co-residents. Like other large Hsp70s, GRP170 possesses an N-terminal NBD that very closely resembles that of BiP, except that it lacks ATPase activity and does not possess the highly conserved arginine on the NBD that interacts with DnaJ proteins. It has been unclear as to whether the long C-terminal extension on these proteins encodes a SBD, although the recent crystal structure of yeast See1p suggests they do [23]. Interestingly, not all tissues are equally affected by BAP mutations in either MSS patients or the knock-out mouse, arguing that GRP170 may compensate in some tissues as has been observed in yeast [24]. Indeed, over-expression of GRP170 in the BAP knock-out mouse prevented neurodegeneration [25]. Therefore, it is possible that the relative abundance of these two proteins varies by tissue leading to larger effects with the loss of BAP in some tissues or that some BiP clients are more reliant on one NEF than another.
2.2 ER localized DnaJ-proteins
DnaJ proteins stimulate the ATPase activity of their Hsp70 partner, thereby stabilizing the binding of Hsp70s to substrates [26]. In mammals, seven resident ER proteins with J domains inside the ER lumen (ERdjs) have been described (Figure 1). The individual ERdjs are present at widely different concentrations and in total are present in substoichiometric quantities compared to BiP [27]. This is consistent with recent data demonstrating that ERdjs bind to substrates, recruit BiP, and after ensuring that a series of functional interactions occur between the two proteins, it is released well before protein folding is completed [28,29]. Since BiP is involved in both folding nascent proteins and targeting misfolded ones for degradation, and because some ERdj proteins can bind directly to substrate proteins, it is reasonable to postulate that the various ERdjs control the diverse functions of BiP.
Figure 1. Characteristics of ERdj proteins.
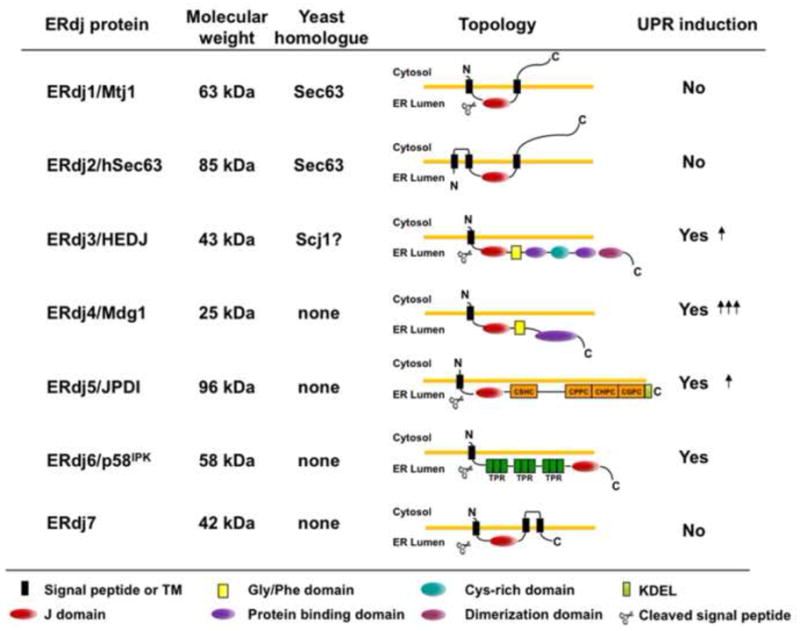
Seven DnaJ-like proteins have been identified in the mammalian ER. DnaJ proteins can be divided into three subgroups based on the similarities with E. coli DnaJ, the founding member of this family [77]. Type I proteins contain all three domains: an N-terminal J domain, a glycine/phenylalanine rich domain (yellow square), and a substrate binding domain, which is intersected with a cysteine-rich Zn2+ binding domain (light blue circle); type II proteins contain the first two domains and often bind to unfolded substrates, although this region is more poorly defined; and type III proteins have only a J domain, which can occur anywhere in the protein. In the mammalian ER most of the ERdjs are type III except for ERdj3 (type I) and ERdj4 (type II). ERdj5 contains four thioredoxin domains (orange squares) and ERdj6 has three tetratricopeptide domains (dark green squares). The detailed mechanism of their function can be found in the text.
