Abstract
The tumor suppressor p53 induces potent anti-proliferative responses in stressed cells; in unstressed cells this ability of p53 is restrained by Hdm2. Expression of Hdm2 is also induced by p53, thereby establishing feedback inhibition. Regulation of the p53-Hdm2 interaction and the feedback inhibition of p53 are not well understood. Here we show that the p53-Hdm2 interaction in unstressed cells is promoted by Siva1, which, like Hdm2, is the product of a p53 target gene. Siva1 binds to both p53 and Hdm2 through distinct regions and enhances Hdm2-mediated p53 ubiquitination and degradation. Siva1 strongly inhibits p53-mediated gene expression and apoptosis. In xenograft mouse models, down-regulation of Siva1 markedly inhibits tumor formation due to the activation of p53. Upon DNA damage, the interactions of Siva1 with both p53 and Hdm2 are diminished. These results identify Siva1 as an important adaptor promoting p53 degradation via Hdm2. Siva1 may be part of the negative feedback loop that inhibits p53 activity at the end of a non-lethal stress response.
Introduction
The p53 tumor suppressor activates anti-proliferative processes in response to a wide range of stresses including DNA damage and oncogene activation.1, 2 The potent anti-proliferative effect of p53 makes its tight regulation a central issue in higher organisms. An elaborate collection of cellular factors strictly restrains p53 function in unstressed cells, permitting cellular survival and proliferation. These factors activate p53 to provoke an appropriate response to the stress signal, and terminate p53 activation after a non-lethal stress, preventing cellular damage. p53 is primarily regulated at the level of protein stability. In unstressed cells, p53 levels are low due to rapid ubiquitination and proteasomal degradation mediated by E3 ligases. The principle E3 for p53 is Mdm2 (murine double minute, also known as Hdm2 for the human protein),3–5 the importance of which is underscored by the observation that the early embryonic lethality in mice with Mdm2 deficiency can be completely rescued by simultaneous inactivation of p53.6,7 Mdm2 is itself an unstable protein, and its stabilization in unstressed cells requires the adaptor protein Daxx and the de-ubiquitinase Hausp.8 Under stress conditions, p53 is activated mainly via the inhibition of Mdm2.9 For example, DNA damage leads to destabilization of Mdm2 through phosphorylation mediated by ATM (ataxia-telangiectasia, mutated), while oncogene activation causes inhibition of Mdm2 through the tumor suppressor Arf. Upon activation, p53 induces expression of a large number of genes that regulate apoptosis and the cell cycle progression. One such p53 target is the Mdm2 gene;10 this establishes a negative feedback loop that decreases the level of p53 after a non-lethal stress.
This potent anti-proliferative effect of p53 simultaneously provides a critical brake in tumor development and makes it a primary target for oncogenic mutations. Mutations in the p53 gene itself are found in half of all examined human tumors. In tumors retaining wild type p53, the function of p53 is often compromised due to alterations in its regulators and/or effectors. That p53 is mainly controlled by a single master regulator, Mdm2, makes the inhibition of the Mdm2-p53 interaction an attractive approach for re-activating p53 in p53 wild type tumors.11 Nutlin-3, a small compound that inhibits the p53-Mdm2 interaction, has shown promise in treating p53 wild type tumors in animal models.12 However, Mdm2 does not function alone, and other proteins have been implicated in Mdm2-mediated p53 ubiquitination and degradation, including Yin-Yang1,13 gankyrin,14 and Daxx.8 To date, the regulation of the p53-Mdm2 interaction and the negative feedback for p53 are not completely understood.
Siva1 was originally identified as a protein associated with the cytoplasmic tail of CD27 conveying an apoptotic signal.15 Ectopically expressed Siva1 also binds to Bcl-XL and inhibits Bcl-XL-mediated protection against UV radiation-induced apoptosis.16 Siva1 is induced by p53,17 and is also reported to participate in p53-dependent apoptosis in cerebella granule neurons.18 In this study, we show that Siva1 is a crucial regulator for the p53-Hdm2 interaction. Siva1 potently inhibits p53-dependent gene expression and apoptosis. Furthermore, down-regulation of Siva1 leads to marked suppression of tumor formation. Siva1 interacts with both p53 and Hdm2, and facilitates Hdm2-mediated ubiquitination and degradation of p53. This function of Siva1 appears to require its oligomerization and is disrupted by DNA damage signals. These results reveal Siva1 as an important mediator for the Hdm2-p53 interaction. In addition, Siva1 may also be an integral component of the negative feedback mechanism for p53 inhibition.
Results
Siva1 interacts with and de-stabilizes p53
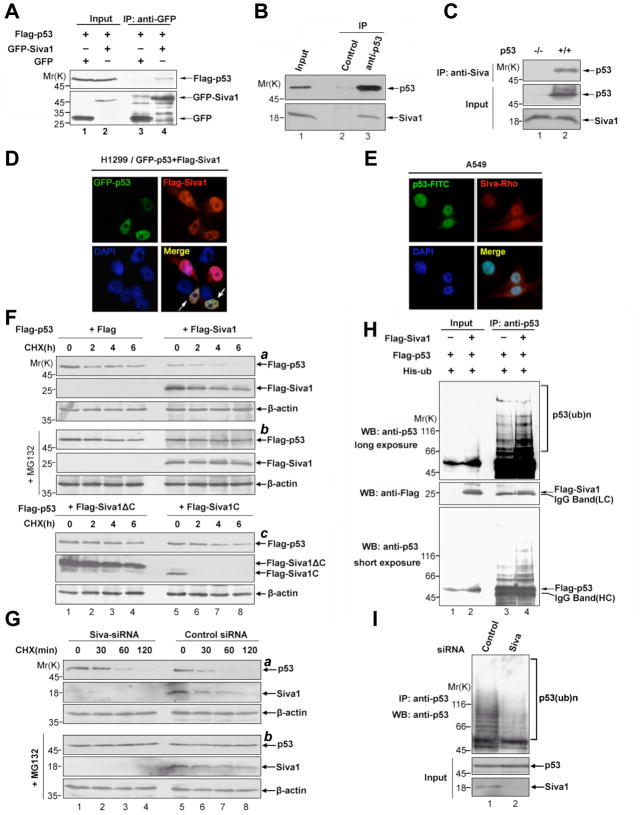
Given that Siva1 is implicated in p53-mediated apoptosis, we investigated whether Siva1 physically interacts with p53. We expressed Flag-tagged p53 together with either GFP-tagged Siva1 or GFP in human lung cancer H1299 cells, which lack endogenous p53. Using an anti-GFP antibody for immunoprecipitation, Flag-p53 was found to associate with GFP-Siva1, but not GFP (Figure 1A), indicating a specific interaction between ectopically expressed p53 and Siva1. We subsequently examined the interaction between endogenous Siva1 and p53. Endogenous Siva1 was immunoprecipitated by an anti-p53 antibody, but not by a control antibody, from lysates of the p53 wild type human pulmonary epithelial A549 cells (Figure 1B, lane 3 versus 2). Reciprocally, p53 was immunoprecipitated by an anti-Siva antibody from the lysates of the p53 wild type colon cancer HCT116 cells; the specificity of this co-immunoprecipitation was shown by the lack of p53 signal when the p53-null HCT116 cells were used (Figure 1C). The interaction of Siva1 and p53 was further examined by immunostaining assays. Ectopically expressed GFP-Siva1 and Flag-p53 were co-localized to the nucleus of H1299 cells (Figure 1D). Similarly, endogenous Siva1 accumulated in the nucleus of A549 cells, co-localizing with endogenous p53 (Figure 1E).
