Abstract
Various combinations of subunits assemble to form the NMDA-type glutamate receptor (NMDAR), generating diversity in its functions. Here we review roles of the unique NMDAR subunit, NR3A, which acts in a dominant-negative manner to suppress receptor activity. NR3A-containing NMDARs display striking regional and temporal expression specificity, and, unlike most other NMDAR subtypes, they have a low conductance, are only modestly permeable to Ca2+, and pass current at hyperpolarized potentials in the presence of magnesium. While glutamate activates triheteromeric NMDARs composed of NR1/NR2/NR3A subunits, glycine is sufficient to activate diheteromeric NR1/NR3A-containing receptors. NR3A dysfunction may contribute to neurological disorders involving NMDARs, and the subunit offers an attractive therapeutic target given its distinct pharmacological and structural properties.
1. Introduction
N-methyl-D-aspartate receptors (NMDARs) are activated by glutamate, the most common excitatory neurotransmitter in the central nervous system, and are essential for the proper development of cortical circuitry and synaptic function (Cull-Candy et al., 2001; Lau and Zukin, 2007; McBain and Mayer, 1994; Perez-Otano and Ehlers, 2004). Dysfunction of NMDARs has been implicated in a variety of pathological conditions including schizophrenia (Mueller and Meador-Woodruff, 2004), neurodegenerative disease (Bonuccelli and Del Dotto, 2006; Fan and Raymond, 2007; Wenk, 2006), stroke (Martin et al., 1998), white-matter injury (Karadottir et al., 2005; Micu et al., 2006; Salter and Fern, 2005), and neuropathic pain (Dubner and Ruda, 1992).
Great diversity exists in NMDAR subtypes. The NMDAR is thought to be a tetrameric transmembrane channel (Laube et al., 1998; Rosenmund et al., 1998; Ulbrich and Isacoff, 2007) composed of combinations of the obligatory NR1 subunit with NR2 and/or NR3 subunits (Furukawa et al., 2005; Schorge and Colquhoun, 2003). The NR1 subunit exists as eight functional splice variants (Durand et al., 1993; Durand et al., 1992; Hollmann et al., 1993; Mori and Mishina, 1995; Sugihara et al., 1992), while separate genes produce four types of NR2 (NR2A-D) and two types of NR3 (NR3A-B) subunits (Ciabarra et al., 1995; Meguro et al., 1992; Mishina et al., 1993; Monyer et al., 1992; Moriyoshi et al., 1991; Nishi et al., 2001; Sucher et al., 1995). Each receptor subtype exhibits temporal and regional specificity (Monyer et al., 1994; Sheng et al., 1994) and unique functional properties (Monyer et al., 1992; Paoletti and Neyton, 2007) which are determined by the specific combination of subunits assembled to form the heteromer. For instance, subunit composition determines receptor properties such as glutamate affinity, receptor desensitization, and pharmacological sensitivity (Cull-Candy and Leszkiewicz, 2004; Paoletti and Neyton, 2007). A major goal in neuroscience is to characterize the contributions of individual subunit types to NMDAR function. Such knowledge will increase our understanding of how NMDARs contribute to normal brain development, as well as guide therapeutic strategies for treating pathologies associated with NMDAR dysfunction.
While attention has focused on the role of NR2 subunits in neural function, much less is known about how the more recently identified NR3 subunits, NR3A and NR3B, modify NMDAR functions. This is surprising, given that NR3 subunits act in a novel, dominant-negative manner to suppress NMDAR activity (Ciabarra et al., 1995; Sucher et al., 1995) (Matsuda et al., 2002; Nishi et al., 2001). Interest in NR3A has grown with the observations that the subunit influences dendritic spine density (Das et al., 1998; Roberts et al., 2009), synapse maturation (Roberts et al., 2009), memory consolidation (Roberts et al., 2009), cell survival (Nakanishi et al., 2009), and may be involved in certain neuropathologies (Karadottir et al., 2005; Micu et al., 2006; Mueller and Meador-Woodruff, 2004; Salter and Fern, 2005; Zhang et al., 2002).
Although NR3A was identified over a decade ago, there have been few efforts to consolidate knowledge of its functions and possible disease roles (although see (Cavara and Hollmann, 2008; Eriksson et al., 2007a). Here we provide a timely primer of NR3A, with a limited discussion of NR3B, to summarize the major findings surrounding this receptor subunit and its suggested roles in neuropathologies. Given the large number of disorders associated with NMDAR dysfunction, and the paucity of studies that have examined the putative contribution of NR3 subtypes, we anticipate that its unique structure, expression patterns, and function will make it an important therapeutic target.
2. NR3A amino acid sequence and structure
NR3A was identified through sequence and structure homology to other GRIN (Glutamate Receptor Ionotropic N-methyl-D-aspartate) genes, and originally termed ‘χ-1’ or ‘NMDAR-like’ (NMDAR-L) (Ciabarra et al., 1995; Sucher et al., 1995). The GRIN3A gene localizes to human chromosome 9q34.1, and consists of 9 exons spanning 1115 amino acids (Andersson et al., 2001) (Figure 1). The high level of sequence homology between human and rodent NR3A (93%) indicates that its function is likely similar between mammalian species (Andersson et al., 2001; Eriksson et al., 2002). Most consensus sites are conserved between rodents and humans, although the lack of gene equivalents in D. melanogaster and C. elegans suggests that evolutionary changes may have necessitated the emergence of NR3 subtypes in vertebrates (Matsuda et al., 2002). By sequence alignment, NR3A shares the greatest homology with NR3B (GRIN3B; 57%), having low identity with NR1 (27%) and NR2 subunits (24–29%) as well as non-NMDA glutamate receptors (23%) (Andersson et al., 2001).
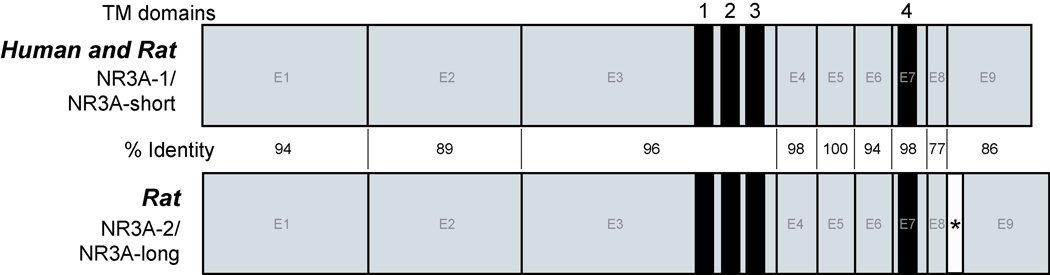
Figure 1.
Schematic diagram of cDNA structures for human and rat NR3A. Both human and rat have NR3A-1 isoforms, also known as NR3A-short (or -s). Rats also have an NR3A-2 isoform, also known as NR3A-long (or -l). Exons 1–9 appear in blue filled boxes (E1-E9). Predicted transmembrane (TM) domains 1–4 are indicated, and region of highest homology is between TM 1 & 4. Lowest homology is found in the C-terminus, which corresponds to the area following TM4. Location of the alternative splice variant (20-amino acid insert) in rodents is denoted in white with an asterisk (*). Percent identity in exon 9 is between NR3A-short sequences, and does not include the 20-amino acid insert found in rat.
NR3A shares structural features with other ionotropic glutamate receptor subunits (Figure 2), including a 4-pass transmembrane topology which forms the channel pore, a large extracellular N-terminal ligand-binding domain with a putative signal peptide and multiple glycosylation consensus sites, and a cytoplasmic tail. Like other NMDAR subunits, the intracellular C-terminus of NR3A contains sites potentially phosphorylated by protein kinases A and C (PKA, PKC), protein tyrosine kinase (PTK), and calcium/calmodulin-dependent kinase II (CaMKII) (Andersson et al., 2001; Ciabarra et al., 1995; Eriksson et al., 2002; Nishi et al., 2001; Sucher et al., 1995), suggesting that phosphorylation could play critical roles in regulating NR3A trafficking, signaling, and channel properties (Chen and Roche, 2007). NR3A also contains a motif with two vicinal cysteine residues, CC(Y/K)G(Y/F)CID(I/L)L, required for redox modulation (Andersson et al., 2001; Sucher et al., 1995; Sullivan et al., 1994), as well as an intracellular RXR motif which may serve as an endoplasmic reticulum (ER) retention signal (Perez-Otano et al., 2001) .
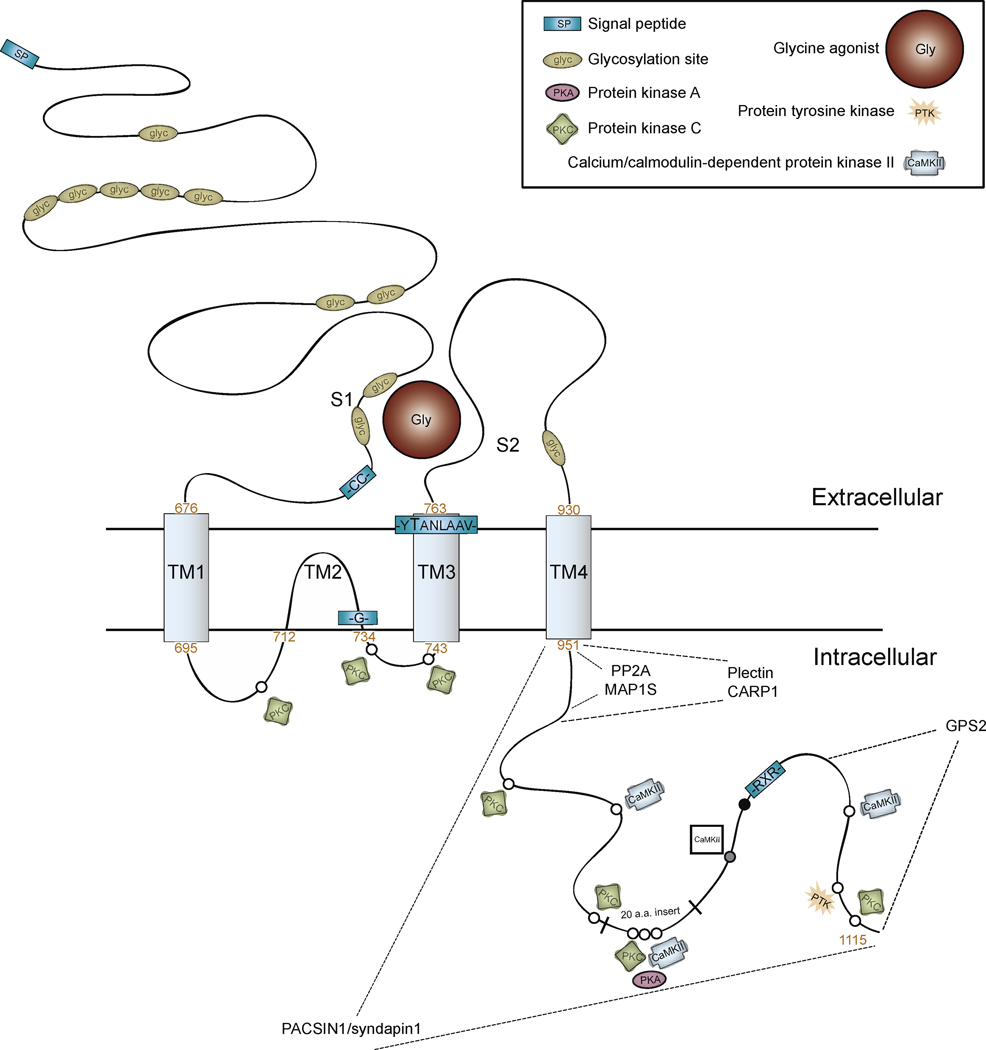
Figure 2.
Putative transmembrane topology of rat NR3A. Predicted sites are shown for signal peptide (SP), glycosylation (glyc), phosphorylation (PKC, CaMKII, PTK), and protein binding (indicated by dotted lines for PACSIN1/syndapin1, MAP1S, PP2A, plectin, CARP1, and GPS2). Sequence motifs (-CC-, -G-, -YTANLAAV-, -RXR-), transmembrane regions (TM1-TM4) and the extracellular ligand-binding domain/glycine binding pocket (S1-S2) are noted. Predicated kinase phosphorylation sites are indicated by open circles (○). A polyproline motif is indicated by a single black circle (●). CaMKII site predicted for human but not rat is indicated within a square symbol. MAP1 S and PP2A binding sites are overlapping and found just intracellular to TM4. TM2 and TM3 segments are thought to form the channel pore. Alternative splicing in rodents but not humans produces a twenty amino acid insert (-SRWRRWTCKTEGDSELSLFP-). This area contains potential phosphorylation sequences for PKA, PKC, and CaMKII. Most sites have been proposed by sequence analysis and are not verified. Amino acid numbers appear in orange. Figure based on sequence data from (Andersson et al., 2001).
Despite similarities to other NMDAR subunits, NR3A exhibits a number of distinctive features. As with other NMDAR subunits, the NR3A amino acid sequence encodes an extracellular bi-lobed domain (split into S1 and S2 segments by membrane domains TM1, TM2, and TM3) that determines ligand binding specificity. But unlike NR2 subunits, which bind glutamate, the S1 region of the extracellular N-terminus and the S2 region of the extracellular loop between TM3 and TM4 form a glycine binding pocket, a clamshell-like structure (Paas, 1998; Yao et al., 2008; Yao and Mayer, 2006). Additionally, the second membrane pore loop (TM2) and the third membrane pore loop (TM3) in both NR3A and NR3B are substantially different from other NMDAR subunits. In NR1/NR2 NMDARs, an ion selectivity filter at TM2 is formed by an interaction between asparagine (N) residues of NR1 (N-site) and NR2 subunits (N+1 site) (Kuner et al., 1996). In NR3A subunits, however, the N-site is replaced by a glycine (G) (Matsuda et al., 2002; Nishi et al., 2001; Sucher et al., 1995), and this residue apparently has little influence upon ion selectivity (Wada et al., 2006). While NR3A-containing NMDARs apparently lack the N and N+1 site selectivity filter, they appear to have a novel construction of the outer vestibule that can influence ionic currents. Unlike NR1/NR2 NMDARs, the TM3 domain between NR1 and NR3A subunits has a symmetrical configuration that forms a ring of threonines. This ring constricts the external vestibule and may account for the observed reductions in Ca2+ permeability, ionic currents, and magnesium (Mg2+) sensitivity of NR3A-containing NMDARs (Wada et al., 2006). Another distinct characteristic is that the C-termini of NR3A and NR3B lack consensus sequences for PDZ domain protein-binding (Eriksson et al., 2007a; Matsuda et al., 2002), prominent features of other glutamate receptor subunits that permit stable anchoring to the postsynaptic density. Finally, with a calculated molecular weight of ~125kD, NR3A is the most heavily glycosylated glutamate receptor subunit (Ciabarra and Sevarino, 1997). The functional significance of this robust glycosylation is unclear, but it may alter channel gating kinetics (Chazot et al., 1995; Covarrubias et al., 1989).