2.2.1 ERdj1/Mtj1p
ERdj1/Mtj1p is a transmembrane ER protein with a cytosolic domain that associates with the translating ribosomes and inhibits further protein synthesis if BiP is not bound to the lumenal J domain [30]. This unique translational checkpoint ensures that BiP is present to associate with newly synthesized polypeptides entering the ER. Unlike many of the other ERdj proteins, ERdj1 is not induced by the UPR, in keeping with the fact that translation is lower during ER stress. BiP is also required to gate the translocon during early stages of translocation in order to maintain the unique environment of the ER [31]. This activity requires an integral membrane ERdj protein [32], although the identity of this protein has not been determined.
2.2.2 ERdj2/Sec63
ERdj2 is a second translocon-associated ERdj protein and resembles the overall structure of Sec63p [33], which plays an essential role in posttranslational protein translocation in yeast [34]. ERdj2 possesses three membrane spanning domains, a lumenal J domain, and a cytosolic coiled domain. In mammals, where proteins are primarily translocated cotranslationally, it is not entirely clear what ERdj2 is doing, particularly since it is a relatively abundant protein [27]. Mutations in ERdj2 are linked to polycystic liver disease, where it may contribute to the cotranslational processing of proteins involved in biliary cell growth [35].
2.2.3 ERdj3/HEDJ
ERdj3 was independently identified in canine pancreatic microsomes as a homolog of yeast Scj1p [36], as HEDJ, an Hsp40 co-chaperone involved in Shiga toxin trafficking [37], and as a component of the chaperone complex bound to unassembled immunoglobulin heavy chains [38]. ERdj3 is up-regulated during ER stress and binds directly to a number of both nascent unfolded proteins and proteins unable to fold [39]. BiP exists in a complex with a subset of ER molecular chaperones, which includes ERdj3 only when the complex is bound to the substrate [38]. This strongly suggests that ERdj3 may help recruit BiP to substrates. ERdj3 complements cell wall synthesis and ERAD defects in yeast mutant for Ydj1 and Hlj1, two cytosolic DnaJ proteins [40], suggesting that ERdj3 plays a role in degradation. However, its association with nascent proteins that are folding and the requirement of chaperones to keep ERAD substrates soluble might be more compatible with ERdj3 playing a major role in protein folding.
2.2.4 ERdj4/MDG-1
ERdj4 is membrane-anchored via an uncleaved signal sequence and the remainder of the protein is within the ER lumen [41,42]. ERdj4 is the least abundant ERdj protein [27], but it is highly up-regulated by ER stress [41,43]. ERdj4 is induced in response to expression of two misfolded variants of surfactant protein SP-C, which cause interstitial lung disease, and associates with the misfolded SP-C mutants [44]. Over-expression of ERdj4 reduced the half-life of mutant SP-C, whereas reduction of ERdj4 levels significantly slowed its degradation, suggesting that ERdj4 plays a role in the degradation of these ERAD substrates.
2.2.5 ERdj5/JPDI
In addition to having a J domain, ERdj5 has four thioredoxin-like domains that contribute to its reductase activity [45,46]. Over-expression of ERdj5 promotes the degradation of the non-glycosylated mutant SP-C proteins, suggesting that it may reduce BiP substrates and enhance their ability to be unfolded for retrotranslocation [44]. A two hybrid screen identified ERdj5 as an EDEM interacting protein and found that it also played a role in the degradation of two glycoproteins; the NHK variant of α1-antitrypsin and IgM J chain [47]. Reductases must be kept in a reduced form to break disulfide bonds in substrate proteins. This function could be provided by a recently discovered flavoprotein protein ERFAD [48], which interacts with ERdj5, SEL1L, and OS-9; known components of the ERAD machinery for glycoproteins [49].