Figure 1. Siva1 physically interacts with and destabilizes p53.
(A) Ectopically-expressed Siva1 and p53 interact with each other in vivo. H1299 cells were transfected with Flag-p53 and either GFP-Siva1 or vector. Cells were treated with 20 μM MG132 for 6 h, and cell lysates were incubated with anti-GFP antibody. The lysates and immunoprecipitated proteins were analyzed by Western blotting.
(B)&(C) Interaction between endogenous Siva1 and p53. (B) Lysates from A549 cells treated with 20 μM MG132 for 6 h were immunoprecipitated with anti-p53 or a control antibody (anti-HA) and followed by Western blotting. (C) Lysates from HCT116 p53−/− and p53+/+ cells treated with 20 μM MG132 for 6 h were immunoprecipitated with anti-Siva antibody, and both lysates and immunoprecipitates were analyzed by Western blotting
(D) Colocalization of ectopically expressed Siva1 and p53. H1299 cells transfected with GFP-p53 and Flag-Siva1 were immunostained with rhodamine-conjugated anti-Flag antibody and DAPI.
(E) Colocalization of endogenous Siva1 and p53. A549 cells were stained with rhodamine-conjugated anti-Siva1, FITC-conjugated anti-p53 antibodies, and DAPI.
(F) Overexpression of Siva1, but not Siva1ΔC or Siva1C, destabilizes p53. H1299 cells were transfected with Flag-p53 and the indicated Flag-Siva1 or its derivatives. Cells were treated with (part b) or without (part a, c) MG132 (20 μM) for 1 h, and then treated with CHX (50 μg/ml) for indicated periods of time. Protein levels were analyzed by Western blotting using anti-Flag antibody. Expression of endogenous actin is shown as a loading control.
(G) siRNA-mediated down-regulation of Siva1 leads to p53 stabilization. HCT116 (p53+/+) cells transfected with siRNA-Siva were treated with or without MG132 (20 μM) for 1 h, and were treated again with CHX (50 μg/ml) for indicated periods of time. Levels of p53 and Siva1 were analyzed by Western blotting.
(H) Siva1 enhances p53 ubiquitination. H1299 cells were co-transfected with Flag-p53, His-ub, and either Flag-Siva1 or vector control. Cells were treated with MG132 for 6 h, and whole-cell lysates were subjected to immunoprecipitation with anti-p53 antibody.
(I) Knock-down of Siva1 causes a decrease in p53 polyubiquitination. HCT116 cells transfected with Siva-siRNA or scramble-siRNA were treated with MG132 (20 μM). Cell lysates were immunoprecipitated with anti-p53 antibodies, followed by Western blotting.
To define the region of Siva1 that interacts with p53, we constructed a panel of Siva1 deletion mutants (Supplementary Figure S1A). These Siva1 mutants were expressed in H1299 cells and tested for their interaction with recombinant GST-p53 protein in an in vitro pull down assay. Both the N-terminal region (Siva1N) and the middle DDHR region (Siva1DDHR) of Siva1 were able to associate with p53 (Supplementary Figure S1B, lanes 2, 3, and 5). In contrast, the C-terminal region (Siva1C), which consists of a B-Box-like domain and a zinc finger domain, was dispensable for the interaction (lane 4). Thus, Siva1 binds to p53 through two separate regions. We also mapped the region of p53 that interacts with Siva1 using a panel of p53 deletion mutations (Supplementary Figure S1C). Two p53 mutants that contain the DNA binding domain (DBD), p53ΔTA and p53Δ C, associated with Siva1, while a mutant p53 lacking a DBD (p53ΔDBD) did not (Supplementary Figure S1D). Therefore, p53 likely binds to Siva1 via its DBD domain.
We noticed that when co-expressed with p53, Siva1 significantly decreased the steady-state level of p53 (Figure 1F, part a, top panel, lanes 5 versus 1). A cycloheximide chase assay revealed that Siva1 significantly shortened the half-life of p53 (Figure 1F, part a, top panel, lanes 1–4 versus lanes 5–8). This inhibitory effect of Siva1 on p53 level was diminished when cells were treated with the proteasome inhibitor MG132 (Figure 1F, part b, top panel, lanes1–4 versus lanes 5–8), suggesting that Siva1 promoted p53 degradation in the proteasome. Moreover, the full length Siva1, not merely the p53-interacting domains, was needed for this inhibitory activity because neither Siva1C nor Siva1ΔC could de-stabilize p53 (Figure 1F, part c, top panel, lanes1–4 and lanes 5–8). To assess the effect of endogenous Siva1 on p53 level, we used siRNA to knock down the expression of Siva1 in HCT116 (p53+/+) cells. Knockdown of Siva1 noticeably prolonged the half-life of endogenous p53 (Figure 1G, panel a, lanes1–4 versus lanes 5–8). Again, this effect of Siva1 was diminished when cells were treated with MG132 (Figure 1G, panel b, lanes1–4 versus lanes 5–8). The effect of Siva1 on p53 half-life was indeed correlated with its effect on p53 ubiquitination. Over-expression of Siva1 led to enhanced ubiquitination of co-expressed p53 in H1299 cells (Figure 1H, lane 4 versus lane 3), while Siva1 knockdown via siRNA decreased ubiquitination of endogenous p53 in HCT116 (p53+/+) cells (Figure 1I, lane 2 versus lane 1). Together, these data show that Siva1 destabilizes p53 by enhancing its ubiquitination and proteasomal degradation.