Alternative mRNA splicing is another means to modify NR3A functions. A 60-base pair insert encoding an additional 20 amino acid sequence at the intracellular C-terminus has been described in rodents, resulting in short (NR3A-s or NR3A-1) and long (NR3A-l or NR3A-2) forms of NR3A (Eriksson et al., 2002; Sasaki et al., 2002; Sun et al., 1998) (Figure 1 and Figure 2). The long splice variant NR3A-l, which contains putative PKA, PKC, and CaMKII phosphorylation sites, is apparently lacking in human NR3A (Andersson et al., 2001; Eriksson et al., 2002).
3. Developmental and regional expression
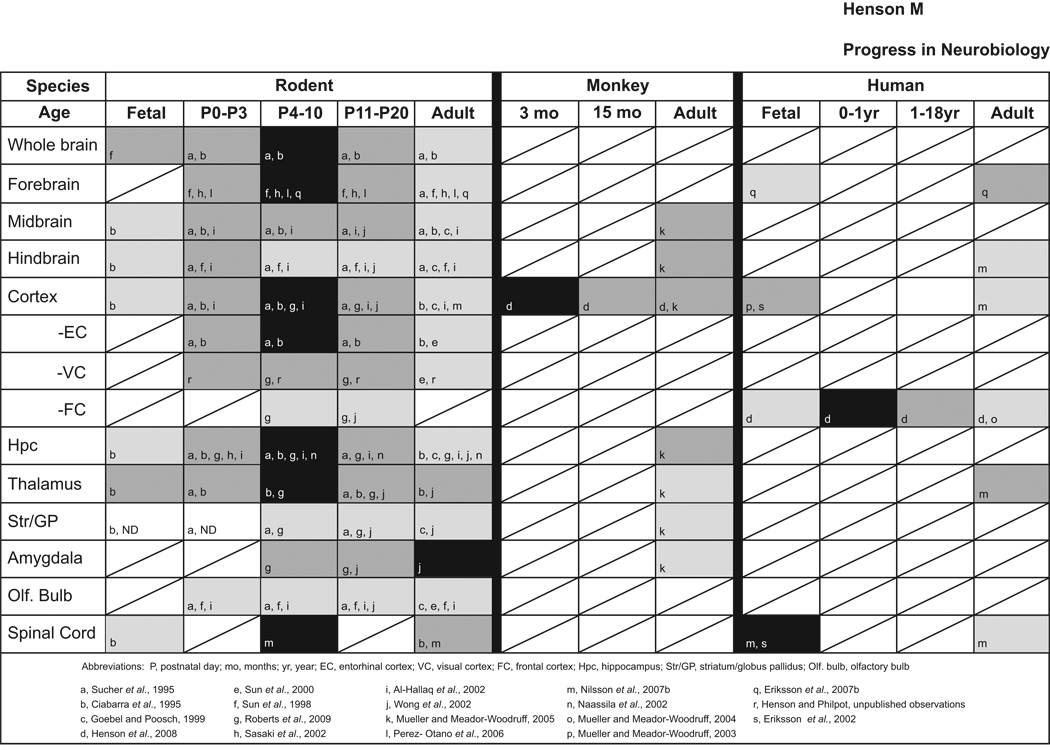
NR3A levels change dramatically over development and exhibit a unique spatiotemporal expression pattern when compared to other NMDAR subunits. While NR3A expression is grossly similar across mammalian species, there are some notable differences. Understanding these distribution patterns will help predict how prospective NR3A agonists and antagonists are likely to affect neural function. Because the existing data on NR3A expression patterns are sometimes contradictory, the information is summarized here in tabular format to present an overview of the consensus findings from brain tissue (Table 1).
Table 1.
Developmental profile of NR3A expression patterns in rodent, monkey, and human brain. Consensus results of mRNA and protein expression data are depicted as having high (dark), moderate (gray), weak (light gray), and non-detectable (ND) levels of expression. Boxes with diagonal lines indicate no data available to date. Letters within boxes denote references as matched below. Most studies have been conducted in rodents and reveal a general pattern of low prenatal presence of NR3A that increases dramatically in the first two postnatal weeks, and declines into adulthood. When both isoforms were reported, only data from expression of NR3A-short sequences were considered. Note that this schematic is meant to present a general overview of NR3A expression, but the schematic is unable to depict data reported from specific subregions/cell layers (for these details, see cited manuscripts).
 |
3.1 Ontogenetic profiles
Overall, NR3A expression is low embryonically, peaks during early postnatal life, and diminishes to much lower levels in adulthood. This expression profile is observed in many regions of the brain and in different species (Table 1), suggesting that it may play a specific role during postnatal neural development amongst mammals (Al-Hallaq et al., 2002; Bendova et al., 2009; Ciabarra et al., 1995; Das et al., 1998; Eriksson et al., 2002; Eriksson et al., 2007b; Goebel and Poosch, 1999; Henson et al., 2008; Mueller and Meador-Woodruff, 2003, 2004, 2005; Naassila and Daoust, 2002; Nilsson et al., 2007b; Perez-Otano et al., 2006; Roberts et al., 2009; Sasaki et al., 2002; Sucher et al., 1995; Sun et al., 1998; Sun et al., 2000; Wong et al., 2002). Despite having a roughly similar expression profile across species, future studies with a finer level of analysis may reveal important differences. Further heterogeneity of NR3A expression across species and brain regions could also arise from differential expression of its two splice variants (discussed above).
In contrast to NR3A, NR3B levels are low around postnatal day (P) 10 and gradually increase into adulthood within the neocortex, hippocampus, striatum, cerebellum, brainstem, and spinal cord (Andersson et al., 2001; Bendel et al., 2005; Eriksson et al., 2002; Matsuda et al., 2002; Nishi et al., 2001; Wee et al., 2008) (Figure 3A). Given the functional similarities and high sequence homology between NR3A and NR3B (57%) (Andersson et al., 2001), it is surprising that these dominant-negative subunits have distinct temporal expression patterns in many of the same brain regions.
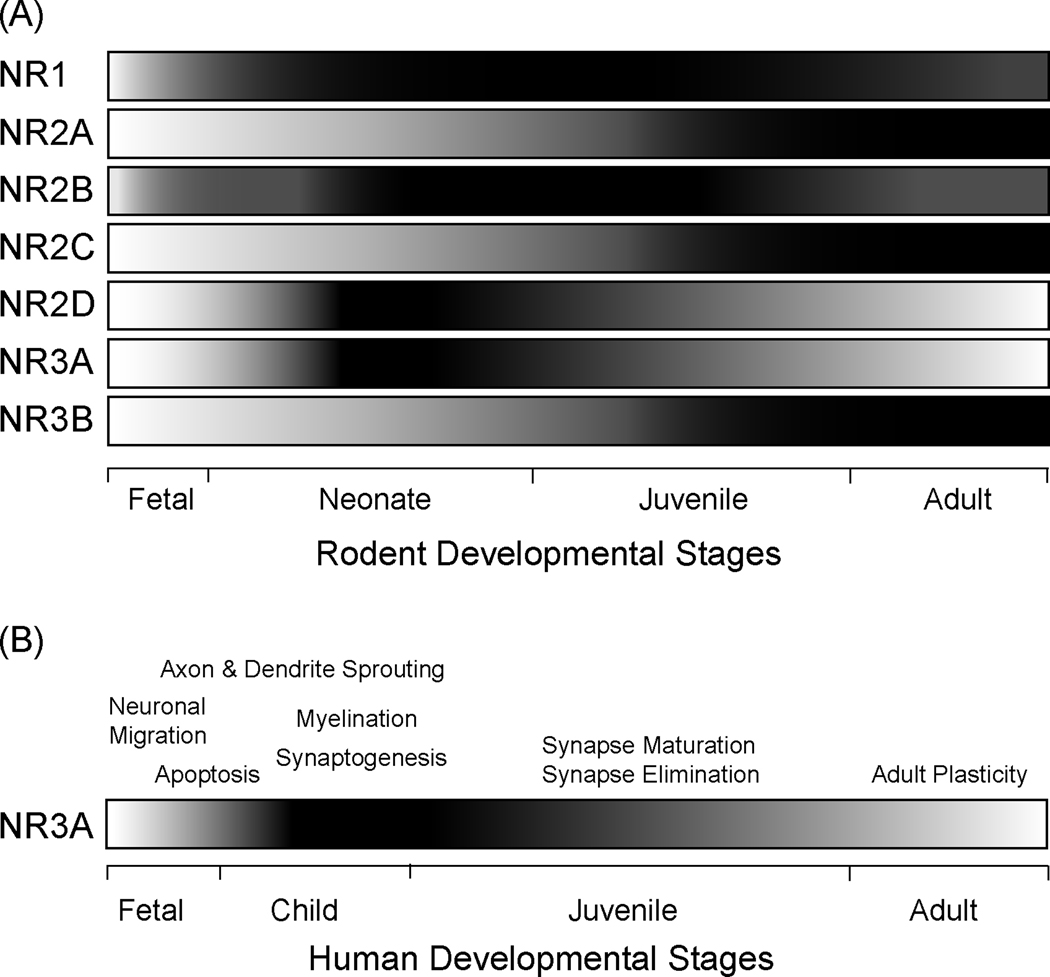
Figure 3.
(A) Schematic representation of NMDAR subunit expression in the developing rat brain. The gray scale gradient shows the differences of each subunit relative to maximum, with the darkest regions reflecting the strongest expression. NR3A appears to be expressed in similar temporal fashion to NR2D, with subunits peaking between P7 and P14. This is contrasted with NR3B, NR2A, and NR2C, which increase developmentally and peak in the third postnatal week. Adapted with permission from (Lujan et al., 2005). (B) Schematic representation of NR3A expression profile and potential roles in the developing human central nervous system. Several key developmental processes in the brain characterize the early postnatal timeframe, when NR3A is maximally expressed: axon and dendrite sprouting, synaptogenesis, myelination, cell death, synapse maturation and elimination (de Graaf-Peters and Hadders-Algra, 2006). The gray scale gradient illustrates the changes in NR3A expression levels, with the darkest regions (child/juvenile stages) reflecting the strongest expression, and low expression seen in the lightest regions (fetal and adult life).
Each NMDAR subunit (NR1, NR2A-D, and NR3A-B) exhibits a unique ontogenetic profile (Ciabarra et al., 1995; Dunah et al., 1996; Fukaya et al., 2005; Laurie et al., 1997; Matsuda et al., 2002; Monyer et al., 1994; Sheng et al., 1994; Sun et al., 1998; Watanabe et al., 1992) (Figure 3A), consistent with its functional “signature” (Al-Hallaq et al., 2002; Goebel and Poosch, 1999). The NR1 subunit is obligatory for NMDAR function and is present throughout life. NR3A is developmentally expressed following a time course most similar to NR2D; both are prominent in the first two postnatal weeks in overlapping brain regions (Ciabarra et al., 1995; Dunah et al., 1996; Laurie et al., 1997; Monyer et al., 1994; Sun et al., 1998; Watanabe et al., 1992). The developmental decreases of NR3A and NR2D expression are in contrast to that of NR3B, NR2A, and NR2C, whose expression levels increase developmentally and peak during the third postnatal week (Fukaya et al., 2005; Matsuda et al., 2002; Monyer et al., 1994; Sheng et al., 1994) (Figure 3A). While both NR3A and NR2B are strongly expressed early in development, NR3A (but not NR2B) levels exhibit a pronounced reduction during postnatal maturation.
3.2 Cellular, laminar, and subcellular localization
NR3A and NR3B are expressed by multiple neuronal cell types, including interneurons, pyramidal cells, motor neurons, trigeminal neurons, retinal ganglion and amacrine cells (Ishihama et al., 2005; Ishihama and Turman, 2006; Matsuda et al., 2002; Mueller and Meador-Woodruff, 2005; Nishi et al., 2001; Paarmann et al., 2005; Sucher et al., 2003). Although NR3A is present in oligodendrocytes, it does not seem to be expressed in astrocytes (Ishihama et al., 2005; Karadottir et al., 2005; Matsuda et al., 2002; Paarmann et al., 2005; Perez-Otano et al., 2006; Salter and Fern, 2005). NR3B localization has been observed in most of the cells located in adult cerebral cortex, hippocampus, striatum, cerebellum, and lumbar spinal cord. This ubiquitous expression of NR3B parallels that of NR1 and suggests a role for NR3B in adult NMDAR function (Wee et al., 2008). Data from NR3B knockout mice will be required to determine the extent to which NR3B influences NMDAR-mediated transmission in the adult central nervous system.
Because of the inside-out pattern in which cortical lamination occurs, the high levels of NR3A observed in layer 5 have raised the possibility that NR3A may play a role in establishing early cortical circuits, perhaps by modulating Ca2+ influx and cell vulnerability to excitotoxicity (Mueller and Meador-Woodruff, 2005; Sucher et al., 1995). NR3B also shows predominant expression in layer 5, but the implications of this laminar specificity in the adult have not yet been investigated.
Ultrastructurally, NR3A colocalizes with NR1 and NR2 subunits at postsynaptic membranes of asymmetric (excitatory) synapses (Perez-Otano et al., 2006; Wong et al., 2002), although it appears to be more abundant at perisynaptic and extrasynaptic sites than at the postsynaptic density (PSD) in both juvenile and adult animals (Perez-Otano et al., 2006). The localization of NR3A contrasts sharply with the PSD-centric localization of NR2A, but is similar to the more lateral and extrasynaptic positioning of NR2B-containing receptors (Groc et al., 2006; Racca et al., 2000). Biochemical fractionation studies confirm the ultrastructural data, and show that NR3A is associated with membranous fractions, such as ER, Golgi, endosomes, and synaptic membranes, but is not as enriched in PSDs as NR1 or NR2 subunits (Perez-Otano et al., 2006). Future experiments are needed to determine (1) whether NR3A-containing receptors shift from synaptic to extrasynaptic sites during development, as has been suggested for other NMDAR subtypes (Kohr, 2006), or (2) whether their peri/extrasynaptic localization indicates a graded and reciprocal organization of different NMDAR subtypes within the PSD that could be fitted for sensing the varied patterns of glutamate release.