2.2.6 ERdj6/p58IPK
ERdj6 was originally reported to be a cytosolic protein that negatively regulated PERK phosphorylation [50], and the loss of p58IPK sensitized mice to ER stress [51]. Further experiments revealed that this protein possessed an ER targeting sequence and was translocated into the ER lumen where it binds to unfolded substrates via its tetratricopeptide repeat domain and acts as a co-chaperone for BiP [29,52]. Modulating ERdj6 levels did not affect the degradation of several model proteins, suggesting that it may reduce the burden of unfolded and misfolded proteins by promoting protein folding in the ER lumen [29,52].
2.2.7 ERdj7
Recent mass spectrophometric analysis of canine pancreatic microsomes identified the newest member of this family [53]. ERdj7 is a resident ER protein, with two transmembrane regions and a lumenal J domain. Its function is currently not known. However, since it is not up-regulated by ER stress, and reducing ERdj7 levels did not induce the UPR [53], it is unlikely that it plays a role in protein folding or degradation.
3. Disposing of BiP substrates
3.1 Recognition
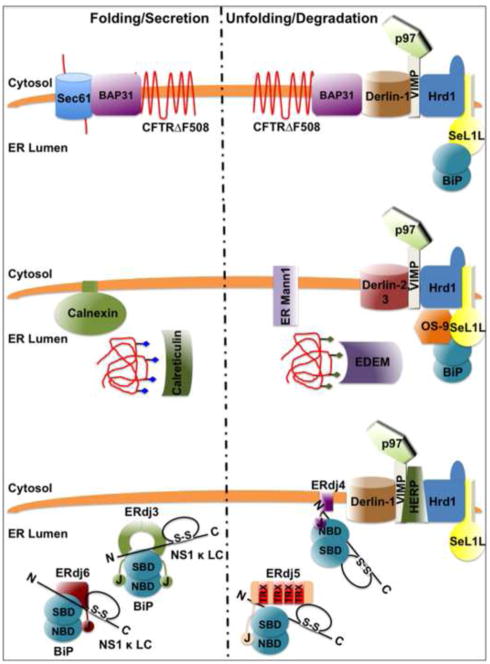
The disposal of unfolded proteins requires they be recognized as such and directed through a protein channel to the cytosol for degradation by the 26S proteasome [54]. For glycoproteins this involves changes in the glycan structure that allow the substrate to move from association with calnexin/calreticulin (folding) to EDEM (degradation) [9]. This transition is less clear for BiP substrates that often do not have N-linked glycans, but a number of recent studies described above suggest that this could be achieved by switching from ERdj3/6 (folding) to ERdj4/5 (degradation). There are no data to address how this switching might occur, but since studies with ERdj3/6 revealed that these proteins can leave the unfolded protein once it has successfully recruited BiP and induced its tight binding to the substrate [28, 29], it is conceivable that this provides the opportunity for another ERdj to bind to the substrate and target it for degradation. As ERdj5 was shown to interact with EDEM [47] and over-expressed ERdj4 and ERdj5 interact with p97 [44], a component of the ERAD machinery [55], this is an appealing model (Figure 2).
Figure 2. Models for the putative movement of ERAD substrates from sites of folding within the ER to sites of unfolding and degradation.