Siva1 binds to Hdm2 and enhances Hdm2-mediated p53 degradation
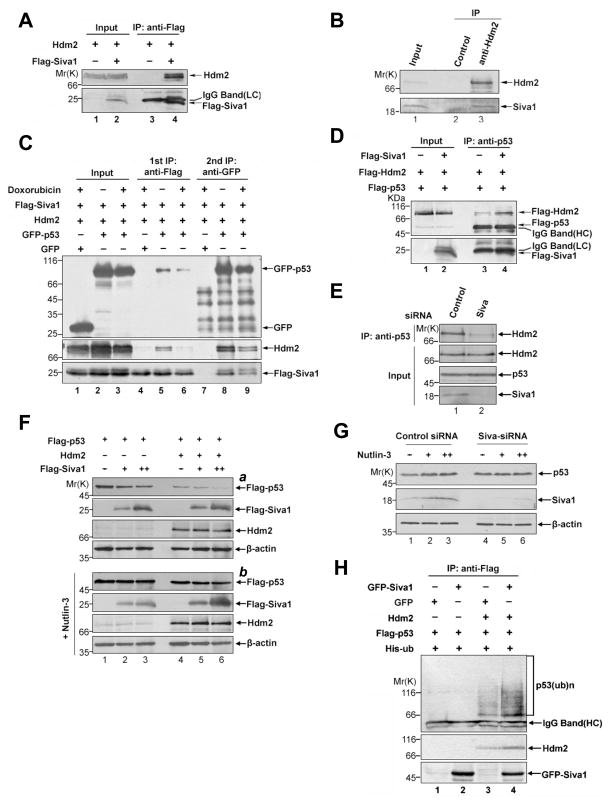
To investigate the mechanism by which Siva1 inhibits p53 stability, we first tested whether Siva1 is an E3 ubiquitin ligase. Siva1 contains a RING finger-like domain. However, Siva1 did not exhibit E3 ubiquitin ligase activity in an in vitro ubiquitination assay (data not shown). We then tested whether Siva1 might function as an adaptor linking p53 to an E3 ligase. A co-immunoprecipitation assay revealed a strong interaction between Siva1 and the principle p53 E3, Hdm2, when both were co-expressed in p53−/− Mdm2−/− mouse embryonic fibroblast cells (MEFs) (Figure 2A). Similarly, endogenous Siva1 and Hdm2 associated with each other in the osteosarcoma U2OS cells (Figure 2B). To define the region of Siva1 responsible for Hdm2 binding, we performed a GST pull-down assay using purified recombinant proteins. Hdm2 interacted with the C-terminal region of Siva1, rather than the N-terminal and the middle DDHR regions of Siva1 that bind to p53 (Supplementary Figure S2A). Siva1, on the other hand, interacted with the middle region of Hdm2 (aa 245–338) consisting of an acidic domain and a zinc finger domain (Supplementary Figure S2B and S2C). Despite interacting with Hdm2, Siva1 did not alter either the Hdm2 stability (Supplementary Figure S3A) or its ubiquitination (Supplementary Figure S3B).
Figure 2. Siva1 binds to Hdm2 and enhances Hdm2-mediated p53 degradation.
(A) Association of ectopically-expressed Siva1 and Hdm2. p53−/−mdm2−/− MEF cells were transfected with Hdm2 and either Flag-Siva1 or vector control. After treatment with 20 μM MG132 for 6 h, whole cell lysates were prepared and immunoprecipitated with anti-Flag antibody. The lysates and immunoprecipitated proteins were analyzed by Western blotting.
(B) Association of endogenous Siva1 and Hdm2. Whole-cell extracts from U2OS cells treated with 20 μM MG132 for 6 h were immunoprecipitated with anti-Hdm2 or a control antibody (anti-HA), and followed by Western analysis using antibody against Hdm2 and Siva.
(C) Siva1, p53 and Hdm2 form a ternary complex. Flag-Siva1, Hdm2, GFP and GFP-p53 were transfected into p53−/−Mdm2−/− MEF cells in the indicated combinations. Cells were treated with 20 μM MG-132 for 2 h and with 4 μM doxorubicin or left untreated for an additional 6 h. Cell lysates were immunoprecipitated with EzviewTM Red anti-Flag M2 Affinity Gel (lanes 4–6). Flag-Siva1 and the associated proteins were eluted with 3XFlag peptide. Twenty percent of the eluent was subject to Western analysis using indicated antibodies. The remaining eluent was used for secondary immunoprecipitation with anti-GFP antibody (lanes 7–9).
(D) Overexpression of Siva1 enhances Hdm2-p53 interaction. H1299 cells were transfected with Flag-Hdm2, Flag-p53, and either Flag-Siva1 or the vector control. Transfected cells were treated with MG132 for 6 h. The association of Flag-p53 and Flag-Hdm2 was analyzed by immunoprecipitation assay with anti-p53 antibody.
(E) Knockdown of Siva1 diminishes the Hdm2-p53 interaction. The blot (top panel) shown in Figure 1I was re-probing with antibody against Mdm2. Input was equivalent to 10% of the whole cell lysates used for Co-IP.
(F) Siva1 affects p53 steady-state level in H1299 cells. H1299 cells were co-transfected with a fixed amount of Flag-p53 (0.1 μg), increasing amount of Flag-Siva1 (0, 0.1 and 0.3 μg), and together with either GFP-Hdm2 or GFP as indicated, then treated with (part b) or without (part a) Nutlin-3 (10 μM) for 24 h. Cell lysates were analyzed by Western blotting. Expression of β-actin was shown as a loading control.