4. NR3 subunits exert a dominant-negative effect upon NMDAR function
The properties of typical NMDARs containing NR2A or NR2B subunits include channel block by Mg2+ at hyperpolarized potentials, high permeability to Ca2+, and a subunit-specific complement of protein binding partners (Kennedy et al., 2005; Matute et al., 1997). These properties are critical to many forms of plasticity (Gustafsson and Wigstrom, 1988; Madison et al., 1991) and profoundly influence the wiring of the nervous system and memory processes (Bliss and Collingridge, 1993; Constantine-Paton et al., 1990). Receptors containing NR3A or NR3B subunits have unique properties that differ from conventional NR1/NR2 heteromers. In heterologous expression systems, NR3 subunits assemble into two functional receptor combinations: (1) a heterodimer of NR1 and NR3 subunits that unexpectedly forms an excitatory glycine receptor, and (2) a heterotrimeric complex of NR1, NR2, and NR3 subunits that forms an NMDAR with novel properties and attenuated currents compared to NR1/NR2 NMDARs (Chatterton et al., 2002; Ciabarra et al., 1995; Das et al., 1998; Sucher et al., 1995).
4.1 Triheteromeric NR3A-containing glutamate receptors
NR3A-containing NMDARs only sense glutamate when glutamate-binding NR2 subunits are included (Ciabarra et al., 1995; Perez-Otano et al., 2001; Sucher et al., 1995). When NR1, NR2A, and NR3 subunits are co-expressed in heterologous expression systems, single-channel recordings reveal two populations of NMDA-evoked currents: one characterized by a large conductance resembling the prototypical channel response of NR1/NR2A heteromers, and a second population of smaller conductance, presumably due to channels containing NR3 (Das et al., 1998; Perez-Otano et al., 2001; Sasaki et al., 2002; Ulbrich and Isacoff, 2008) (Figure 4B, C). The putative NR3-containing receptors exhibit dramatically reduced single-channel open probabilities and longer mean open times (Perez-Otano et al., 2001; Sasaki et al., 2002), but see (Das et al., 1998).
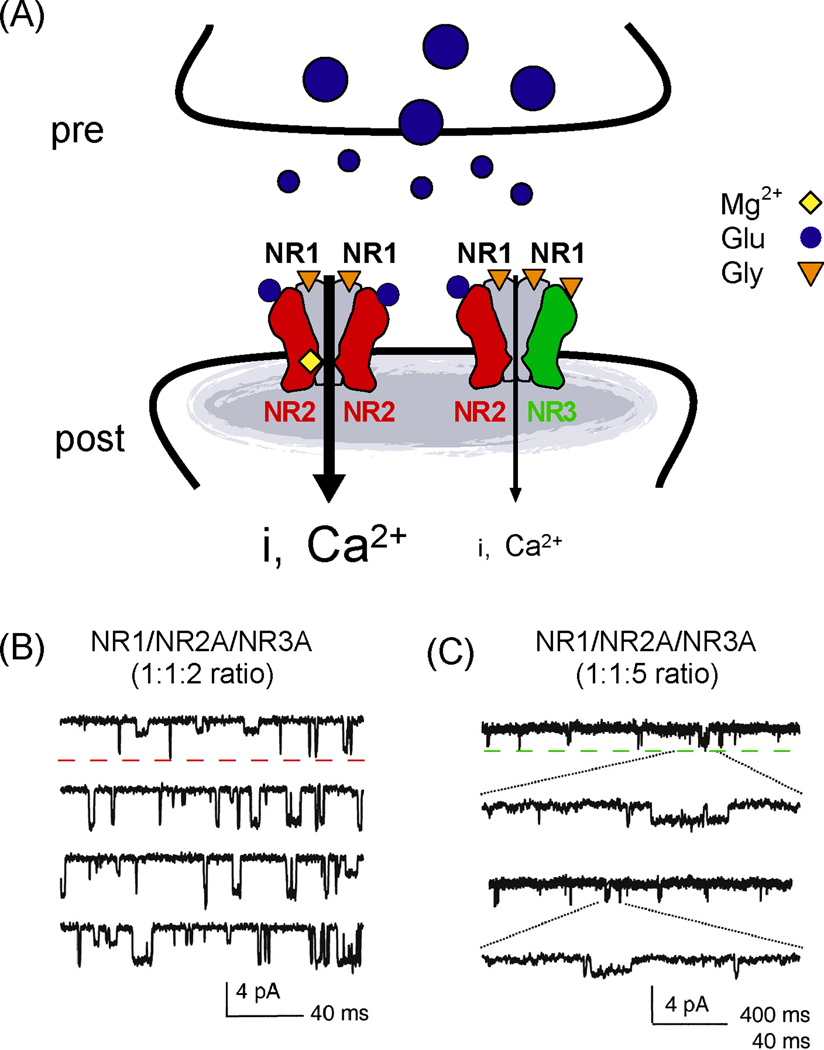
Figure 4.
The NR3 subfamily decreases NMDAR-mediated neurotransmission. (A) Model demonstrating the influence of the NR3 subfamily on current flux through NMDARs. NMDARs containing the glutamate-binding NR2 subunits (red) are highly permeable to Ca2+ and dependent upon postsynaptic depolarization due to the Mg2+ block. Conversely, NMDARs containing glycine-binding NR3 subunits (green) flux less current (“i”), are less permeable to Ca2+, and are less sensitive to Mg2+ block. This is demonstrated in panels (B) and (C) by single-channel recordings (used with permission from (Sasaki et al., 2002). These recordings from outside-out patches are taken from oocytes expressing NR1/NR2A/NR3A from injections of cRNA in a (B) 1:1:2 ratio or (C) 1:1:5 ratio and demonstrate the dominant-negative effects of the NR3 subunits on NMDAR-mediated neurotransmission. Dotted red line in (B) indicates high conductance state of a putative NR1/NR2A-receptor, while dotted green line in (C) indicates low conductance state of a putative NR1/NR2A/NR3A-receptor.
Along with generally decreasing ionic conductance through NMDARs, inclusion of NR3 subunits alters two of the most prominent properties of traditional NMDARs (NR1/NR2 heteromers). First, it causes a five- to ten-fold decrease in Ca2+ permeability, with the variation of the estimates likely reflecting differences between the recombinant systems employed (Das et al., 1998; Matsuda et al., 2002; Perez-Otano et al., 2001; Sasaki et al., 2002; Tong et al., 2008) (Figure 4). Second, NR3A-containing NMDARs are also almost completely insensitive to Mg2+ block at hyperpolarized potentials (Sasaki et al., 2002), but see (Nishi et al., 2001). Both the reduced Mg2+ sensitivity and Ca2+ permeability of NR3A-NMDARs have recently been confirmed in neuronal cultures or hippocampal slices from transgenic mice overexpressing NR3A and knockout mice lacking NR3A (Roberts et al., 2009; Tong et al., 2008) (Table 2). As previously discussed in Section 2, the observed physiological characteristics of NR3A-containing NMDARs most likely result from the formation of a narrow constriction of the outer vestibule along the TM3 pore-forming region of the NR3 subunits.
Table 2.
NR3A loss-of-function and gain-of-function studies demonstrate reciprocal effects in mutant mice.
| Measurement | NR3A-KO | NR3A-OE | References |
|---|---|---|---|
| Spine number and synapse size | ↑ | ↓ | Das et al., 1998; Roberts et al., 2009 |
|
| |||
| Magnitude of NMDAR-mediated currents | ↑ | ↓ |
Das et al., 1998; Perez-Otano et al., 2001; Sasaki et al., 2002; Tong et al., 2008; Roberts et al., 2009 |
|
| |||
| NMDAR calcium permeability | ↑ | ↓ |
Perez-Otano et al., 2001; Sasaki et al., 2002; Sucher et al., 2003; Tong et al., 2008 |
|
| |||
| Hyperpolarization-mediated block of NMDAR currents by magnesium |
↑ | ↓ | Sasaki et al., 2002; Tong et al., 2008; Roberts et al., 2009 |
|
| |||
| - LTP (3×100 Hz) | Early developmental onset |
↓ | Roberts et al., 2009 |
| - LTD (1 Hz; 15 min) | No change | No change | |
|
| |||
| Cell death after ischemic-hypoxic insults | ↑ | ↓ | Nakanishi et al., 2009 |
|
| |||
| Spatial memory (Morris water maze) | ND | ↓ | Roberts et al., 2009 |
| Long-term memory (food preference and object recognition) | ND | ↓ | Roberts et al., 2009 |
|
| |||
| Sensorimotor gating (prepulse inhibition to startle) | ↑ in males; no change in females |
No change | Brody et al., 2005 |
Abbreviations: NR3A-KO, NR3A knockout mouse; NR3A-OE, NR3A overexpressor mouse; LTP, long-term potentiation; LTD, long-term depression; Hz, hertz; ND, no data.
4.2 Diheteromeric NR3A-containing excitatory glycine receptors
The physiological significance of NR1/NR3 excitatory glycine receptors remains an open question because of conflicting reports in vitro, and this subunit combination has yet to be documented in vivo. Despite evidence that NR1/NR3 complexes are targeted to the plasma membrane in heterologous systems (Madry et al., 2007; Perez-Otano et al., 2001), early reports failed to record NMDAR-mediated responses to glutamate or NMDA application in the absence of NR2 subunits (Ciabarra et al., 1995; Das et al., 1998; Perez-Otano et al., 2001; Sucher et al., 1995). Instead of exhibiting a responsiveness to glutamate, NR1/NR3 complexes were found to be sensitive to glycine application (Chatterton et al., 2002). This finding is consistent with the observation that the NR3A ligand binding domain has very high affinity for glycine, but is relatively insensitive to glutamate (Nilsson et al., 2007a; Yao and Mayer, 2006). Glycine application to NR1/NR3 diheteromers evokes a large inward current, indicating that these receptors form an excitatory glycine receptor. The ion permeability of NR1/NR3 diheteromers was similar to that of NR3-containing triheteromers, in that the currents were relatively impermeable to Ca2+ and insensitive to Mg2+ block (Chatterton et al., 2002; Das et al., 1998; Nishi et al., 2001; Perez-Otano et al., 2001; Sasaki et al., 2002). Additionally, while both NR1/NR3A and NR1/NR3B receptors form in Xenopus oocytes (Chatterton et al., 2002), NR3 subunits fail to assemble with NR1 subunits in HEK cells unless both NR3A and NR3B are present (Smothers and Woodward, 2007).
Chatterton and colleagues (2002) reported that bath application of glycine to cerebrocortical cultures elicited a bursting response, which they interpreted as evidence that excitatory glycine receptors exist in the brain. Furthermore, single-channel recordings from outside-out patches from these cultured neurons exhibited channel properties similar to NR1/NR3 receptors in heterologous systems. However, this bursting response to glycine can be found in cultures in which NR1 subunits, and hence all NMDARs, are lacking (Matsuda et al., 2003). Given that the presence of NR1 is required for NR3 subunits to exit the ER and form stable complexes in the plasma membrane (Perez-Otano et al., 2001), these data indicate that the glycine-induced bursting may be independent of NR3 subunits. The existence of glycinergic NR1/NR3 receptors was also questioned by studies finding no evidence for excitatory glycine currents in cultured neurons from mice genetically engineered to overexpress NR3A (Tong et al., 2008) (Table 2).
The high affinity of NR1/NR3 receptors for glycine suggests that, if found in vivo, this receptor combination might be saturated by physiological concentrations of glycine (Yao and Mayer, 2006). Thus, these receptors would serve to keep neurons in a more depolarized state and, depending upon their subcellular localization, could have a profound influence upon synaptic transmission and action potential firing.
5. NR3A pharmacology
5.1 Agonists
The study of NR3A pharmacology is still in its infancy. However, it is clear that NR3A shares more commonalities with NR1 than it does with NR2 subunits. For instance, NR1 binds glycine (Hirai et al., 1996; Yao and Mayer, 2006) whereas NR2 subunits bind glutamate (Laurie et al., 1997), and receptors formed by NR1 and NR3A bind glycine with much higher affinity than do NR1/NR2 receptors (Chatterton et al., 2002). Studies of the rodent ligand binding domain and human full-length protein confirm that glycine binds NR3A with much higher affinity than it binds NR1 [rodent NR3A, Kd = 40 nM, 650 times less than that for NR1 (Yao and Mayer, 2006), and human NR3A, Kd = 535 nM (Nilsson et al., 2007a)]. The lower affinity for glycine of human NR3A compared to rodent NR3A could be due to species differences or differences between the full-length protein and the truncated soluble ligand binding domain. In both human and rodent, NR3A is likely saturated by its preferred endogenous ligand, glycine, at physiological concentrations (Yao and Mayer, 2006). The NR3A ligand binding domain has a low affinity for glutamate (Kd = 9.6 mM) (Yao and Mayer, 2006), and the presumptive activation of NR1/NR2/NR3A triheteromeric receptors by glutamate or NMDA is almost certainly due to the binding of these agonists to NR2 subunits.
D-serine is a potent glycine site agonist of NMDARs (Kleckner and Dingledine, 1988), and accordingly, binding assays demonstrate that D-serine also binds human and rat NR3A with high affinity (Nilsson et al., 2007a; Yao and Mayer, 2006). One electrophysiological study indicated that D-serine behaves as a functional antagonist of NR1/NR3A receptors (Chatterton et al., 2002). This differs from other reports, and may be due to the fact that D-serine produces a rapid desensitization resembling an antagonist block (Yao and Mayer, 2006). Another possibility is that assembly of NR3A with other subunits (e.g. NR1) produces allosteric interactions that alter ligand binding properties (Laurie and Seeburg, 1994). The ligand binding properties of NR3A, and if/how they differ when NR3A co-assembles with other NMDAR subunits have yet to be determined. Until these limitations are overcome, caution should be used when interpreting whether NR3A-containing NMDARs are present or absent based on the effects of pharmacological agonists/antagonists such as D-serine (Li and Han, 2006).
While NR3A shares more attributes with NR1 than with NR2 subunits, there are nonetheless striking pharmacological differences between NR1 and NR3A subunits (Awobuluyi et al., 2007; Nilsson et al., 2007a; Yao and Mayer, 2006). For example, NR1 binds the agonist ACPC with higher affinity than ACBC, while the converse is true for NR3A (Yao and Mayer, 2006). Moreover, neither the partial agonist D-cycloserine nor the antagonist 7-chlorokynurenic acid displace glycine binding to human NR3A expressed in HEK cells (Nilsson et al., 2007a), in contrast to what is observed for the binding of glycine to NR1. The differences between NR1 and NR3A subunits raise the exciting possibility that NR3A-specific agonists or antagonists could be generated in the future to target diseases of NMDAR dysfunction.