Top: Membrane proteins, like CFTR F508, bind BAP31, which cycles between sites of protein folding near the Sec61 translocon and the ERQC where it binds Derlin1 to promote retrotranslocation of ERAD substrates. Middle: Glycosylated proteins (blue diamonds) undergo cycles of binding and release from the lectins calnexin and calreticulin as they fold. If they fail to do so, their polysaccharide chains (green diamonds) are modified by ER mannosidase I, which prevents the unfolded protein from rebinding calnexin/calreticulin and allows it to bind EDEM, a soluble lectin. EDEM targets the protein to Sel1 and OS-9, which associate with a complex of Derlin 2/3, p97 and presumably the Hrd1 E3 ligase, promoting its retrotranslocation and degradation. Bottom: Non-glycosylated lumenal ERAD substrates, like the NS1 κ LC, bind BiP, which can promote their folding when the substrate is associated with ERdj3/ERdj6 or reduce and unfold the substrate when it is bound to ERdj4 or ERdj5. The fact that a number of the ERdjs are soluble and bind to both substrates and components of the folding machinery or degradation complexes may provide a mechanism for moving from one ER function to the other.
3.2 Retrotranslocation
ERAD substrates pass through a protein channel in the ER membrane and are deposited in the cytosol for degradation [56]. While the proteins that form the “retrotranslocon” or “dislocase” channel remain controversial, in S. cerevisiae, the multi-pass integral membrane protein Der1p is believed to be a component [57,58]. Mammalian cells possess three Der1p homologues, Derlin 1–3 [59–61]. Studies revealed that NHK interacted with Derlin-2 and -3, but not with Derlin-1 [61], while recent studies on two non-glycosylated soluble BiP substrates [10] and the transmembrane MHC heavy chain [59,60] revealed that they interacted with Derlin-1. Cross-linking studies revealed that Derlin 1 interacts with Hrd1, an E3 ligase, and Herp, which contains a ubiquitin-like domain [62] and binds to both ubiquitylated ERAD substrates and the 26S proteasome [10]. Reducing Herp levels inhibited the degradation of two BiP substrates, but had no affect on NHK turnover. Together these data suggest that either distinct retrotranslocon channels exist for the disposal of different types of substrates or that a single channel exists but the various substrates are dependent on different components of the channel.
3.3 Degradation
Once in the retrotranslocon, the protein is pulled through the channel by the Cdc48p/p97 ATPase complex, which uses the energy of hydrolysis to extract the substrate [63]. On the cytosolic face of the ER, the substrate interacts with an E3 ubiquitin ligase, such as Hrd1 [64] or gp78 [65], is ubiquitylated, and ultimately degraded by the proteasome. E3s are usually quite specific for directly the ubiquitylation of a single or small group of target proteins [66]. Given the fact that any protein synthesized in the secretory pathway could fail to fold, it is noteworthy that only two ERAD associated E3 have been identified, which show broad substrate specificity, suggesting that the identification of misfolded ER proteins by these E3s may be conceptually quite different than the use of ubiquitylation to regulate biological processes. Unlike retrotranslocon components and Herp, there is no evidence that the two broad spectrum E3s have a preference for a type of substrate.
4. ER Structure
Although the ER consists of a continuous membrane, it has different structural and functional compartments [67]. The ER is classified into rough (RER) and smooth (SER) ER based on the presence or absence of bound ribosomes respectively. These two types of ER are present in different amounts depending on cell type. For example, Ig secreting plasma cells possess mostly RER, whereas hepatocytes, which are involved in lipid synthesis and detoxification have an abundant SER. One topic of great interest is how the different ER domains are maintained and what functions are carried out in each of them.
Whether these domains or regions of the ER have different roles in ER quality control is an area of active research. For example, a study designed to analyze the proteome of the secretory pathway showed that proteins involved in ERAD, such as p97 and Derlin1 are enriched in the SER, whereas proteins involved in translocation, modification and folding were largely present in the RER fraction [68]. A number of other chaperones, co-chaperones and components of the degradation machinery were not uncovered in this study, probably due to their low abundance in cells not experiencing ER stress. This knowledge will be essential to determining whether protein folding and degradation occur in a compartmentalized manner.