(G) Siva1 affects p53 steady-state level via Hdm2. HCT116 cells were transfected with either Siva-siRNA or scramble-siRNA (control), and were then treated with increasing amount of Nutlin-3 (0, 5, 10 μM) for 24h. Cell lysates were analyzed by Western blotting. Expression of β-actin was shown as a loading control.
(H) Siva1 increases p53 polyubiquitination. p53−/−Mdm2−/− MEF cells were transfected with Flag-p53, His-ubiquitin, Hdm2, GFP-Siva1, GFP in the indicated combinations. Transfected cells were grown in medium containing MG132 (20 μM) for another 6 h. Flag-p53 was immunoprecipitated using anti-Flag antibody and analyzed by Western blotting using anti-p53 antibody.
The observation that Siva1 interacts with p53 and Hdm2 through non-overlapping regions is consistent with the notion that Siva1 may promote the p53-Hdm2 interaction. To test this possibility, we first examined whether Siva1, p53 and Hdm2 form a ternary complex by a sequential immunoprecipitation assay. These three proteins were co-expressed in p53−/−Mdm2−/− MEF cells. An initial immunoprecipitation assay using an anti-Flag antibody against Flag-Siva1 pulled down both Hdm2 and GFP-p53 (Figure 2C, lane 5). The immunocomplexes were eluted and GFP-p53 was subsequently precipitated by an anti-GFP. Hdm2 and Flag-Siva1 were present in the anti-GFP-p53 precipitates (Figure 2C, lane 8), indicating that these three proteins form a ternary complex. Next, we tested whether Siva1 enhances the p53-Hdm2 interaction. Hdm2 and p53 were both expressed in H1299 cells with and without Siva1. The transfected cells were treated with the proteasome inhibitor MG132 to block protein degradation, thus equalizing the levels of p53 and Hdm2 in the lysates between cell groups. The Hdm2-p53 interaction was significantly enhanced when Siva1 was present (Figure 2D, lane 4 versus lane 3). Conversely, knockdown of endogenous Siva1 by siRNA in HCT116 (p53+/+) cells led to a significant decrease in the interaction of endogenous Hdm2 and p53 (Figure 2E, lane 2 versus lane 1). These results showed that Siva1 enhances the interaction between Hdm2 and p53.
To determine whether Siva1 promotes Hdm2-induced p53 degradation, p53 and Siva1 were co-transfected into H1299 cells in the presence or absence of pcDNA3-Hdm2. Siva1 dramatically decreased the levels of p53 in the presence, but not in the absence, of Hdm2 (Figure 2F, panel a, lanes 4–6 versus 1–3). In the absence of ectopically expressed Hdm2, the residual effect of Siva1 on p53 was likely mediated by endogenous Hdm2 in these cells. We thus used p53−/−Mdm2−/− MEF cells and found that Siva1 decreased p53 levels in these cells only in the presence of ectopically expressed Hdm2 (Supplementary Figure S3C). To confirm that Siva1 promotes p53 degradation in an Hdm2-dependent manner, we used Nutlin-3, which disrupts the p53-Hdm2 association. In the presence of Nutlin-3, overexpression of Siva1 did not decrease the p53 levels (Figure 2F, panel b), and knockdown of endogenous Siva1 failed to elevate p53 levels (Figure 2G, lanes 5, 6 versus lanes 2, 3). Consistent with the effect of Siva1 on Hdm2-mediated p53 degradation, over-expression of Siva1 strongly enhanced p53 polyubiquitination in an Hdm2-dependent manner (Figure 2H). Together, these data show that Siva1 enhances Hdm2 interaction with p53 and promotes Hdm2-mediated p53 ubiquitination and degradation.
Role of Siva1 oligomerization in p53 inhibition
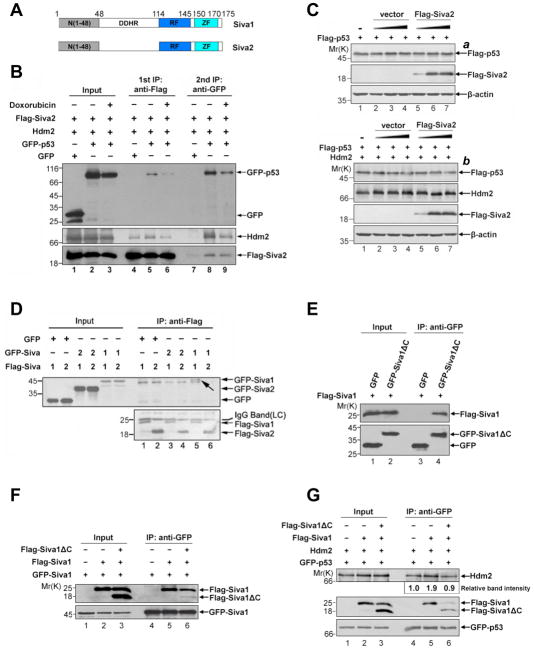
Siva2 is a short splicing variant of Siva1 that lacks the DDHR region (Figure 3A).19 To assess the contribution of this region to the function of Siva1, we assessed the properties of Siva2. Like Siva1, Siva2 associates with both p53 and Hdm2 (Supplementary Figure S4A and S4B), and forms a ternary complex with them (Figure 3B). An in vitro GST pull-down assay using purified recombinant proteins demonstrated that Siva2, like Siva1, enhanced the interaction between p53 and Hdm2 (Supplementary Figure S4C, top panel, lanes 11 and 12 versus lane 10). However, Siva2 did not affect p53 protein stability even in the presence of Hdm2 (Figure 3C, panel a versus panel b), nor did it enhance p53 ubiquitination (Supplementary Figure S4D). In an effort to assess the function of the DDHR domain, we found that Siva1, but not Siva2, formed homo-oligomers (Figure 3D, lane 5 versus 4). Siva2 did not interact with Siva1 either (lanes 3 and 6). To confirm that oligomerization of Siva1 is needed for enhancing Hdm2-mediated p53 degradation, we used Siva1Δ C, which retained the DDHR region but lacked the Hdm2-binding region. Indeed, Siva1ΔC interacted with Siva1 (Figure 3E) and decreased the oligomerization of Siva1 (Figure 3F, upper panel, lane 6 versus 5). To test whether Siva1Δ C affects the ability of Siva1 to enhance p53-Hdm2 interaction, we tested the Hdm2-p53 interaction in the presence of Siva1 alone or Siva1 plus Siva1ΔC. As expected, the Hdm2-p53 interaction was enhanced when Siva1 was present (Figure 3G, lane 5 versus lane 4). However, Siva1ΔC diminished this effect of Siva1 (lane 6 versus 5). Together, these results suggest that Siva1 oligomerization may be important for p53 inhibition.