5.2 Antagonists
Very little is known about how NR3A-containing receptors can be pharmacologically blocked. Studies to date have focused on the binding of NR1 antagonists to NR3A or have used electrophysiological measures to determine how NR1/NR3A or NR1/NR2/NR3A heteromers respond to typical NMDAR antagonists. When NR1/NR3A diheteromers are expressed in heterologous systems, they exhibit little electrophysiological block by the classic NMDAR antagonists APV, MK-801, or memantine (Chatterton et al., 2002). Triheteromeric receptors containing NR1/NR2/NR3A are more difficult to isolate electrophysiologically because they must be distinguished from either NR1/NR2 or NR1/NR3A receptors. However, this has been possible using single-channel recordings, which showed that the small conductance NR1/NR2A/NR3A channel is blocked by D-APV (200 µM) (Sasaki et al., 2002). Interestingly, NR1/NR2/NR3A triheteromeric receptors may also be blocked by antagonists targeting NR2 subunits, as receptors composed of NR1/NR2B/NR3A triheteromers and NR1/NR2B diheteromers are similarly blocked by high concentrations of the NR2B-antagonist ifenprodil (Smothers and Woodward, 2003). One might predict that, compared to NR1/NR2B diheteromers, NR1/NR2B/NR3A triheteromers might be less sensitive to lower concentrations of ifenprodil where NR2B-specificity is greater, as previous studies have shown that the magnitude of ifenprodil block depends on the number of NR2B subunits contained within the NMDAR complex (Hatton and Paoletti, 2005).
Studies of the ligand binding domain demonstrate that NR3A subunits interact with classical NMDAR antagonists. Six of eight NR1 subunit antagonists exhibit more than a 100-fold larger affinity for NR1 subunits compared to NR3A subunits (Yao and Mayer, 2006). For example, kynurenic acid binds the NR1 ligand binding domain with high affinity (Kd= ~53 µM) but only weakly binds the NR3A binding domain (Kd = ~15 mM) (Yao and Mayer, 2006). These observations provide further evidence that NR1 and NR3A have distinct ligand binding properties and increase the likelihood that specific agonists/antagonists could be designed to target NR3A selectively. However, the extremely high affinity of NR3A for glycine indicates that the design of competitive antagonists would require a particularly high affinity compound. Thus, it might be more feasible to design non-competitive antagonists that act via allosteric interactions with NR3A-containing NMDARs.
6. Intracellular binding partners
NMDAR-mediated effects can be initiated directly via ionic flow through the channel pore, and can also be shaped by intracellular partners tethered to the NMDAR complex. To date, few studies have examined interactions of NR3A with other proteins, but the existing reports have identified possible associations between NR3A and protein phosphatases, cytoskeletal proteins, and an adaptor protein implicated in receptor trafficking.
One specific C-terminal binding partner of NR3A is the catalytic subunit of protein phosphatase 2A (PP2A) (Chan and Sucher, 2001);(Ma and Sucher, 2004) (Figure 2). PP2A plays important roles at postsynaptic sites, including dephosphorylating NR1 subunits on serine 897 (NR1-S897). Dephosphorylation of NR1-S897 by PP2A attenuates NMDAR single-channel currents and reduces permeability of the receptors to Ca2+. Importantly, activation of NMDARs causes the physical uncoupling between NR3A and PP2A, decreasing PP2A activity at the synapse. By influencing NR1 phosphorylation through its interaction with PP2A, NR3A may provide a feedback mechanism to modulate NMDAR function based on the history of synaptic activity (Chan and Sucher, 2001). Intriguingly, brains from schizophrenic patients have increased dephosphorylation of NR1-S897 (Emamian et al., 2004) that might be related to overexpression of NR3A (Ma and Sucher, 2004), providing a potential mechanism that could contribute to the NMDAR hypofunction observed in schizophrenia.
In addition to modulating the activity of a phosphatase important to NMDARs, one intracellular NR3A binding partner appears to be crucial for the removal of nascent NMDARs during synapse maturation. NR3A binds PACSIN1/syndapin1, a neuron-specific intracellular adaptor (Perez-Otano et al., 2006). PACSIN1 binds selectively to the C-terminus of NR3A (Figure 2) through tertiary structure interactions with its NPF (Asn/Pro/Phe) motifs, recruiting a larger complex with the endocytic proteins dynamin and clathrin and facilitating the rapid endocytosis of NR3A-containing NMDARs. Disruption of this interaction results in the synaptic accumulation of NR3A-containing NMDARs. The PACSIN1-mediated removal of NR3A-containing NMDARs is activity-dependent, providing a mechanism for regulated receptor replacement that could drive the functional maturation of synaptic NMDARs during postnatal development and the associated changes in the properties of synaptic plasticity.
Additional putative NR3A binding partners are beginning to be identified which could alter its localization, trafficking, and signaling (Figure 2), but these interactions were only shown in vitro and need to be extensively studied and verified. NR3A may interact with a cytoskeletal protein (MAP1S/C19ORF5), a scaffolding protein (plectin), a cell cycle and apoptosis regulatory protein (CARP-1), and a regulator of G-protein signaling (GPS2/AMF1) (Eriksson et al., 2007a; Eriksson et al., 2007b). Confirmation of these associations would position NR3A to play roles in intracellular processes such as trafficking and targeting of NR3A-containing receptors (MAP1S and plectin), PKC activation (plectin), and suppression of the MAPK pathway (GPS2/AMF1). Other NR3A binding partners will undoubtedly be identified, which may link NR3A to more signaling pathways. Notably, there is a conspicuous absence of NR3A interaction with the postsynaptic scaffolding protein PSD-95 (Eriksson et al., 2007a; Matsuda et al., 2002), which may explain the labile synaptic expression of NR3A-containing NMDARs (Perez-Otano et al., 2006). Alternatively, the clustering and stabilization of NR3A-containing receptors near synaptic sites may rely on NR2 anchoring with other membrane-associated guanylate kinase family scaffolding proteins, such as SAP102, which are present at high levels in developing synapses (Perez-Otano et al., 2006; Sans et al., 2000; Wong et al., 2002).
7. Assembly, trafficking, and targeting of NR3-containing NMDARs
Developmental regulation of the membrane expression of different NMDAR subtypes shapes synaptic and behavioral plasticity (Isaac et al., 1995; Liao et al., 1995; Philpot et al., 2001; Quinlan et al., 1999; Roberts et al., 2009; Rumpel et al., 1998; Sawtell et al., 2003; Tang et al., 1999; Wu et al., 1996). NMDARs can rapidly translocate in and out of synaptic regions by lateral diffusion in the plane of the membrane and by endo/exocytic trafficking. It is increasingly recognized that regulation of trafficking contributes to the synapse-specific and activity-dependent control of NMDAR expression at individual synapses, by favoring local receptor removal, insertion, or exchange (Lavezzari et al., 2004; Montgomery et al., 2005; Scott et al., 2004; Washbourne et al., 2004).
Although peak expression of rodent NR3A occurs in the second postnatal week, its expression levels and association with NR1 subunits may nonetheless be fairly limited compared to that of NR2 subunits. Immunoprecipitation experiments found that, despite the majority of NR3A associating with the obligatory NR1 subunit, NR3A subunits are likely to contribute only a small fraction of the total receptor complex composition (Al-Hallaq et al., 2002). Specifically at age P10, approximately 80% of NR3A subunits are associated with NR1, but less than 10% of NR1, NR2A, and NR2B are bound to NR3A, and this association decreases further over development. Strikingly, NR3A genetic deletion studies demonstrate that even a low NR3A abundance is sufficient to have profound consequences on synapse formation and NMDAR-mediated transmission during early postnatal development (Das et al., 1998; Perez-Otano et al., 2001; Roberts et al., 2009; Sasaki et al., 2002; Tong et al., 2008).
Protein degradation, fluorescence recovery after photobleaching, and fluorescence resonance energy transfer experiments suggest a central role for NR1 in the folding and assembly of NMDAR heteromers in the endoplasmic reticulum (ER) (Atlason et al., 2007; Schuler et al., 2008). Homomeric NR3 complexes fail to exit the ER, presumably because the C-terminus of NR3A/NR3B carries a putative ER retention signal (RXR) (Figure 2). For forward secretory trafficking and functional insertion into the plasma membrane to occur, the RXR motif is likely masked by heteromeric assembly with NR1 subunits (Matsuda et al., 2003; Perez-Otano et al., 2001; Schuler et al., 2008; Scott et al., 2001; Standley et al., 2000). However, NR3 subunits may be rapidly degraded, even in the presence of NR1, resulting in inefficient assembly mechanisms or extremely rapid turnover of NR3A-containing NMDARs lacking NR2 subunits (Atlason et al., 2007).
Once at the plasma membrane surface, the abundance and synaptic targeting of NR3A-containing NMDARs appears to be highly regulated by synaptic activation of NMDARs. In experiments using cultured hippocampal neurons transfected with GFP-tagged NR3A, intrinsic neuronal activity induces the movement of NR3A-containing NMDARs from membrane and synaptic locations to intracellular compartments (Perez-Otano et al., 2006).
Endocytic removal of NR3A can be facilitated by bath application of NMDA and prevented by disruption of synaptic activity (TTX) or antagonism of NMDARs with APV. Furthermore, removal relies upon a selective interaction with PACSIN1/syndapin1, a specialized accessory protein involved in clathrin-mediated endocytosis and enriched at synapses (discussed in more detail in Section 6). The synaptic expression of PACSIN1 is developmentally regulated and exhibits a reciprocal expression pattern to that of NR3A (Perez-Otano et al., 2006), supporting a role for PACSIN1 in the developmental downregulation of NR3A subunits.
Another regulator of NR3A synaptic targeting is the protein phosphatase PP2A, which binds NR3A directly and influences the dephosphorylation state of NR1 subunits (Chan and Sucher, 2001; Ma and Sucher, 2004). PACSIN1 and PP2A binding domains on the C-terminus of NR3A overlap (Figure 2), making it unlikely that these proteins would bind concurrently (Chan and Sucher, 2001; Perez-Otano et al., 2006). Interestingly, NMDAR activation disrupts the PP2A-NR3A protein interaction, and may unmask binding sites for PACSIN1, enabling NR3A endocytosis.
Finally, PDZ-domain binding motifs, which provide areas of attachment for prototypical PSD proteins and support the postsynaptic anchoring of other NMDAR subunits, are absent in the C-termini of NR3A and NR3B (Eriksson et al., 2007a; Matsuda et al., 2002). Perhaps as a consequence, these subunits are more loosely attached to the PSD than NR2 subunits (Perez-Otano et al., 2006; Roberts et al., 2009). Despite this emerging set of data, and in contrast to the large literature on NR2 subunits, little is yet known about how and where NR3 proteins traffic.
8. Influence of NR3A subunits in the developing brain
NR3A is expressed in the right place at the right time to regulate spine and synapse development. It is unclear, however, if the influence of NR3A is to serve as a ‘synaptic brake’ to limit synapse/spine formation or if it serves as a ‘synaptic marker’ to promote the elimination of spines. These two hypotheses are not mutually exclusive and NR3A may influence both processes.
8.1 Synaptic brake hypothesis
NR3A-containing receptors may act collectively as a synaptic brake. One possibility is that at early stages, when their expression is highest, they limit synapse formation by raising the threshold required for synaptic activity to induce synaptic maturation. Later in development, activity-dependent removal of NR3A by PACSIN1/syndapin1 would relieve this brake and allow the replacement of immature synaptic NMDARs by mature subtypes that are more stably anchored to PSDs. Synapse maturation would then proceed with the recruitment of other synaptic scaffolds and signaling complexes, and ensuing changes in spine morphology.
Synaptic plasticity mechanisms, such as LTP (long-term potentiation) and LTD (long-term depression), influence the formation and stabilization of synapses and spines (Yuste and Bonhoeffer, 2001) and are critically dependent upon Ca2+ entry via NMDARs (Lisman, 1989). The dominant-negative effect of NR3A on current and Ca2+ flux has recently been shown to limit synaptic plasticity (Roberts et al., 2009), providing a mechanism for NR3A control of spine and synapse density. It will be interesting to determine whether NR3A knockout mice display changes in synaptic plasticity that are causally related to the changes in spine formation, or vice versa.
While Das and coworkers (1998) reported increased spine density and enlarged spines in the cortex of P19 NR3A knockout mice, Tong et al. (2008) did not observe a related increase in the frequency or amplitude of AMPAR mEPSC in the same brain region in P10-P13 mice. However, the NMDAR/AMPAR ratio was enhanced, reflecting an increase in the NMDAR component. This sequence of events makes it tempting to speculate that larger NMDAR currents early in development favor enhanced LTP and subsequent increases in spine density/size in NR3A-deficient mice. If true, the early developmental presence of NR3A may serve as a synaptic brake to prevent the premature strengthening of synapses, and thus ensure a well-coordinated strengthening and stabilization of only appropriate synapses in response to experience. Early release of this brake in NR3A-null mutants would lead to enhanced spine formation and maturation of inappropriate synapses.
8.2 Synaptic elimination hypothesis
Another hypothesis consistent with the phenotype of NR3A-null mice is that NR3A serves as a tag to label weak synapses for elimination. In this scenario, coordinated or strong synaptic activity could drive NR3A out of the synapse, possibly via activity- and NMDAR-dependent synaptic removal of NR3A. Synapses with weak or uncoordinated activity would retain NR3A and, hence, be targeted for elimination. NR3A may also facilitate the elimination of immature synapses by recruiting the machinery necessary to exclude NMDARs from synapses, one of the steps thought to underlie synapse elimination and spine loss, via its interaction with PACSIN1/syndapin1 (Perez-Otano et al., 2006).
Another possible link between NR3A and spine elimination was suggested by the discovery of a family of proteins termed “takusans” that are upregulated in NR3A-null mice and appear to influence the formation of dendritic spines (Tu et al., 2007). Overexpression of takusans enhances spine density, AMPAR expression, and PSD-95 clustering. Considering that the expression of takusans is reciprocally regulated in development when compared to NR3A subunits, NR3A may foster spine elimination by limiting the synaptic expression of takusan proteins. No direct interaction between these proteins has been observed, and future studies will be needed to investigate any causal relationship between NR3A and takusan expression.