4.1 Quality Control Compartments in the ER
Since nascent proteins that have not yet folded are likely to closely resemble proteins that are unable to fold, it is not clear how the ER can distinguish between these two states so that newly synthesized proteins are not constantly being targeted for degradation. Hence, it would make sense to separate these two functions physically. This compartmentalization of the ER could be constitutive, as supported by the proteomics study [68], or a putative quality control compartment(s) could appear when misfolded proteins accumulate. The fact that the ER structure changes upon expression of ERAD substrates [69] or inhibition of proteolysis [70] might be more compatible with a transient or poorly developed structure under normal folding conditions. The existence of a dedicated ER quality control compartment (ERQC) is supported by studies on two calnexin substrates; asialoglycoprotein receptor and the heavy chain of class 1 MHC [70,71]. These proteins localized to a perinuclear region when proteasomal degradation was inhibited, which contained Sec61 and calnexin but not BiP or PDI. These results beg the questions as to whether the over-expression of these ERAD substrates simply causes a compartment that is always present to be expanded allowing its detection, and if BiP substrates were similarly over-expressed would this ERQC contain proteins like BiP and PDI and exclude calnexin, which is in keeping with other data suggesting that the disposal of nonglycosylated proteins relies on different ERAD components than those used for glycosylated proteins [10].
4.2 Maintenance of redox state in the ER
As the folding of nascent proteins in the ER is often accompanied by the formation of intra- and interchain disulfide bonds, whereas the retrotranslocation of proteins that fail to fold may require reducing disulfide bonds on partially folded substrates, the ability to maintain discrete regions of the ER with oxidizing versus reducing environment would appear to allow these opposing processes to occur efficiently [72]. The formation of disulfide bonds is an enzyme catalyzed relay in which a disulfide bond is transferred from oxidized Ero1 to a member of the protein disulfide isomerase family (PDI) to the substrate protein [73]. This cascade can also work in reverse with disulfide bonds on the substrate protein being transferred to a reduced PDI protein, the presence of which is dependent on the redox state of Ero1. In human cells, there are two Ero1 paralogs, Ero1-Lα and Ero1-Lβ [73]. Ero1-Lα is constitutively expressed, while Ero1-Lβ is induced by the UPR, like yeast Ero1p [74]. In addition to maintaining an oxidized redox environment in the ER, the Ero1-PDI interaction influences many processes such as protein folding and degradation [75].
Although overall the ER is an oxidizing environment, it is possible that there are regions that differ in their redox microenvironment to promote protein folding in an oxidized microenvironment and protein unfolding in a reduced microenvironment. There are several ways by which this could be achieved. For example, Ero1 could be localized to areas of protein synthesis, near the translocon channel to promote PDI oxidation and hence disulfide bond formation, or different Ero1 isoforms may localize to specific areas in the ER. For instance, Ero1α could be expressed throughout the ER, but the UPR inducible Ero1β could localize to a specific area or compartment in the ER leading to areas of higher oxidation. Additionally, the diversity of ER oxidoreductases in mammalian cells (>19 have been identified [75]) may influence different redox environments within the ER, as it is possible that they have very different redox potentials, arguing that some may serve primarily as oxidases and others as reductases and that their sub-organellar localization is critical to the processes of folding versus degradation. Understanding how these enzymes are regulated and whether they promote different environments in the ER will lead us to a better understanding of how proteins can be both folded and misfolded within the same organelle and whether these reactions occur in a compartmentalized manner.