Figure 3. Role of Siva1 oligomerization in p53 inhibition.
(A) Schematic representation of Siva1 and Siva2. The positions of amino-acid residues are indicated.
(B) Formation of a Siva2, p53 and Hdm2 complex. Flag-Siva2, Hdm2, GFP-p53 and GFP were expressed into p53−/−Mdm2−/− MEF cells with the indicated combinations. Cells were treated with 20 μM MG-132 for 2 h, and with or without doxorubicin (4 μM) for an additional 6 h. Cell lysates were immunoprecipitated with EzviewTM Red anti-Flag M2 Affinity Gel (lanes 4, 5 and 6). Flag-Siva2 and the associated proteins were eluted using 3XFlag peptide. 20% of the eluent was subject to Western analysis using indicated antibodies; the remaining eluent was used for secondary immunoprecipitation with anti-GFP antibody.
(C) Siva2 does not affect p53 stability. p53−/−Mdm2−/− MEF cells were transfected with Flag-p53 and an increasing amount (0, 0.3 and 0.6 μg) of Flag-Siva2, in the presence (lower part) or absence (upper part) of Hdm2.
(D) Siva1, but not Siva2, forms oligomers. p53−/−Mdm2−/− MEF cells were transfected with GFP, and GFP- and Flag-tagged Siva1 (1) and Siva2 (2) plasmids as indicated. Whole cell lysates were immunoprecipitated with anti-Flag antibody, followed by immunoblotting with anti-Flag and anti-GFP antibodies. The coprecipitated GFP-Siva1 is indicated by an arrow (upper panel, lane 5).
(E) Siva1ΔC was able to interact with Siva1. H1299 cells were co-transfected with Flag-Siva1 and GFP-Siva1ΔC. Whole cell lysates were immunoprecipitated with anti-GFP antibody, followed by immunoblotting with anti-Flag and anti-GFP antibodies.
(F) Siva1ΔC reduces the oligomerization of Siva1. GFP-Siva1, Flag-Siva1 and Flag-Siva1ΔC were expressed into p53−/−Mdm2−/− MEF cells with the indicated combinations. Whole cell lysates were immunoprecipitated with anti-GFP antibody. The lysates and immunoprecipitated proteins were analyzed by Western blotting.
(G) Siva1ΔC affects the Siva1-enhanced p53-Hdm2 interaction in a dominant-negative manner. GFP-p53, Hdm2, Flag-Siva1 and Flag-Siva1Δ C were expressed in p53−/−Mdm2−/− MEF cells in the indicated combinations and were further treated with MG132 for 6 h. Cell lysates were incubated with anti-GFP antibody for Co-IP. The lysates and immunoprecipitated proteins were analyzed by Western blotting.
Siva1-p53 and Siva1-Hdm2 interactions are regulated by DNA damage signaling
The interaction between p53 and Hdm2 is disrupted by stresses such as DNA damage, enabling p53 activation.9 We investigated whether Siva1 dissociates from Hdm2 and p53 upon DNA damage. When Siva1 and p53 were ectopically expressed in H1299 cells, their interaction was significantly weakened when the cells were treated with the genotoxic agent doxorubicin (Figure 4A, lane 4 versus lane 3). The interaction between endogenous Siva1 and p53 in A549 cells was also diminished upon DNA damage (Figure 4B, upper panel, lane 4 versus lane 3). Similarly, the interactions between both ectopically expressed and endogenous Siva1 and Hdm2 were decreased upon doxorubicin treatment (Figures 4C and 4D). Consistently, DNA damage signaling weakened the Siva1-p53-Hdm2 complex (Figure 2C, lane 6 versus lane 5). The disruption of Siva1-p53 and Siva1-Hdm2 interactions occurred when cells were treated with MG132, which blocked Mdm2 and p53 degradation and equalized the levels of these two proteins under unstressed and stressed conditions. Thus, the dissociation of Siva1 and p53 and Hdm2 reflected a change in their intrinsic affinity rather than in protein levels. The dissociation of Siva1 from p53 and Hdm2 likely enables p53 activation on DNA damage.
Figure 4. Disruption of the p53-Siva1-Hdm2 complex by DNA damage signaling.
(A) & (B) Doxorubicin reduces the interaction between Siva1 and p53. (A) H1299 cells were co-transfected with Flag-Siva1 and GFP-p53. After 24 h, cells were treated with 20 μM MG-132 for 2 h and further treated with or without 4 μM doxorubicin for an additional 6 h. The whole cell lysates were prepared for immunoprecipitation with anti-Flag M2 beads, and followed by immunoblotting with anti-Flag and anti-GFP antibodies. (B) A549 cells were treated with 20 μM MG-132 for 2 h, and further treated with or without 4 μM doxorubicin for an additional 6 h, and the whole cell lysates were then immunoprecipitated with anti-p53 antibodies, followed by immunoblotting with anti-p53 and anti-Siva antibodies.
(C) & (D) Doxorubicin reduces the interaction between Siva1 and Hdm2. (C) p53−/−Mdm2−/− MEF cells were co-transfected with Hdm2 and either Flag vector or Flag-Siva1. After 24 h, the cells were treated with 20 μM MG-132 for 2 h, followed by treating with or without 4 μM doxorubicin for an additional 6 h. Whole cell lysates were collected and immunoprecipitated with anti-Flag antibody, followed by immunoblotting with anti-Flag and anti-Hdm2 antibodies. (D) U2OS cells were treated with 20 μM MG-132 for 2 h, followed by treatment with or without 4 μM doxorubicin for 6 h. Whole cell lysates were immunoprecipitated with either anti-Siva antibody or a control antibody, followed by immunoblotting with anti-Hdm2 and anti-Siva antibodies.