9. NR3A in disease
9.1 Schizophrenia
A prominent theory is that impaired glutamatergic transmission contributes to the pathophysiology of schizophrenia (Olney and Farber, 1995), and several lines of evidence invoke an important role of NMDAR hypofunction (Coyle et al., 2003; Olney et al., 1999; Tamminga, 1998; Tsai et al., 1998): (1) NMDAR antagonists, such as PCP, exacerbate symptoms in patients with schizophrenia and produce symptoms nearly indistinguishable from schizophrenia in normal individuals (Javitt and Zukin, 1991). Genetic or pharmacological downregulation of NMDAR function in rodents also triggers schizophrenic-related behaviors (Moghaddam and Jackson, 2003; Mohn et al., 1999). (2) NMDAR co-agonists, such as glycine and D-serine, can improve cognitive functioning and alleviate negative symptoms associated with schizophrenia (Goff et al., 1999; Goff et al., 1995; Heresco-Levy et al., 1996; Heresco-Levy et al., 1998; Javitt, 2002; Javitt et al., 1994; Millan, 2005; Tsai et al., 1998). (3) Many candidate schizophrenia genes influence NMDAR signaling and expression (Chowdari et al., 2002; Harrison and Weinberger, 2005; Martucci et al., 2003; Moghaddam, 2003; Straub et al., 2002). For example, neuregulin, the protein product of a major schizophrenia susceptibility gene, alters the surface expression of synaptic NMDARs (Gu et al., 2005; Ozaki et al., 1997; Stefansson et al., 2004). (4) More recently, imaging studies have provided the first in vivo evidence for reduced NMDAR binding in medication-free schizophrenic patients (Pilowsky et al., 2006).
Although the mechanistic basis for NMDAR hypofunction in schizophrenia remains unknown, it could be caused by a change in NMDAR subunit composition. To date there is little consensus on alterations in NMDAR subunits that might contribute to the NMDAR hypofunction observed in schizophrenia (Akbarian et al., 1996; Meador-Woodruff and Healy, 2000; Moghaddam, 2003). However, a recent study demonstrated that NR3A mRNA levels are increased within layer 5 in subregions of the dorsolateral prefrontal cortex in schizophrenics (Mueller and Meador-Woodruff, 2004). This finding raises the possibility that inappropriate expression of NR3A could exacerbate this subunit’s unique ability to suppress receptor activity, and underlie NMDAR hypofunction. Consequences of aberrant expression would be predicted to have a larger impact in the mature brain, where NR3A is remarkably downregulated in most brain regions. Four further observations support a role for elevated NR3A in schizophrenia: (1) Schizophrenic brains exhibit decreased phosphorylation of NR1 at serine 897, which can be reversed by antipsychotics (Emamian et al., 2004). Because NR3A subunits form a signaling complex with PP2A, which can dephosphorylate serine 897, overexpression of NR3A could contribute to the reductions in NR1 phosphorylation in schizophrenia (Chan and Sucher, 2001). (2) Alterations in NR3A levels impair prepulse inhibition (Brody et al., 2005), a measure of sensorimotor gating that is also impaired in many schizophrenia patients (Braff et al., 2001). (3) Spine density is decreased in the DLPFC of schizophrenia patients (Glantz and Lewis, 2000, 2001), a region where NR3A levels are elevated (Mueller and Meador-Woodruff, 2004), and elevated NR3A levels have been shown to decrease spine formation (Roberts et al., 2009). (4) Impaired working memory in schizophrenia could be explained by deficits in NMDAR function, potentially resulting from NR3A overexpression, in layer 5 pyramidal neurons in the DLPFC (Fellous and Sejnowski, 2003; Sanchez-Vives and McCormick, 2000; Sucher et al., 1995; Wong et al., 2002). Although it is now clear that NR3A levels are not changed in a wholesale manner in the schizophrenic DLPFC (Henson et al., 2008), the above data underscore a need to further examine regional changes in NR3A expression, protein interactions, and signaling in schizophrenics and to evaluate if/how they contribute to the pathophysiology of schizophrenia.
9.2 White matter injury
The dogma that NMDARs are expressed only in neurons within the CNS has been overturned, and increasing evidence indicates not only that NMDARs are expressed in glia, but also that glial NMDARs contain the NR3A subunit (Paoletti and Neyton, 2007). Three recent studies highlighted the role of NMDARs in oligodendrocyte damage (Karadottir et al., 2005; Micu et al., 2006; Salter and Fern, 2005), reporting that oligodendrocytes and the myelin sheaths they form are damaged by excitotoxicity in a number of acute and chronic disorders, including ischemic stroke, cerebral palsy, traumatic neural injury, and multiple sclerosis (Matute, 2006; Matute et al., 2007) (Figure 5). Both Ca2+ entry and subsequent oligodendrocyte damage could be attenuated by NMDAR antagonists, indicating that white matter damage was caused at least in part by NMDAR activation (Karadottir et al., 2005; Micu et al., 2006; Salter and Fern, 2005). These studies demonstrated that NMDAR currents in oligodendrocytes failed to show rectification at hyperpolarized potentials due to Mg2+ block, mimicking the behavior of NR3A- or NR2C-containing NMDARs. This suggests that these NMDARs are positioned to respond to glutamate activation, even in the absence of strong depolarization. Further, all three studies showed that NR3A levels are high in myelin. Despite the low Ca2+ permeability of NR3A-containing NMDARs, high levels of NR3A-containing NMDARs and/or insufficient Ca2+ buffering might predispose oligodendrocytes to cell death mediated by elevations of glutamate associated with ischemic stroke or other excitotoxic events. If so, the development of NR3A-selective antagonists could be of therapeutic benefit for preventing white matter excitotoxic damage, with less severe adverse side effects than broad spectrum NMDAR antagonists.
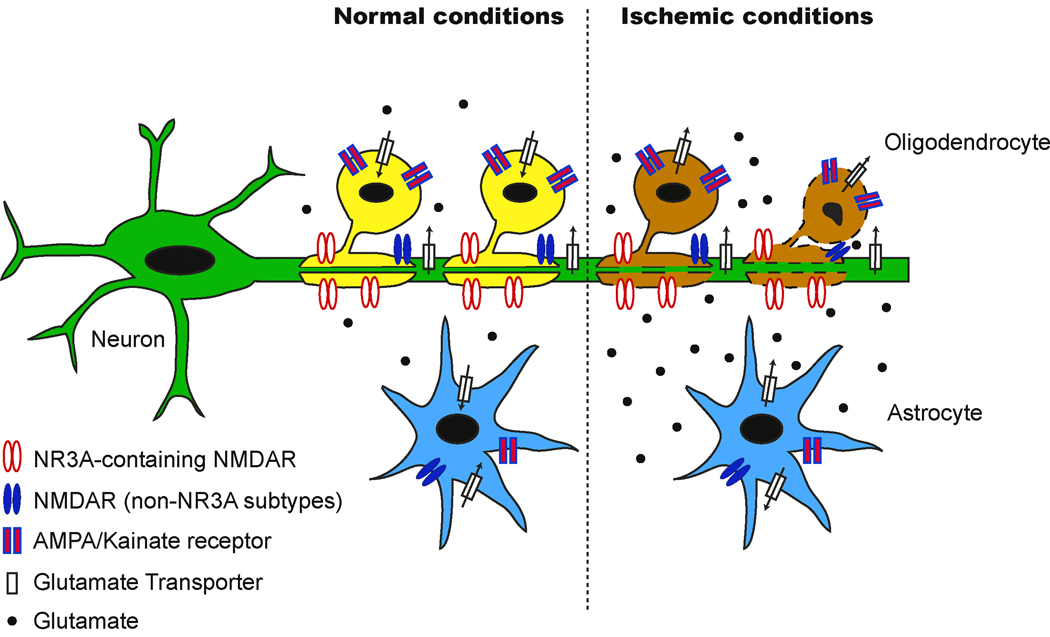
Figure 5.
NR3A-containing NMDARs may contribute to white matter damage with ischemia. High concentrations of extracellular glutamate caused by reversal of glutamate transport during ischemic conditions can make oligodendrocytes particularly vulnerable to injury. Unlike traditional NMDARs, NR3A-containing receptors may be preferentially activated by glutamate because they are found at high levels in oligodendrocytes and they are insensitive to magnesium block. The resulting calcium influx through NR3A-NMDARs may be sufficient to cause excitotoxic damage to oligodendrocyte processes, ultimately resulting in cell death (indicated by brown color). Modified with permission from (Matute, 2006).
9.3 Diseases of neuronal excitotoxicity
A large number of neurological disorders are associated with increased excitotoxicity, including Huntington’s disease (Fan and Raymond, 2007), Parkinson’s disease (Bonuccelli and Del Dotto, 2006), Alzheimer’s disease (Wenk, 2006), chronic alcohol exposure (Crews et al., 1998; Lovinger, 1993), and neuropathic pain (Dubner and Ruda, 1992). Other disorders associated with excitotoxicity include acute brain injuries such as stroke (Martin et al., 1998), epilepsy (Fujikawa, 2005; Meldrum, 1993), and traumatic injury (Arundine and Tymianski, 2004). Excessive activation of NMDARs is thought to contribute to excitotoxicity because the depolarization-dependent activation of Ca2+ entry mediated by NMDARs can more readily induce cell death compared to other forms of glutamatergic activation (Abdrachmanova et al., 2002; Choi, 1992; Rothman and Olney, 1995). While excessive glutamate activity triggers NMDAR-mediated cell death in most of these acute and chronic disorders, disruption of cellular metabolic processes can also cause depolarization-induced activation of NMDARs by shifting ionic balances (Zeevalk and Nicklas, 1992).
The proposed role of NMDAR-mediated excitotoxicity in neuronal disorders has fostered intense research into the therapeutic benefits of NMDAR antagonists, but most efforts have yielded disappointing results due to harmful side effects of the antagonists (Hoyte et al., 2004; Lipton, 2006). Because NMDARs are crucial for so many neuronal functions, including learning and memory (Bear, 1996), and because too little NMDAR activation can also lead to apoptotic cell death (Ikonomidou et al., 1999), successful blockade of NMDAR-mediated excitotoxicity must carefully attenuate NMDAR functions without eliminating them (Lipton, 2006). NR3A provides an attractive target for accomplishing this subtle manipulation of NMDAR functions. One intriguing idea is that overexpression of NR3A might attenuate NMDAR-mediated cell death by reducing Ca2+ permeability of existing NMDARs. However, such an approach would have to carefully titrate the degree of NR3A overexpression, as apoptosis can also be triggered by a dramatic decrease in NMDAR currents (Lipton and Nakanishi, 1999).
A neuroprotective role for NR3A has recently been explored further by Nakanishi and coworkers (2009), who used in vivo models of hypoxic-ischemic insults as well as retinal cultures to examine the effects of NR3A on cell death. Using NR3A knockout and transgenic overexpressing mice, these authors have provided convincing evidence that ischemic-induced neuronal damage is extensive in the absence of NR3A, while the presence of NR3A reduces cell loss (Table 2). Other recent observations support their findings, namely that retinal ganglion cells are relatively invulnerable to NMDAR-mediated excitotoxicity in contrast to many other neuronal classes (Ullian et al., 2004), and only the retinal cell types with high NR3A content exhibit attenuated calcium responses to NMDA (Nakanishi et al., 2009; Sucher et al., 2003). Consistent with the idea that NR3A might offer neuroprotective benefits, high levels of NR3A expression during early brain development might explain why excitotoxicity is not more prevalent at ages before the maturation of inhibitory circuitry. Perhaps it is not a coincidence that NR3A levels diminish during development (Wong et al., 2002) as inhibition increases (Coyle and Yamamura, 1976). These tantalizing correlations raise the possibility that exogenous introduction of NR3A might be useful to treat a variety of neurological disorders by preventing inappropriate cell death without producing deleterious side effects.
9.4 Mental retardations associated with improper dendritic spine development
Dendritic spines are the major site for excitatory synaptic connections on neurons. Changes in their number, density, and/or shape have been implicated in a number of mental retardations including Fragile X, Rett, and Down syndromes (Carlisle and Kennedy, 2005; Chechlacz and Gleeson, 2003; Fiala et al., 2002; Irwin et al., 2000; Kaufmann and Moser, 2000; Newey et al., 2005), but the molecular basis for these spine abnormalities remains unknown. Activation of NMDARs is required both for the bidirectional changes in synaptic strength thought to underlie learning and memory (Malenka and Bear, 2004) and for the activity-dependent growth or retraction of spines which may help encode enduring changes in synapses (Engert and Bonhoeffer, 1999; Maletic-Savatic et al., 1999; Nagerl et al., 2004). Therefore, dysregulation of NMDARs might contribute to abnormalities in spine number or shape that could be prevented (or reverted) by normalizing NMDAR activity. Even if NMDAR dysfunction did not play a causal role, modifying NMDAR function could potentially ameliorate conditions arising from abnormal spine development. Indeed, one of the most striking phenotypes of mice lacking NR3A is an approximate three to five-fold increase in spine density (Das et al., 1998). Studies in mice that lack or overexpress NR3A should help elucidate links between spine abnormalities and deficits in synaptic plasticity and learning, and hence evaluate if targeted and carefully titrated exogenous modulation of NR3A could be of therapeutic value in certain forms of mental retardation.
9.5 Other links of NR3A to clinical issues
NR3A is likely involved in other neurological conditions, yet possible roles for NR3A may have gone unrecognized. NR3A was identified relatively recently, and, because of this, it was not incorporated into many tests, such as gene array analyses, that could have detected its involvement in disease. In addition to the direct and indirect suggestions of NR3A involvement in neurological disorders mentioned above, we list here other possible links of NR3A to disease. (1) Decreased NR3A mRNA and protein levels have been reported in patients with bipolar disorder (Mueller and Meador-Woodruff, 2004). (2) Chronic low-level lead exposure during development is associated with cognitive impairments in young children (Bellinger et al., 1991), and decreased levels of NR3A mRNA are reported in rodent hippocampus after lead exposure (Zhang et al., 2002). (3) Genetic analyses of NR3A in humans have identified a common missense variation (Val362Met) that is associated with a strikingly different prefrontal cortex activation during auditory target processing (Gallinat et al., 2007).
While these are just a few additional examples of how NR3A might be involved in neurological disorders, the recognized involvement of NR3A is likely to increase as disease-related changes in NR3A expression levels, polymorphisms, or function are evaluated. The above studies also provide direct evidence for a functional role for NR3A in the adult human brain, and further support the idea that modulation of NR3A in humans might alter neurological functions. Development of specific agonists and antagonists of NR3A may thus be of therapeutic value for a variety of neurological disorders. Intriguingly, exposure of rodents to weak magnetic fields alters NR3A mRNA levels, raising the possibility that there may even be non-invasive and non-pharmacological means to modify NR3A levels in humans (Hirai and Yoneda, 2004).