5. Concluding remarks
We have progressed significantly in understanding how ER proteins are handled as they fold and how they are dispose of if they fail. Clearly these processes are more specific and controlled than initially envisioned. In other words, not all ERAD substrates are created and degraded equally. They are folded, transported and degraded by different proteins and maybe even in different regions of the ER. If separate regions of the ER do exist, ERAD substrates would need to move from a “folding” to a “degradation” subcompartment (Figure 2). Interestingly, a recent study found that p97, Derlin-1, BiP, and BAP31, an ER sorting protein, all localized to the ERQC in response to inhibiting degradation via proteasome inhibitors or expression of a p97 dominant-negative mutant [76]. Similarly, EDEM binds to glycoproteins once the N-linked glycans are trimmed to a form that is no longer recognized by calnexin and takes them to SEL1L, which is bound to other ERAD components including Derlin 2 and 3 [9]. Since BiP’s interaction with substrates is mediated by ERdj proteins, it is tempting to suggest that they may play a role in escorting misfolded BiP substrates from sites of folding to sites of unfolding and retrotranslocation. Whether these various co-chaperones and “escort” proteins reside in specific areas of the ER or move between them is an area of active research. Understanding how BiP substrates “live and die” will have implications not only in understanding essential biological processes, like antibody production and secretion, but may also provide insights into possible intervention strategies for protein folding diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–9. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 2.Werner ED, Brodsky JL, McCracken AA. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci U S A. 1996;93:13797–801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarosch E, Lenk U, Sommer T. Endoplasmic reticulum-associated protein degradation. Int Rev Cytol. 2003;223:39–81. doi: 10.1016/s0074-7696(05)23002-4. [DOI] [PubMed] [Google Scholar]
- 4.Hendershot LM. BiP is a master regulator of ER function. Mt Sinai J Med. 2004;71:289–97. [PubMed] [Google Scholar]
- 5.Molinari M, Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–3. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- 6.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 7.Hampton RY. ER-associated degradation in protein quality control and cellular regulation. Curr Opin Cell Biol. 2002;14:476–82. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- 8.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–57. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehara K, Kawaguchi S, Ng DT. The EDEM and Yos9p families of lectin-like ERAD factors. Semin Cell Dev Biol. 2007;18:743–50. doi: 10.1016/j.semcdb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–54. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–84. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513–24. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Awad W, Estrada I, Shen Y, Hendershot LM. BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc Natl Acad Sci U S A. 2008;105:1164–9. doi: 10.1073/pnas.0702132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung KT, Shen Y, Hendershot LM. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem. 2002;277:47557–63. doi: 10.1074/jbc.M208377200. [DOI] [PubMed] [Google Scholar]
- 15.Awe K, Lambert C, Prange R. Mammalian BiP controls posttranslational ER translocation of the hepatitis B virus large envelope protein. FEBS Lett. 2008;582:3179–84. doi: 10.1016/j.febslet.2008.07.062. [DOI] [PubMed] [Google Scholar]
- 16.Anttonen AK, Mahjneh I, Hamalainen RH, Lagier-Tourenne C, Kopra O, Waris L, et al. The gene disrupted in Marinesco-Sjogren syndrome encodes SIL1, an HSPA5 cochaperone. Nat Genet. 2005;37:1309–11. doi: 10.1038/ng1677. [DOI] [PubMed] [Google Scholar]
- 17.Senderek J, Krieger M, Stendel C, Bergmann C, Moser M, Breitbach-Faller N, et al. Mutations in SIL1 cause Marinesco-Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet. 2005;37:1312–4. doi: 10.1038/ng1678. [DOI] [PubMed] [Google Scholar]
- 18.Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, et al. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–79. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 19.Zhao L, Longo-Guess C, Harris BS, Lee JW, Ackerman SL. Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat Genet. 2005;37:974–9. doi: 10.1038/ng1620. [DOI] [PubMed] [Google Scholar]
- 20.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–28. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- 22.Weitzmann A, Volkmer J, Zimmermann R. The nucleotide exchange factor activity of Grp170 may explain the non-lethal phenotype of loss of Sil1 function in man and mouse. FEBS Lett. 2006;580:5237–40. doi: 10.1016/j.febslet.2006.08.055. [DOI] [PubMed] [Google Scholar]