(E) ATM regulates the p53-Hdm2-Siva1 complex. HCT116 cells were transfected with a control siRNA or an ATM-specific siRNA. After 72 h, cells were treated with 20 μM MG-132 for 2 h and with 4 μM doxorubicin for an additional 6 h. The whole cell lysates were prepared for immunoprecipitation with anti-Hdm2 or anti-p53 antibody, and followed by immunoblotting with indicated antibodies (left part). Input (right part) was equivalent to 10% of the whole cell lysate used for Co-IP.
ATM plays a central role in responses to various forms of DNA damage.20 To investigate how DNA damage regulates the p53-Siva1-Hdm2 complex, ATM expression was knocked down by siRNA (Figure 4E, lanes 7, 8 versus lanes 5, 6). In ATM siRNA-treated cells, the interaction of Siva1 and p53 was no longer affected by doxorubicin treatment (Figure 4E, part a, bottom panel, lanes 3, 4 versus lanes 1, 2). Similarly, the Siva1-Hdm2 interaction was also less affected in ATM siRNA-treated cells compared to control siRNA-treated cells (Figure 4E, part b, bottom panel, lanes 3, 4 versus lanes 1, 2). Together, these data suggest that DNA-damage signals disrupt the p53-Hdm2-Siva1 complex through the activation of ATM.
Siva1 suppresses p53-mediated gene expression and apoptosis
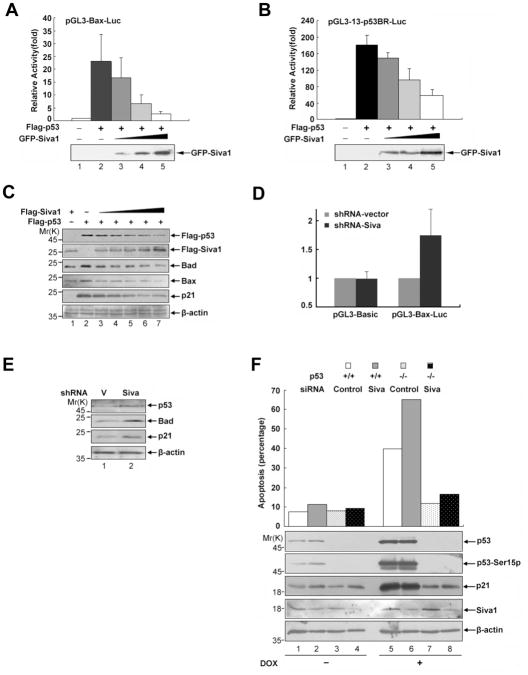
To determine the functional consequence of Siva1-mediated p53 inhibition, we tested the effect of Siva1 on p53-mediated gene expression. We used two p53-responsive reporter plasmids, pGL3-bax-Luc and pGL3-13-p53BR-Luc, in which the luciferase gene is controlled by the Bax promoter and an array of 13 consecutive synthetic p53 binding sites, respectively. When p53 was co-transfected with either reporter plasmid in H1299 cells, it potently activated luciferase expression (Figures 5A, B, columns 2). However, when Siva1 was expressed at the same time, it diminished p53-mediated reporter gene expression in a dose-dependent manner (Figures 5A, B, columns 3–5). To study the effect of Siva1 on the expression of endogenous p53-controlled genes, p53 was introduced into the H1299 cells alone or together with Siva1. The level of p53 declined as the dosage of Siva1 increased, and the expression of known p53 target genes, including Bad, Bax and p21, decreased accordingly (Figure 5C, lanes 3–7). Conversely, when Siva1 expression was knocked down by shRNA, the ability of p53 to activate the pGL3-Bax-Luc reporter (Figure 5D) and the expression of endogenous p21 and Bad (Figure 5E) was significantly enhanced.
Figure 5. Siva1 suppresses p53-mediated gene expression and apoptosis.
(A) & (B) Overexpression of Siva1 inhibits the transcriptional activity of p53. H1299 cells were transfected with Flag-p53 (0.1 μg) together with either pGL3-Bax-Luc (A) or pGL3-13-p53BR-Luc (B) in the absence or presence of increasing amounts of pGFP-Siva1 (0.1, 0.3 and 0.6 μg). The total amount of plasmid DNA per transfection was kept constant with pGFP-C1. Each transfection was performed in triplicate. Results are shown as fold induction of the firefly luciferase activity compared with that in control cells transfected with pGFP-C1 alone. Error bars indicate standard variations. Successful expression of GFP-Siva1 was detected by Western blotting.
(C) H1299 cells were co-transfected with a constant amount of Flag-p53 (0.1 μg) and an increasing amount of Flag-Siva1 (0.1, 0.2, 0.3, 0.4 and 0.5 μg). At 24 h post-transfection, whole-cell lysates were prepared and subjected to immunoblotting analysis with the indicated antibodies. Beta-actin was used as a control for equal loading.
(D) & (E) Knock-down of Siva expression enhances the p53 transcriptional ability. (D) U2OS cells stably expressing shRNA-vector or shRNA-Siva were transfected with either pGL3-Basic or pGL3-Bax-Luc. Each transfection was performed in triplicate. Results are shown as fold induction of the firefly luciferase activity compared with that in control cells transfected with shRNA-vector. Error bars indicate standard deviation. (E) U2OS cells stably expressing shRNA-vector or shRNA-Siva were analyzed for the expression of the indicated p53 target genes Bad and p21. β-actin was used as a control for equal loading.
(F) Knockdown of Siva sensitizes p53-dependent apoptosis. HCT116 (p53+/+) and HCT116 (p53−/−) cells were transfected with either a control siRNA or Siva siRNA. The cells were treated with or without 4 μM doxorubicin for 20 h. Apoptosis was quantified by FACS analysis of sub-G1 DNA content. Levels of p53 and Siva1 are shown and percentages of apoptosis are indicated.
We also assessed the effect of Siva1 on p53-induced apoptosis. The expression of Siva1 was knocked down in HCT116(p53+/+) and HCT116(p53−/−) cells. Doxorubicin induced apoptosis in a p53-dependent manner because it killed HCT116(p53+/+) cells but not HCT116(p53−/−) cells (Figure 5F, column 5 versus column 7). When Siva1 was knocked down in HCT116(p53+/+) cells, these cells showed heightened sensitivity to doxorubicin-induced apoptosis (column 6 versus column 5). These results confirm that Siva1 inhibits the function of p53 in inducing apoptosis.