10. Concluding remarks
The use of many NMDAR agonists and antagonists had been abandoned because of their limited therapeutic value in neurological disorders, largely due to their severe side effects. Interest has peaked again because of tentative success at developing a new generation of non-competitive (Lipton, 2006) and subunit-selective (Paoletti and Neyton, 2007) NMDAR antagonists. The NR3A subunit represents a promising new therapeutic target, due to its unique dominant-negative suppression of NMDAR responses combined with its temporal and regional distribution. However, before targeting of NR3A begins to hold any true therapeutic potential, more research is needed. First, thorough studies of the temporal and regional specificity of NR3A in humans need to be performed, as expression profiles of NR3A may differ dramatically from rodents. Second, understanding how NR3A is trafficked to the plasma membrane, especially to synaptic sites, needs to be refined before effects of modifying NR3A expression levels or function can be predicted. Third, the role of NR3A at both extrasynaptic and synaptic sites in mammalian cells needs to be rigorously examined, as most studies to date have used heterologous expression systems. Fourth, parsing the reciprocal effects from gain-of-function and loss-of-function analyses will clarify how NR3A affects cellular morphology, neuronal circuit function, and cognition. Finally, specific agonists and antagonists of NR3A must be generated, a prospect that appears feasible given the functional and pharmacological differences between NR3A and other NMDAR subunits. The NR3A subunit has been receiving exponentially increasing interest recently, and this continued trend is likely to drive the identification of new NR3A functions and drug discovery.
Acknowledgments
Work that contributed to this review was supported by grants from NARSAD (BDP, ACR, IPO), an NICHD training grant T32-HD40127 (ACR), UTE project CIMA, Gobierno de Navarra, Spanish Ministry of Science SAF2006-10025 and CSI2008-00005 (IPO), the National Science Foundation grant# 0822969 (BDP), National Eye Institute R01EY018323 (BDP), UNC Charles Lee Raper Dissertation Fellowship (MAH), and Society for Neuroscience/NINDS Neuroscience Scholars Program (MAH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abdrachmanova G, Teisinger J, Vyklicky L., Jr Axotomy-induced changes in the properties of NMDA receptor channels in rat spinal cord motoneurons. J Physiol. 2002;538:53–63. doi: 10.1113/jphysiol.2001.012794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Sucher NJ, Bradley D, Tafazzoli A, Trinh D, Hetrick WP, Potkin SG, Sandman CA, Bunney WE, Jr, Jones EG. Selective alterations in gene expression for NMDA receptor subunits in prefrontal cortex of schizophrenics. J Neurosci. 1996;16:19–30. doi: 10.1523/JNEUROSCI.16-01-00019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hallaq RA, Jarabek BR, Fu Z, Vicini S, Wolfe BB, Yasuda RP. Association of NR3A with the N-methyl-D-aspartate receptor NR1 and NR2 subunits. Mol Pharmacol. 2002;62:1119–1127. doi: 10.1124/mol.62.5.1119. [DOI] [PubMed] [Google Scholar]
- Andersson O, Stenqvist A, Attersand A, von Euler G. Nucleotide sequence, genomic organization, and chromosomal localization of genes encoding the human NMDA receptor subunits NR3A and NR3B. Genomics. 2001;78:178–184. doi: 10.1006/geno.2001.6666. [DOI] [PubMed] [Google Scholar]
- Arundine M, Tymianski M. Molecular mechanisms of glutamate-dependent neurodegeneration in ischemia and traumatic brain injury. Cell Mol Life Sci. 2004;61:657–668. doi: 10.1007/s00018-003-3319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlason PT, Garside ML, Meddows E, Whiting P, McIlhinney RA. N-Methyl-D-aspartate (NMDA) receptor subunit NR1 forms the substrate for oligomeric assembly of the NMDA receptor. J Biol Chem. 2007;282:25299–25307. doi: 10.1074/jbc.M702778200. [DOI] [PubMed] [Google Scholar]
- Awobuluyi M, Yang J, Ye Y, Chatterton JE, Godzik A, Lipton SA, Zhang D. Subunit-specific roles of glycine-binding domains in activation of NR1/NR3 N-methyl-D-aspartate receptors. Mol Pharmacol. 2007;71:112–122. doi: 10.1124/mol.106.030700. [DOI] [PubMed] [Google Scholar]
- Bear MF. A synaptic basis for memory storage in the cerebral cortex. Proc. Natl. Acad. Sci. USA. 1996 doi: 10.1073/pnas.93.24.13453. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger D, Sloman J, Leviton A, Rabinowitz M, Needleman HL, Waternaux C. Low-level lead exposure and children's cognitive function in the preschool years. Pediatrics. 1991;87:219–227. [PubMed] [Google Scholar]
- Bendel O, Meijer B, Hurd Y, von Euler G. Cloning and expression of the human NMDA receptor subunit NR3B in the adult human hippocampus. Neurosci Lett. 2005;377:31–36. doi: 10.1016/j.neulet.2004.11.064. [DOI] [PubMed] [Google Scholar]
- Bendova Z, Sumova A, Mikkelsen JD. Circadian and developmental regulation of N-methyl-d-aspartate-receptor 1 mRNA splice variants and N-methyl-d-aspartate-receptor 3 subunit expression within the rat suprachiasmatic nucleus. Neuroscience. 2009;159:599–609. doi: 10.1016/j.neuroscience.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Bliss TVP, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Bonuccelli U, Del Dotto P. New pharmacologic horizons in the treatment of Parkinson disease. Neurology. 2006;67:S30–S38. doi: 10.1212/wnl.67.7_suppl_2.s30. [DOI] [PubMed] [Google Scholar]
- Braff DL, Geyer MA, Swerdlow NR. Human studies of prepulse inhibition of startle: normal subjects, patient groups, and pharmacological studies. Psychopharmacology (Berl) 2001;156:234–258. doi: 10.1007/s002130100810. [DOI] [PubMed] [Google Scholar]
- Brody SA, Nakanishi N, Tu S, Lipton SA, Geyer MA. A developmental influence of the N-methyl-D-aspartate receptor NR3A subunit on prepulse inhibition of startle. Biol Psychiatry. 2005;57:1147–1152. doi: 10.1016/j.biopsych.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Carlisle HJ, Kennedy MB. Spine architecture and synaptic plasticity. Trends Neurosci. 2005;28:182–187. doi: 10.1016/j.tins.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Cavara NA, Hollmann M. Shuffling the deck anew: how NR3 tweaks NMDA receptor function. Mol Neurobiol. 2008;38:16–26. doi: 10.1007/s12035-008-8029-9. [DOI] [PubMed] [Google Scholar]
- Chan SF, Sucher NJ. An NMDA receptor signaling complex with protein phosphatase 2A. J Neurosci. 2001;21:7985–7992. doi: 10.1523/JNEUROSCI.21-20-07985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, Tong G, Lipton SA, Zhang D. Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature. 2002;415:793–798. doi: 10.1038/nature715. [DOI] [PubMed] [Google Scholar]
- Chazot PL, Cik M, Stephenson FA. An investigation into the role of N-glycosylation in the functional expression of a recombinant heteromeric NMDA receptor. Mol Membr Biol. 1995;12:331–337. doi: 10.3109/09687689509072435. [DOI] [PubMed] [Google Scholar]
- Chechlacz M, Gleeson JG. Is mental retardation a defect of synapse structure and function? Pediatr Neurol. 2003;29:11–17. doi: 10.1016/s0887-8994(03)00152-8. [DOI] [PubMed] [Google Scholar]
- Chen BS, Roche KW. Regulation of NMDA receptors by phosphorylation. Neuropharmacology. 2007;53:362–368. doi: 10.1016/j.neuropharm.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW. Excitotoxic cell death. J. Neurobiol. 1992;23:1261–1276. doi: 10.1002/neu.480230915. [DOI] [PubMed] [Google Scholar]
- Chowdari KV, Mirnics K, Semwal P, Wood J, Lawrence E, Bhatia T, Deshpande SN, B KT, Ferrell RE, Middleton FA, Devlin B, Levitt P, Lewis DA, Nimgaonkar VL. Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum Mol Genet. 2002;11:1373–1380. doi: 10.1093/hmg/11.12.1373. [DOI] [PubMed] [Google Scholar]
- Ciabarra AM, Sevarino KA. An anti-chi-1 antibody recognizes a heavily glycosylated protein in rat brain. Brain Res Mol Brain Res. 1997;46:85–90. doi: 10.1016/s0169-328x(96)00275-6. [DOI] [PubMed] [Google Scholar]
- Ciabarra AM, Sullivan JM, Gahn LG, Pecht G, Heinemann S, Sevarino KA. Cloning and characterization of chi-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J Neurosci. 1995;15:6498–6508. doi: 10.1523/JNEUROSCI.15-10-06498.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantine-Paton M, Cline HT, Debski E. Patterned activity, synaptic convergence, and the NMDA receptor in developing visual pathways. Ann. Rev. Neurosci. 1990;13:129–154. doi: 10.1146/annurev.ne.13.030190.001021. [DOI] [PubMed] [Google Scholar]
- Covarrubias M, Kopta C, Steinbach JH. Inhibitors of asparagine-linked oligosaccharide processing alter the kinetics of the nicotinic acetylcholine receptor. J Gen Physiol. 1989;93:765–783. doi: 10.1085/jgp.93.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle JT, Tsai G, Goff D. Converging evidence of NMDA receptor hypofunction in the pathophysiology of schizophrenia. Ann N Y Acad Sci. 2003;1003:318–327. doi: 10.1196/annals.1300.020. [DOI] [PubMed] [Google Scholar]
- Coyle JT, Yamamura HI. Neurochemical aspects of the ontogenesis of cholinergic neurons in the rat brain. Brain Res. 1976;118:429–440. doi: 10.1016/0006-8993(76)90310-3. [DOI] [PubMed] [Google Scholar]
- Crews FT, Steck JC, Chandler LJ, Yu CJ, Day A. Ethanol, stroke, brain damage, and excitotoxicity. Pharmacol Biochem Behav. 1998;59:981–991. doi: 10.1016/s0091-3057(97)00538-8. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE. 2004;2004:re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, Dikkes P, Conner DA, Rayudu PV, Cheung W, Chen HS, Lipton SA, Nakanishi N. Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature. 1998;393:377–381. doi: 10.1038/30748. [DOI] [PubMed] [Google Scholar]
- de Graaf-Peters VB, Hadders-Algra M. Ontogeny of the human central nervous system: what is happening when? Early Hum Dev. 2006;82:257–266. doi: 10.1016/j.earlhumdev.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Dubner R, Ruda MA. Activity-dependent neuronal plasticity following tissue injury and inflammation. Trends Neurosci. 1992;15:96–103. doi: 10.1016/0166-2236(92)90019-5. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Yasuda RP, Wang YH, Luo J, Davila-Garcia M, Gbadegesin M, Vicini S, Wolfe BB. Regional and ontogenic expression of the NMDA receptor subunit NR2D protein in rat brain using a subunit-specific antibody. J Neurochem. 1996;67:2335–2345. doi: 10.1046/j.1471-4159.1996.67062335.x. [DOI] [PubMed] [Google Scholar]
- Durand GM, Bennett MV, Zukin RS. Splice variants of the N-methyl-D-aspartate receptor NR1 identify domains involved in regulation by polyamines and protein kinase C. Proc Natl Acad Sci U S A. 1993;90:6731–6735. doi: 10.1073/pnas.90.14.6731. [published erratum appears in Proc Natl Acad Sci U S A 1993 Oct 15;90(20):9739] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand GM, Gregor P, Zheng X, Bennett MV, Uhl GR, Zukin RS. Cloning of an apparent splice variant of the rat N-methyl-D-aspartate receptor NMDAR1 with altered sensitivity to polyamines and activators of protein kinase C. Proc Natl Acad Sci U S A. 1992;89:9359–9363. doi: 10.1073/pnas.89.19.9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian ES, Karayiorgou M, Gogos JA. Decreased phosphorylation of NMDA receptor type 1 at serine 897 in brains of patients with Schizophrenia. J Neurosci. 2004;24:1561–1564. doi: 10.1523/JNEUROSCI.4650-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Nilsson A, Froelich-Fabre S, Akesson E, Dunker J, Seiger A, Folkesson R, Benedikz E, Sundstrom E. Cloning and expression of the human N-methyl-D-aspartate receptor subunit NR3A. Neurosci Lett. 2002;321:177–181. doi: 10.1016/s0304-3940(01)02524-1. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Nilsson A, Samuelsson H, Samuelsson EB, Mo L, Akesson E, Benedikz E, Sundstrom E. On the role of NR3A in human NMDA receptors. Physiol Behav. 2007a;92:54–59. doi: 10.1016/j.physbeh.2007.05.026. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Samuelsson H, Samuelsson EB, Liu L, McKeehan WL, Benedikz E, Sundstrom E. The NMDAR subunit NR3A interacts with microtubule-associated protein 1S in the brain. Biochem Biophys Res Commun. 2007b;361:127–132. doi: 10.1016/j.bbrc.2007.06.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan MM, Raymond LA. N-methyl-D-aspartate (NMDA) receptor function and excitotoxicity in Huntington's disease. Prog Neurobiol. 2007;81:272–293. doi: 10.1016/j.pneurobio.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Fellous JM, Sejnowski TJ. Regulation of persistent activity by background inhibition in an in vitro model of a cortical microcircuit. Cereb Cortex. 2003;13:1232–1241. doi: 10.1093/cercor/bhg098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harris KM. Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res Brain Res Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- Fujikawa DG. Prolonged seizures and cellular injury: understanding the connection. Epilepsy Behav. 2005;7 Suppl 3:S3–S11. doi: 10.1016/j.yebeh.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Fukaya M, Hayashi Y, Watanabe M. NR2 to NR3B subunit switchover of NMDA receptors in early postnatal motoneurons. Eur J Neurosci. 2005;21:1432–1436. doi: 10.1111/j.1460-9568.2005.03957.x. [DOI] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Gallinat J, Gotz T, Kalus P, Bajbouj M, Sander T, Winterer G. Genetic variations of the NR3A subunit of the NMDA receptor modulate prefrontal cerebral activity in humans. J Cogn Neurosci. 2007;19:59–68. doi: 10.1162/jocn.2007.19.1.59. [DOI] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA. Dendritic spine density in schizophrenia and depression. Arch Gen Psychiatry. 2001;58:203. doi: 10.1001/archpsyc.58.2.203. [DOI] [PubMed] [Google Scholar]