- 23.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–79. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Tyson JR, Stirling CJ. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–52. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao L, Rosales C, Seburn K, Ron D, Ackerman SL. Alteration of the Unfolded Protein Response Modifies Neurodegeneration in a Mouse Model of Marinesco-Sjogren Syndrome. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991;88:2874–8. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weitzmann A, Baldes C, Dudek J, Zimmermann R. The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum - a quantitative approach. FEBS J. 2007;274:5175–87. doi: 10.1111/j.1742-4658.2007.06039.x. [DOI] [PubMed] [Google Scholar]
- 28.Jin Y, Awad W, Petrova K, Hendershot LM. Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 2008;27:2873–82. doi: 10.1038/emboj.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–72. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dudek J, Greiner M, Muller A, Hendershot LM, Kopsch K, Nastainczyk W, Zimmermann R. ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat Struct Mol Biol. 2005;12:1008–14. doi: 10.1038/nsmb1007. [DOI] [PubMed] [Google Scholar]
- 31.Hamman BD, Hendershot LM, Johnson AE. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–58. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- 32.Alder NN, Shen Y, Brodsky JL, Hendershot LM, Johnson AE. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J Cell Biol. 2005;168:389–99. doi: 10.1083/jcb.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer HA, Grau H, Kraft R, Kostka S, Prehn S, Kalies KU, et al. Mammalian Sec61 is associated with Sec62 and Sec63. J Biol Chem. 2000;275:14550–7. doi: 10.1074/jbc.275.19.14550. [DOI] [PubMed] [Google Scholar]
- 34.Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995;92:9643–6. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmermann R, Muller L, Wullich B. Protein transport into the endoplasmic reticulum: mechanisms and pathologies. Trends Mol Med. 2006;12:567–73. doi: 10.1016/j.molmed.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Bies C, Blum R, Dudek J, Nastainczyk W, Oberhauser S, Jung M, et al. Characterization of pancreatic ERj3p, a homolog of yeast DnaJ-like protein Scj1p. Biol Chem. 2004;385:389–95. doi: 10.1515/BC.2004.043. [DOI] [PubMed] [Google Scholar]
- 37.Yu M, Haslam RH, Haslam DB. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J Biol Chem. 2000;275:24984–92. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- 38.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–69. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol Biol Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.]Vembar SS, Jin Y, Brodsky JL, Hendershot LM. the mammalian HSP40 ERdj3 requires its HSP70 interaction and substrate binding properties to complement various yeast HSP40-dependent functions. J Biol Chem. 2009 doi: 10.1074/jbc.M109.000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen Y, Meunier L, Hendershot LM. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J Biol Chem. 2002;277:15947–56. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- 42.Kurisu J, Honma A, Miyajima H, Kondo S, Okumura M, Imaizumi K. MDG1/ERdj4, an ER-resident DnaJ family member, suppresses cell death induced by ER stress. Genes Cells. 2003;8:189–202. doi: 10.1046/j.1365-2443.2003.00625.x. [DOI] [PubMed] [Google Scholar]
- 43.Prols F, Mayer MP, Renner O, Czarnecki PG, Ast M, Gassler C, et al. Upregulation of the cochaperone Mdg1 in endothelial cells is induced by stress and during in vitro angiogenesis. Exp Cell Res. 2001;269:42–53. doi: 10.1006/excr.2001.5294. [DOI] [PubMed] [Google Scholar]
- 44.Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol Biol Cell. 2008;19:2620–30. doi: 10.1091/mbc.E07-07-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosoda A, Kimata Y, Tsuru A, Kohno K. JPDI, a novel endoplasmic reticulum-resident protein containing both a BiP-interacting J-domain and thioredoxin-like motifs. J Biol Chem. 2003;278:2669–76. doi: 10.1074/jbc.M208346200. [DOI] [PubMed] [Google Scholar]
- 46.Cunnea PM, Miranda-Vizuete A, Bertoli G, Simmen T, Damdimopoulos AE, Hermann S, et al. ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J Biol Chem. 2003;278:1059–66. doi: 10.1074/jbc.M206995200. [DOI] [PubMed] [Google Scholar]