Down-regulation of Siva1 inhibits tumorigenesis
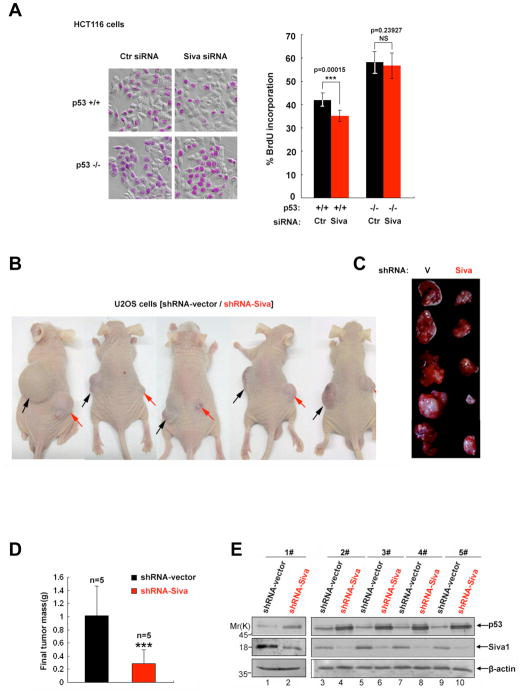
To study the effects of Siva1 on p53-induced inhibition of cellular proliferation, we performed a BrdU incorporation assay. Knockdown of Siva1 led to a noticeable decrease in BrdU incorporation in HCT116(p53+/+) but not in HCT116(p53−/−)cells (Figure 6A), suggesting that Siva1 enhances cell proliferation in a p53-dependent manner. To determine the role of Siva1 in p53-mediated tumor suppression, we used a xenograft animal model. U2OS cells stably expressing Siva shRNA or the vector control were injected subcutaneously into the opposite flanks of the same nude mice. Tumor cells stably expressing the vector control developed larger tumors 5 weeks after injection (Figure 6B, black arrowheads, and Figure 6C, left column). Notably, knockdown of Siva1 resulted in dramatic suppression of tumor growth (Figure 6B, red arrowheads, and Figure 6C, right column). The mean tumor weights were considerably decreased in the Siva shRNA group (n=5 in each group) (Figure 6D). To verify whether this tumor suppression is related to p53 expression, levels of p53 in vector control versus Siva1 knockdown excised tumors were compared. As shown in Figure 6E, increased p53 levels were observed in Siva1 knockdown tumors compared with those in the control samples. Thus, the decrease in tumor size for Siva shRNA-expressing cells is likely due to higher levels of endogenous p53 protein. The dramatic effect of Siva1 down-regulation on tumor formation indicates that Siva1 is a potent inhibitor of p53.
Figure 6. Down-regulation of Siva1 suppresses tumor formation.
(A) HCT116 p53+/+ and HCT116 p53−/− cells were pulsed with BrdU for 1 hour prior to fixation. BrdU immunofluorescence analysis was performed to determine the percentages of cells undergoing DNA synthesis. Cells were imaged in bright field to score total cell number. Error bars indicate standard deviations, *** = p<0.005, NS = no significant difference.
(B–D) Knockdown of Siva expression decreases tumorigenicity. (B) U2OS tumor cells expressing shRNA-vector control and shRNA-Siva (2 × 106 for each side) were subcutaneously inoculated in dorsal left or right flanks of 5-week-old nude mice (n=5) respectively. Representative pictures of tumor bearing nude mice were taken 5 weeks after inoculation. (C) The volumes of all flank tumors excised from 5 mice were compared (n=5). (D) Histograms show data points representing average tumor weights for each treatment.
(E) Siva shRNA-expressing tumor showed higher levels of p53. Paired tumor samples (shRNA-Siva versus shRNA-vector) from five mice were analyzed by Western blotting to evaluate the expression of Siva1 and p53. β-actin was used as a loading control.
Discussion
Mdm2 is the principle p53 antagonist that keeps p53 at a low concentration in unstressed cells to allow for normal growth. In a non-lethal stress response, Mdm2 also limits the duration and intensity of p53 activation.5 This current study reveals Siva1 as an important co-factor for Mdm2 to inhibit p53. Previous studies suggested that Siva1 is a pro-apoptotic protein. However, these studies relied almost exclusively on overexpression of Siva1, which might have generated a squelching effect.15–18 Using multiple approaches, we have shown that Siva1 exhibits potent inhibitory effects on p53 function. Siva1 strongly inhibits p53-mediated gene expression, apoptosis, and cell growth inhibition; in a xenograft tumor model, Siva1 down-regulation leads to a dramatic decrease in tumor formation correlating with the up-regulation of p53 in these tumors.
Siva1 is unique among Hdm2 co-factors. Unlike Daxx and Hausp, Siva1 does not promote Hdm2 stability; rather, Siva1 enhances Hdm2-mediated p53 ubiquitination and degradation. Consistent with this notion, Siva1 binds to both Hdm2 and p53 through non-overlapping regions, enhances Hdm2 interaction with p53, and promotes p53 degradation in an Hdm2-dependent manner. Furthermore, Siva1 mutants that can bind to either p53 or Hdm2, but not both, have no effect on the stability of p53 (Figure 1F). However, simultaneous binding to both p53 and Mdm2 may not be sufficient for enhancing p53 degradation. The short isoform of Siva1, Siva2, forms a ternary complex with p53 and Mdm2 but fails to destabilize p53. We found that Siva1 self-associates through the DDHR region, absent in Siva2. Siva1 oligomerization may allow for the assembly of a higher order complex that facilitates the close proximity of Mdm2 and p53. Alternatively, the Siva1 oligomer may enhance the activation of Mdm2 (Supplementary Figure S4E). The latter is consistent with the observation that the oligomeric form of Mdm2 is more active than the monomeric form.21, 22
The interactions of Siva1 with both p53 and Hdm2 are weakened by DNA-damage signaling, leading to the disruption of the p53-Siva1-Hdm2 complex. This disruption is likely mediated by ATM, the kinase that orchestrates the DNA-damage response (Figure 4). Although Yin Yang1 and gankyrin have been reported to promote Hdm2-mediated p53 ubiquitination,13,14 it is not clear whether the interactions involving Yin Yang1 and gankyrin are affected by DNA-damage signaling. The precise mechanism of the Hdm2-Siva1-p53 complex disruption remains to be determined. Nevertheless, the disruption of this complex likely weakens the effect of Hdm2 on p53 and contributes to p53 activation induced by DNA damage. Thus, the interactions involving Siva1 may be a focal point for p53 regulation. Siva1 is also notable among Hdm2 co-factors in that its expression is induced by p53. This property of Siva1 suggests that it is part of the auto-regulatory feedback loop that restrains p53 after a non-lethal stress response. Stimulating p53 activity through the inhibition of the Mdm2-p53 interaction is an attractive strategy to eradicate tumor cells expressing wild type p53.11 As an important molecule that links Mdm2 with p53, Siva1 may represent an alternative target for cancer therapy.