- Goebel DJ, Poosch MS. NMDA receptor subunit gene expression in the rat brain: a quantitative analysis of endogenous mRNA levels of NR1Com, NR2A, NR2B, NR2C, NR2D and NR3A. Brain Res Mol Brain Res. 1999;69:164–170. doi: 10.1016/s0169-328x(99)00100-x. [DOI] [PubMed] [Google Scholar]
- Goff DC, Tsai G, Levitt J, Amico E, Manoach D, Schoenfeld DA, Hayden DL, McCarley R, Coyle JT. A placebo-controlled trial of D-cycloserine added to conventional neuroleptics in patients with schizophrenia. Arch Gen Psychiatry. 1999;56:21–27. doi: 10.1001/archpsyc.56.1.21. [DOI] [PubMed] [Google Scholar]
- Goff DC, Tsai G, Manoach DS, Coyle JT. Dose-finding trial of D-cycloserine added to neuroleptics for negative symptoms in schizophrenia. Am J Psychiatry. 1995;152:1213–1215. doi: 10.1176/ajp.152.8.1213. [DOI] [PubMed] [Google Scholar]
- Groc L, Heine M, Cousins SL, Stephenson FA, Lounis B, Cognet L, Choquet D. NMDA receptor surface mobility depends on NR2A-2B subunits. Proc Natl Acad Sci U S A. 2006;103:18769–18774. doi: 10.1073/pnas.0605238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Jiang Q, Fu AK, Ip NY, Yan Z. Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J Neurosci. 2005;25:4974–4984. doi: 10.1523/JNEUROSCI.1086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson B, Wigstrom H. Physiological mechanisms underlying long-term potentiation. Trends Neurosci. 1988;11:156–162. doi: 10.1016/0166-2236(88)90142-7. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. image 45. [DOI] [PubMed] [Google Scholar]
- Hatton CJ, Paoletti P. Modulation of triheteromeric NMDA receptors by N-terminal domain ligands. Neuron. 2005;46:261–274. doi: 10.1016/j.neuron.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Henson MA, Roberts AC, Salimi K, Vadlamudi S, Hamer RM, Gilmore JH, Jarskog LF, Philpot BD. Developmental regulation of the NMDA receptor subunits, NR3A and NR1, in human prefrontal cortex. Cereb Cortex. 2008;18:2560–2573. doi: 10.1093/cercor/bhn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heresco-Levy U, Javitt DC, Ermilov M, Mordel C, Horowitz A, Kelly D. Double-blind, placebo-controlled, crossover trial of glycine adjuvant therapy for treatment-resistant schizophrenia. Br J Psychiatry. 1996;169:610–617. doi: 10.1192/bjp.169.5.610. [DOI] [PubMed] [Google Scholar]
- Heresco-Levy U, Javitt DC, Ermilov M, Silipo G, Shimoni J. Double-blind, placebo-controlled, crossover trial of D-cycloserine adjuvant therapy for treatment-resistant schizophrenia. Int J Neuropsychopharmcol. 1998;1:131–135. doi: 10.1017/S1461145798001242. [DOI] [PubMed] [Google Scholar]
- Hirai H, Kirsch J, Laube B, Betz H, Kuhse J. The glycine binding site of the N-methyl-D-aspartate receptor subunit NR1: identification of novel determinants of co-agonist potentiation in the extracellular M3-M4 loop region. Proc Natl Acad Sci U S A. 1996;93:6031–6036. doi: 10.1073/pnas.93.12.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai T, Yoneda Y. Functional alterations in immature cultured rat hippocampal neurons after sustained exposure to static magnetic fields. J Neurosci Res. 2004;75:230–240. doi: 10.1002/jnr.10819. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Boulter J, Maron C, Beasley L, Sullivan J, Pecht G, Heinemann S. Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron. 1993;10:943–954. doi: 10.1016/0896-6273(93)90209-a. [DOI] [PubMed] [Google Scholar]
- Hoyte L, Barber PA, Buchan AM, Hill MD. The rise and fall of NMDA antagonists for ischemic stroke. Curr Mol Med. 2004;4:131–136. doi: 10.2174/1566524043479248. [DOI] [PubMed] [Google Scholar]
- Ikonomidou C, Bosch F, Miksa M, Bittigau P, Vockler J, Dikranian K, Tenkova TI, Stefovska V, Turski L, Olney JW. Blockade of NMDA receptors and apoptotic neurodegeneration in the developing brain. Science. 1999;283:70–74. doi: 10.1126/science.283.5398.70. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Swain RA, Christmon CA, Chakravarti A, Weiler IJ, Greenough WT. Evidence for altered Fragile-X mental retardation protein expression in response to behavioral stimulation. Neurobiol Learn Mem. 2000;74:87–93. [PubMed] [Google Scholar]
- Isaac JTR, Nicoll RA, Malenka RC. Evidence for silent synapses: implications for the expression of LTP and LTD. Neuron. 1995;15:427–434. doi: 10.1016/0896-6273(95)90046-2. [DOI] [PubMed] [Google Scholar]
- Ishihama K, Kogo M, Wakisaka S, Turman JE., Jr Prenatal development of NMDA receptor composition and function in trigeminal neurons. Arch Histol Cytol. 2005;68:321–335. doi: 10.1679/aohc.68.321. [DOI] [PubMed] [Google Scholar]
- Ishihama K, Turman JE., Jr NR3 protein expression in trigeminal neurons during postnatal development. Brain Res. 2006;1095:12–16. doi: 10.1016/j.brainres.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Javitt DC. Glycine modulators in schizophrenia. Curr Opin Investig Drugs. 2002;3:1067–1072. [PubMed] [Google Scholar]
- Javitt DC, Zukin SR. Recent advances in the phencyclidine model of schizophrenia. Am J Psychiatry. 1991;148:1301–1308. doi: 10.1176/ajp.148.10.1301. [DOI] [PubMed] [Google Scholar]
- Javitt DC, Zylberman I, Zukin SR, Heresco-Levy U, Lindenmayer JP. Amelioration of negative symptoms in schizophrenia by glycine. Am J Psychiatry. 1994;151:1234–1236. doi: 10.1176/ajp.151.8.1234. [DOI] [PubMed] [Google Scholar]
- Karadottir R, Cavelier P, Bergersen LH, Attwell D. NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature. 2005;438:1162–1166. doi: 10.1038/nature04302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Kennedy MB, Beale HC, Carlisle HJ, Washburn LR. Integration of biochemical signalling in spines. Nat Rev Neurosci. 2005;6:423–434. doi: 10.1038/nrn1685. [DOI] [PubMed] [Google Scholar]
- Kleckner NW, Dingledine R. Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes. Science. 1988;241:835–837. doi: 10.1126/science.2841759. [DOI] [PubMed] [Google Scholar]
- Kohr G. NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res. 2006;326:439–446. doi: 10.1007/s00441-006-0273-6. [DOI] [PubMed] [Google Scholar]
- Kuner T, Wollmuth LP, Karlin A, Seeburg PH, Sakmann B. Structure of the NMDA receptor channel M2 segment inferred from the accessibility of substituted cysteines. Neuron. 1996;17:343–352. doi: 10.1016/s0896-6273(00)80165-8. [DOI] [PubMed] [Google Scholar]
- Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci. 2007;8:413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- Laube B, Kuhse J, Betz H. Evidence for a tetrameric structure of recombinant NMDA receptors. J. Neurosci. 1998;18:2954–2961. doi: 10.1523/JNEUROSCI.18-08-02954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DJ, Bartke I, Schoepfer R, Naujoks K, Seeburg PH. Regional, developmental and interspecies expression of the four NMDAR2 subunits, examined using monoclonal antibodies. Brain Res Mol Brain Res. 1997;51:23–32. doi: 10.1016/s0169-328x(97)00206-4. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Seeburg PH. Ligand affinities at recombinant N-methyl-D-aspartate receptors depend on subunit composition. Eur J Pharmacol. 1994;268:335–345. doi: 10.1016/0922-4106(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Lavezzari G, McCallum J, Dewey CM, Roche KW. Subunit-specific regulation of NMDA receptor endocytosis. J Neurosci. 2004;24:6383–6391. doi: 10.1523/JNEUROSCI.1890-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Han TZ. Glycine binding sites of presynaptic NMDA receptors may tonically regulate glutamate release in the rat visual cortex. J Neurophysiol. 2006 doi: 10.1152/jn.00980.2006. [DOI] [PubMed] [Google Scholar]
- Liao D, Hessler NA, Malinow R. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature. 1995;375:400–404. doi: 10.1038/375400a0. [DOI] [PubMed] [Google Scholar]
- Lipton SA. Paradigm shift in neuroprotection by NMDA receptor blockade: memantine and beyond. Nat Rev Drug Discov. 2006;5:160–170. doi: 10.1038/nrd1958. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Nakanishi N. Shakespeare in love--with NMDA receptors? Nat Med. 1999;5:270–271. doi: 10.1038/6481. [DOI] [PubMed] [Google Scholar]
- Lisman J. A mechanism for the Hebb and the anti-Hebb processes underlying learning and memory. Proc Natl Acad Sci U S A. 1989;86:9574–9578. doi: 10.1073/pnas.86.23.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovinger DM. Excitotoxicity and alcohol-related brain damage. Alcohol Clin Exp Res. 1993;17:19–27. doi: 10.1111/j.1530-0277.1993.tb00720.x. [DOI] [PubMed] [Google Scholar]
- Lujan R, Shigemoto R, Lopez-Bendito G. Glutamate and GABA receptor signalling in the developing brain. Neuroscience. 2005;130:567–580. doi: 10.1016/j.neuroscience.2004.09.042. [DOI] [PubMed] [Google Scholar]
- Ma OK, Sucher NJ. Molecular interaction of NMDA receptor subunit NR3A with protein phosphatase 2A. Neuroreport. 2004;15:1447–1450. doi: 10.1097/01.wnr.0000132773.41720.2d. [DOI] [PubMed] [Google Scholar]
- Madison DV, Malenka RC, Nicoll RA. Mechanisms underlying long-term potentiation of synaptic transmission. Annu Rev Neurosci. 1991;14:379–397. doi: 10.1146/annurev.ne.14.030191.002115. [DOI] [PubMed] [Google Scholar]
- Madry C, Mesic I, Bartholomaus I, Nicke A, Betz H, Laube B. Principal role of NR3 subunits in NR1/NR3 excitatory glycine receptor function. Biochem Biophys Res Commun. 2007;354:102–108. doi: 10.1016/j.bbrc.2006.12.153. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- Martin LJ, Al-Abdulla NA, Brambrink AM, Kirsch JR, Sieber FE, Portera-Cailliau C. Neurodegeneration in excitotoxicity, global cerebral ischemia, and target deprivation: A perspective on the contributions of apoptosis and necrosis. Brain Res Bull. 1998;46:281–309. doi: 10.1016/s0361-9230(98)00024-0. [DOI] [PubMed] [Google Scholar]
- Martucci L, Wong AH, Trakalo J, Cate-Carter T, Wong GW, Macciardi FM, Kennedy JL. N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) in schizophrenia: TDT and case-control analyses. Am J Med Genet B Neuropsychiatr Genet. 2003;119:24–27. doi: 10.1002/ajmg.b.20014. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Fletcher M, Kamiya Y, Yuzaki M. Specific assembly with the NMDA receptor 3B subunit controls surface expression and calcium permeability of NMDA receptors. J Neurosci. 2003;23:10064–10073. doi: 10.1523/JNEUROSCI.23-31-10064.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Kamiya Y, Matsuda S, Yuzaki M. Cloning and characterization of a novel NMDA receptor subunit NR3B: a dominant subunit that reduces calcium permeability. Brain Res Mol Brain Res. 2002;100:43–52. doi: 10.1016/s0169-328x(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Matute C. Oligodendrocyte NMDA receptors: a novel therapeutic target. Trends Mol Med. 2006;12:289–292. doi: 10.1016/j.molmed.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Matute C, Alberdi E, Domercq M, Sanchez-Gomez MV, Perez-Samartin A, Rodriguez-Antiguedad A, Perez-Cerda F. Excitotoxic damage to white matter. J Anat. 2007;210:693–702. doi: 10.1111/j.1469-7580.2007.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute C, Sanchez-Gomez MV, Martinez-Millan L, Miledi R. Glutamate receptor-mediated toxicity in optic nerve oligodendrocytes. Proc Natl Acad Sci U S A. 1997;94:8830–8835. doi: 10.1073/pnas.94.16.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain CJ, Mayer ML. N-methyl-D-aspartic acid receptor structure and function. Physiol. Rev. 1994;74:723–760. doi: 10.1152/physrev.1994.74.3.723. [DOI] [PubMed] [Google Scholar]
- Meador-Woodruff JH, Healy DJ. Glutamate receptor expression in schizophrenic brain. Brain Res Brain Res Rev. 2000;31:288–294. doi: 10.1016/s0165-0173(99)00044-2. [DOI] [PubMed] [Google Scholar]
- Meguro H, Mori H, Araki K, Kushiya E, Kutsuwada T, Yamazaki M, Kumanishi T, Arakawa M, Sakimura K, Mishina M. Functional characterization of a heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature. 1992;357:70–74. doi: 10.1038/357070a0. [DOI] [PubMed] [Google Scholar]
- Meldrum BS. Excitotoxicity and selective neuronal loss in epilepsy. Brain Pathol. 1993;3:405–412. doi: 10.1111/j.1750-3639.1993.tb00768.x. [DOI] [PubMed] [Google Scholar]
- Micu I, Jiang Q, Coderre E, Ridsdale A, Zhang L, Woulfe J, Yin X, Trapp BD, McRory JE, Rehak R, Zamponi GW, Wang W, Stys PK. NMDA receptors mediate calcium accumulation in myelin during chemical ischaemia. Nature. 2006;439:988–992. doi: 10.1038/nature04474. [DOI] [PubMed] [Google Scholar]
- Millan MJ. N-Methyl-D-aspartate receptors as a target for improved antipsychotic agents: novel insights and clinical perspectives. Psychopharmacology (Berl) 2005;179:30–53. doi: 10.1007/s00213-005-2199-1. [DOI] [PubMed] [Google Scholar]