- 47.Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–72. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 48.Riemer J, Appenzeller-Herzog C, Johansson L, Bodenmiller B, Hartmann-Petersen R, Ellgaard L. A luminal flavoprotein in endoplasmic reticulum-associated degradation. Proc Natl Acad Sci U S A. 2009;106:14831–6. doi: 10.1073/pnas.0900742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–82. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, et al. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci U S A. 2002;99:15920–5. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ladiges WC, Knoblaugh SE, Morton JF, Korth MJ, Sopher BL, Baskin CR, et al. Pancreatic beta-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54:1074–81. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- 52.Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, et al. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell. 2007;18:3681–91. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zahedi RP, Volzing C, Schmitt A, Frien M, Jung M, Dudek J, et al. Analysis of the membrane proteome of canine pancreatic rough microsomes identifies a novel Hsp40, termed ERj7. Proteomics. 2009;9:3463–73. doi: 10.1002/pmic.200800722. [DOI] [PubMed] [Google Scholar]
- 54.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–72. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 55.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 56.Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–70. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–8. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 58.Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996;15:753–63. [PMC free article] [PubMed] [Google Scholar]
- 59.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–7. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 60.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–40. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 61.Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172:383–93. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kokame K, Agarwala KL, Kato H, Miyata T. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J Biol Chem. 2000;275:32846–53. doi: 10.1074/jbc.M002063200. [DOI] [PubMed] [Google Scholar]
- 63.Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–34. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nadav E, Shmueli A, Barr H, Gonen H, Ciechanover A, Reiss Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem Biophys Res Commun. 2003;303:91–7. doi: 10.1016/s0006-291x(03)00279-1. [DOI] [PubMed] [Google Scholar]
- 65.Liang JS, Kim T, Fang S, Yamaguchi J, Weissman AM, Fisher EA, et al. Overexpression of the tumor autocrine motility factor receptor Gp78, a ubiquitin protein ligase, results in increased ubiquitinylation and decreased secretion of apolipoprotein B100 in HepG2 cells. J Biol Chem. 2003;278:23984–8. doi: 10.1074/jbc.M302683200. [DOI] [PubMed] [Google Scholar]
- 66.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 67.Voeltz GK, Rolls MM, Rapoport TA. Structural organization of the endoplasmic reticulum. EMBO Rep. 2002;3:944–50. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–81. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 69.Borgese N, Francolini M, Snapp E. Endoplasmic reticulum architecture: structures in flux. Curr Opin Cell Biol. 2006;18:358–64. doi: 10.1016/j.ceb.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kamhi-Nesher S, Shenkman M, Tolchinsky S, Fromm SV, Ehrlich R, Lederkremer GZ. A novel quality control compartment derived from the endoplasmic reticulum. Mol Biol Cell. 2001;12:1711–23. doi: 10.1091/mbc.12.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spiliotis ET, Pentcheva T, Edidin M. Probing for membrane domains in the endoplasmic reticulum: retention and degradation of unassembled MHC class I molecules. Mol Biol Cell. 2002;13:1566–81. doi: 10.1091/mbc.01-07-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimizu Y, Hendershot LM. Organization of the functions and components of the endoplasmic reticulum. Adv Exp Med Biol. 2007;594:37–46. doi: 10.1007/978-0-387-39975-1_4. [DOI] [PubMed] [Google Scholar]
- 73.Sevier CS, Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008;1783:549–56. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 74.Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, et al. Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J Biol Chem. 2000;275:23685–92. doi: 10.1074/jbc.M003061200. [DOI] [PubMed] [Google Scholar]
- 75.Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–48. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 76.Wakana Y, Takai S, Nakajima K, Tani K, Yamamoto A, Watson P, et al. Bap31 is an itinerant protein that moves between the peripheral endoplasmic reticulum (ER) and a juxtanuclear compartment related to ER-associated degradation. Mol Biol Cell. 2008;19:1825–36. doi: 10.1091/mbc.E07-08-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]