Materials and Methods
Reagents and antibodies
The following antibodies were used in this study: anti-Bad, anti-Bax, and anti-Hdm2 (SMP14) (Santa Cruz Biotechnology, Santa Cruz, CA); anti-Hdm2 (Ab-1, Ab-4) and anti-p53 (Ab-6) (Oncogene, Manhasset, NY); anti-GFP (BD Biosciences, Palo Alto, CA); anti-p21 and anti-Flag (Sigma, St Louis, MO); anti-actin (Abcam, Cambridge, UK), anti-Siva (Santa Cruz); and Siva polyclonal antibody (Abnova, Taiwan). Most of the chemicals and reagents, including DAPI, Doxorubicin and Nutlin-3, were purchased from Sigma (Sigma, St Louis, MO).
Cell cultures, siRNA, and the generation of Siva1 stable cell lines
A549, U2OS, H1299 and p53−/−Mdm2−/− MEF cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. For knockdown of Siva1, both Siva siRNA and Siva-shRNA were used. Siva siRNA (sc-37385) was purchased from Santa Cruz Biotechnology, CA. The shRNA fragment targeting Siva1 5′-cagtgacatgtacgagaaa-3′ was cloned into a retroviral derivative vector pSUPER shRNA and the resultant recombinant vector was introduced into U2OS cells to generate a stable Siva-knockdown cell line using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Stable transfectants were collected separately from shRNA-Siva-transfected pools and shRNA-vector-transfected control pools after culturing in selective media (1 μg/ml puromycin).
Immunoprecipitation and Western blotting
Whole cell lysates were made in lysis buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate and complete protease cocktail) for 2 h, and then pre-cleared by centrifugation. The supernatants were incubated with protein A/G-coupled Sepharose beads coated with indicated antibodies for at least 4 h or overnight at 4°C. Beads were washed twice with lysis buffer, four times with ice-cold PBS and boiled in 2×loading buffer. Protein samples were resolved by SDS-PAGE and transferred onto nitrocellulose membrane, which was blocked in 5% skim milk in TBST and probed with the indicated antibodies.
Indirect immunofluorescence
Cells cultured on coverslips were washed twice with cold PBS, fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 5 min, blocked with 5% BSA and incubated with antibodies as indicated, and were followed by a Rodamine-conjugated anti-rabbit IgG and a FITC-conjugated anti-mouse IgG antibody. The cells were mounted with DAPI reagent (Vector Laboratories, Burlingame, CA) and the images were acquired with an Olympus DP71X microscope.
Tumorigenesis in nude mice
Cells (2×106) were subcutaneously injected into the dorsal flanks of 4- to 5-week-old, male athymic nude mice (Shanghai SLAC Laboratory Animal Co. Ltd.). After 5 weeks, mice were photographed and tumors were excised and weighed. Proteins extracted from these tumors were used for western blot analysis. A total of 5 athymic nude mice were used and all animal experiments were performed strictly in accordance with the local Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Dr. Serge Benichou (Département de Maladies Infectieuses, Institut Cochin, France) for kindly providing GFP-Siva1. Plasmid pcDNA3-Hdm2 was a gift of Dr. Geoffrey M Wahl (The Salk Institute, La Jolla, USA). This research was supported by grants from the National Natural Science Foundation of China (30530200 and 30728003), the Ministry of Science and Technology of China (2002CB713702, 2006CB933300 and 2006CB910300), the Chinese Academy of Sciences (KSCX1-YW-R-57), and the NIH (CA088868). We have no conflicting financial interests.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 3.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 4.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 5.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 6.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 7.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 8.Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8:855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- 9.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 10.Barak Y, Juven T, Haffner R, Oren M. Mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 12.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 13.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo Y, Masuda T, et al. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005;8:75–87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Prasad KV, Ao Z, Yoon Y, Wu MX, Rizk M, Jacquot S, et al. CD27, a member of the tumor necrosis factor receptor family, induces apoptosis and binds to Siva, a proapoptotic protein. Proc Natl Acad Sci USA. 1997;94:6346–6351. doi: 10.1073/pnas.94.12.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue L, Chu F, Cheng Y, Sun X, Borthakur A, Ramarao M, et al. Siva-1 binds to and inhibits BCL-X(L)-mediated protection against UV radiation-induced apoptosis. Proc Natl Acad Sci USA. 2002;99:6925–6930. doi: 10.1073/pnas.102182299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortin A, MacLaurin JG, Arbour N, Cregan SP, Kushwaha N, Callaghan SM, et al. The proapoptotic gene SIVA is a direct transcriptional target for the tumor suppressors p53 and E2F1. J Biol Chem. 2004;279:28706–28714. doi: 10.1074/jbc.M400376200. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs SB, Basak S, Murray JI, Pathak N, Attardi LD. Siva is an apoptosis-selective p53 target gene important for neuronal cell death. Cell Death Differ. 2007;14:1374–1385. doi: 10.1038/sj.cdd.4402128. [DOI] [PubMed] [Google Scholar]
- 19.Py B, Slomianny C, Auberger P, Petit PX, Benichou S. Siva-1 and an alternative splice form lacking the death domain, Siva-2, similarly induce apoptosis in T lymphocytes via a caspase-dependent mitochondrial pathway. J Immunol. 2004;172:4008–4017. doi: 10.4049/jimmunol.172.7.4008. [DOI] [PubMed] [Google Scholar]
- 20.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 21.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.