- Mishina M, Mori H, Araki K, Kushiya E, Meguro H, Kutsuwada T, Kashiwabuchi N, Ikeda K, Nagasawa M, Yamazaki M, et al. Molecular and functional diversity of the NMDA receptor channel. Ann N Y Acad Sci. 1993;707:136–152. doi: 10.1111/j.1749-6632.1993.tb38049.x. [DOI] [PubMed] [Google Scholar]
- Moghaddam B. Bringing order to the glutamate chaos in schizophrenia. Neuron. 2003;40:881–884. doi: 10.1016/s0896-6273(03)00757-8. [DOI] [PubMed] [Google Scholar]
- Moghaddam B, Jackson ME. Glutamatergic animal models of schizophrenia. Ann N Y Acad Sci. 2003;1003:131–137. doi: 10.1196/annals.1300.065. [DOI] [PubMed] [Google Scholar]
- Mohn AR, Gainetdinov RR, Caron MG, Koller BH. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell. 1999;98:427–436. doi: 10.1016/s0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- Montgomery JM, Selcher JC, Hanson JE, Madison DV. Dynamin-dependent NMDAR endocytosis during LTD and its dependence on synaptic state. BMC Neurosci. 2005;6:48. doi: 10.1186/1471-2202-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Mori H, Mishina M. Structure and function of the NMDA receptor channel. Neuropharmacology. 1995;34:1219–1237. doi: 10.1016/0028-3908(95)00109-j. [DOI] [PubMed] [Google Scholar]
- Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- Mueller HT, Meador-Woodruff JH. Expression of the NR3A subunit of the NMDA receptor in human fetal brain. Ann N Y Acad Sci. 2003;1003:448–451. doi: 10.1196/annals.1300.049. [DOI] [PubMed] [Google Scholar]
- Mueller HT, Meador-Woodruff JH. NR3A NMDA receptor subunit mRNA expression in schizophrenia, depression and bipolar disorder. Schizophr Res. 2004;71:361–370. doi: 10.1016/j.schres.2004.02.016. [DOI] [PubMed] [Google Scholar]
- Mueller HT, Meador-Woodruff JH. Distribution of the NMDA receptor NR3A subunit in the adult pig-tail macaque brain. J Chem Neuroanat. 2005;29:157–172. doi: 10.1016/j.jchemneu.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Naassila M, Daoust M. Effect of prenatal and postnatal ethanol exposure on the developmental profile of mRNAs encoding NMDA receptor subunits in rat hippocampus. J Neurochem. 2002;80:850–860. doi: 10.1046/j.0022-3042.2002.00755.x. [DOI] [PubMed] [Google Scholar]
- Nagerl UV, Eberhorn N, Cambridge SB, Bonhoeffer T. Bidirectional activity-dependent morphological plasticity in hippocampal neurons. Neuron. 2004;44:759–767. doi: 10.1016/j.neuron.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Tu S, Shin Y, Cui J, Kurokawa T, Zhang D, Chen HS, Tong G, Lipton SA. Neuroprotection by the NR3A subunit of the NMDA receptor. J Neurosci. 2009;29:5260–5265. doi: 10.1523/JNEUROSCI.1067-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newey SE, Velamoor V, Govek EE, Van Aelst L. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol. 2005;64:58–74. doi: 10.1002/neu.20153. [DOI] [PubMed] [Google Scholar]
- Nilsson A, Duan J, Mo-Boquist LL, Benedikz E, Sundstrom E. Characterisation of the human NMDA receptor subunit NR3A glycine binding site. Neuropharmacology. 2007a;52:1151–1159. doi: 10.1016/j.neuropharm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Nilsson A, Eriksson M, Muly EC, Akesson E, Samuelsson EB, Bogdanovic N, Benedikz E, Sundstrom E. Analysis of NR3A receptor subunits in human native NMDA receptors. Brain Res. 2007b;1186:102–112. doi: 10.1016/j.brainres.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Nishi M, Hinds H, Lu HP, Kawata M, Hayashi Y. Motoneuron-specific expression of NR3B, a novel NMDA-type glutamate receptor subunit that works in a dominant-negative manner. J Neurosci. 2001;21:RC185. doi: 10.1523/JNEUROSCI.21-23-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olney JW, Farber NB. Glutamate receptor dysfunction and schizophrenia. Arch Gen Psychiatry. 1995;52:998–1007. doi: 10.1001/archpsyc.1995.03950240016004. [DOI] [PubMed] [Google Scholar]
- Olney JW, Newcomer JW, Farber NB. NMDA receptor hypofunction model of schizophrenia. J Psychiatr Res. 1999;33:523–533. doi: 10.1016/s0022-3956(99)00029-1. [DOI] [PubMed] [Google Scholar]
- Ozaki M, Sasner M, Yano R, Lu HS, Buonanno A. Neuregulin-beta induces expression of an NMDA-receptor subunit. Nature. 1997;390:691–694. doi: 10.1038/37795. [DOI] [PubMed] [Google Scholar]
- Paarmann I, Frermann D, Keller BU, Villmann C, Breitinger HG, Hollmann M. Kinetics and subunit composition of NMDA receptors in respiratory-related neurons. J Neurochem. 2005;93:812–824. doi: 10.1111/j.1471-4159.2005.03027.x. [DOI] [PubMed] [Google Scholar]
- Paas Y. The macro- and microarchitectures of the ligand-binding domain of glutamate receptors. Trends Neurosci. 1998;21:117–125. doi: 10.1016/s0166-2236(97)01184-3. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Neyton J. NMDA receptor subunits: function and pharmacology. Curr Opin Pharmacol. 2007;7:39–47. doi: 10.1016/j.coph.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Perez-Otano I, Ehlers MD. Learning from NMDA receptor trafficking: clues to the development and maturation of glutamatergic synapses. Neurosignals. 2004;13:175–189. doi: 10.1159/000077524. [DOI] [PubMed] [Google Scholar]
- Perez-Otano I, Lujan R, Tavalin SJ, Plomann M, Modregger J, Liu XB, Jones EG, Heinemann SF, Lo DC, Ehlers MD. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat Neurosci. 2006;9:611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF. Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J Neurosci. 2001;21:1228–1237. doi: 10.1523/JNEUROSCI.21-04-01228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpot BD, Sekhar AK, Shouval HZ, Bear MF. Visual experience and deprivation bidirectionally modify the composition and function of NMDA receptors in visual cortex. Neuron. 2001;29:157–169. doi: 10.1016/s0896-6273(01)00187-8. [DOI] [PubMed] [Google Scholar]
- Pilowsky LS, Bressan RA, Stone JM, Erlandsson K, Mulligan RS, Krystal JH, Ell PJ. First in vivo evidence of an NMDA receptor deficit in medication-free schizophrenic patients. Mol Psychiatry. 2006;11:118–119. doi: 10.1038/sj.mp.4001751. [DOI] [PubMed] [Google Scholar]
- Quinlan EM, Philpot BD, Huganir RL, Bear MF. Rapid, experience-dependent expression of synaptic NMDA receptors in visual cortex in vivo. Nat. Neurosci. 1999;2:352–357. doi: 10.1038/7263. [DOI] [PubMed] [Google Scholar]
- Racca C, Stephenson FA, Streit P, Roberts JD, Somogyi P. NMDA receptor content of synapses in stratum radiatum of the hippocampal CA1 area. J Neurosci. 2000;20:2512–2522. doi: 10.1523/JNEUROSCI.20-07-02512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AC, Diez-Garcia J, Rodriguiz RM, Lopez IP, Lujan R, Martinez-Turrillas R, Pico E, Henson MA, Bernardo DR, Jarrett TM, Clendeninn DJ, Lopez-Mascaraque L, Feng G, Lo DC, Wesseling JF, Wetsel WC, Philpot BD, Perez-Otano I. Downregulation of NR3A-containing NMDARs is required for synapse maturation and memory consolidation. Neuron. 2009;63:342–356. doi: 10.1016/j.neuron.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Rothman SM, Olney JW. Excitotoxicity and the NMDA receptor--still lethal after eight years. Trends Neurosci. 1995;18:57–58. doi: 10.1016/0166-2236(95)93869-y. [DOI] [PubMed] [Google Scholar]
- Rumpel S, Hatt H, Gottmann K. Silent synapses in the developing rat visual cortex: evidence for postsynaptic expression of synaptic plasticity. J. Neurosci. 1998;18:8863–8874. doi: 10.1523/JNEUROSCI.18-21-08863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, Fern R. NMDA receptors are expressed in developing oligodendrocyte processes and mediate injury. Nature. 2005;438:1167–1171. doi: 10.1038/nature04301. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vives MV, McCormick DA. Cellular and network mechanisms of rhythmic recurrent activity in neocortex. Nat Neurosci. 2000;3:1027–1034. doi: 10.1038/79848. [DOI] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos J, 2nd, Hell JW, Wenthold RJ. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–1271. doi: 10.1523/JNEUROSCI.20-03-01260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YF, Rothe T, Premkumar LS, Das S, Cui J, Talantova MV, Wong HK, Gong X, Chan SF, Zhang D, Nakanishi N, Sucher NJ, Lipton SA. Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J Neurophysiol. 2002;87:2052–2063. doi: 10.1152/jn.00531.2001. [DOI] [PubMed] [Google Scholar]
- Sawtell NB, Frenkel MY, Philpot BD, Nakazawa K, Tonegawa S, Bear MF. NMDA receptor-dependent ocular dominance plasticity in adult visual cortex. Neuron. 2003;38:977–985. doi: 10.1016/s0896-6273(03)00323-4. [DOI] [PubMed] [Google Scholar]
- Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci. 2003;23:1151–1158. doi: 10.1523/JNEUROSCI.23-04-01151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler T, Mesic I, Madry C, Bartholomaus I, Laube B. Formation of NR1/NR2 and NR1/NR3 heterodimers constitutes the initial step in N-methyl-D-aspartate receptor assembly. J Biol Chem. 2008;283:37–46. doi: 10.1074/jbc.M703539200. [DOI] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DB, Michailidis I, Mu Y, Logothetis D, Ehlers MD. Endocytosis and degradative sorting of NMDA receptors by conserved membrane-proximal signals. J Neurosci. 2004;24:7096–7109. doi: 10.1523/JNEUROSCI.0780-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- Smothers CT, Woodward JJ. Effect of the NR3 subunit on ethanol inhibition of recombinant NMDA receptors. Brain Res. 2003;987:117–121. doi: 10.1016/s0006-8993(03)03315-8. [DOI] [PubMed] [Google Scholar]
- Smothers CT, Woodward JJ. Pharmacological characterization of glycine-activated currents in HEK 293 cells expressing N-methyl-D-aspartate NR1 and NR3 subunits. J Pharmacol Exp Ther. 2007;322:739–748. doi: 10.1124/jpet.107.123836. [DOI] [PubMed] [Google Scholar]
- Standley S, Roche KW, McCallum J, Sans N, Wenthold RJ. PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron. 2000;28:887–898. doi: 10.1016/s0896-6273(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Steinthorsdottir V, Thorgeirsson TE, Gulcher JR, Stefansson K. Neuregulin 1 and schizophrenia. Ann Med. 2004;36:62–71. doi: 10.1080/07853890310017585. [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, O'Neill FA, Walsh D, Kendler KS. Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow-up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry. 2002;7:542–559. doi: 10.1038/sj.mp.4001051. [DOI] [PubMed] [Google Scholar]
- Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, Wu MK, Yuan JP, Jones EG, Lipton SA. Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J Neurosci. 1995;15:6509–6520. doi: 10.1523/JNEUROSCI.15-10-06509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucher NJ, Kohler K, Tenneti L, Wong HK, Grunder T, Fauser S, Wheeler-Schilling T, Nakanishi N, Lipton SA, Guenther E. N-methyl-D-aspartate receptor subunit NR3A in the retina: developmental expression, cellular localization, and functional aspects. Invest Ophthalmol Vis Sci. 2003;44:4451–4456. doi: 10.1167/iovs.02-1259. [DOI] [PubMed] [Google Scholar]
- Sugihara H, Moriyoshi K, Ishii T, Masu M, Nakanishi S. Structures and properties of seven isoforms of the NMDA receptor generated by alternative splicing. Biochem Biophys Res Commun. 1992;185:826–832. doi: 10.1016/0006-291x(92)91701-q. [DOI] [PubMed] [Google Scholar]
- Sullivan JM, Traynelis SF, Chen HS, Escobar W, Heinemann SF, Lipton SA. Identification of two cysteine residues that are required for redox modulation of the NMDA subtype of glutamate receptor. Neuron. 1994;13:929–936. doi: 10.1016/0896-6273(94)90258-5. [DOI] [PubMed] [Google Scholar]
- Sun L, Margolis FL, Shipley MT, Lidow MS. Identification of a long variant of mRNA encoding the NR3 subunit of the NMDA receptor: its regional distribution and developmental expression in the rat brain. FEBS Lett. 1998;441:392–396. doi: 10.1016/s0014-5793(98)01590-7. [DOI] [PubMed] [Google Scholar]
- Sun L, Shipley MT, Lidow MS. Expression of NR1, NR2A-D, and NR3 subunits of the NMDA receptor in the cerebral cortex and olfactory bulb of adult rat. Synapse. 2000;35:212–221. doi: 10.1002/(SICI)1098-2396(20000301)35:3<212::AID-SYN6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Tamminga CA. Schizophrenia and glutamatergic transmission. Crit Rev Neurobiol. 1998;12:21–36. doi: 10.1615/critrevneurobiol.v12.i1-2.20. [DOI] [PubMed] [Google Scholar]
- Tang YP, Shimizu E, Dube GR, Rampon C, Kerchner GA, Zhuo M, Liu G, Tsien JZ. Genetic enhancement of learning and memory in mice. Nature. 1999;401:63–69. doi: 10.1038/43432. [DOI] [PubMed] [Google Scholar]
- Tong G, Takahashi H, Tu S, Shin Y, Talantova M, Zago W, Xia P, Nie Z, Goetz T, Zhang D, Lipton SA, Nakanishi N. Modulation of NMDA receptor properties and synaptic transmission by the NR3A subunit in mouse hippocampal and cerebrocortical neurons. J Neurophysiol. 2008;99:122–132. doi: 10.1152/jn.01044.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai G, van Kammen DP, Chen S, Kelley ME, Grier A, Coyle JT. Glutamatergic neurotransmission involves structural and clinical deficits of schizophrenia. Biol Psychiatry. 1998;44:667–674. doi: 10.1016/s0006-3223(98)00151-6. [DOI] [PubMed] [Google Scholar]
- Tu S, Shin Y, Zago WM, States BA, Eroshkin A, Lipton SA, Tong GG, Nakanishi N. Takusan: a large gene family that regulates synaptic activity. Neuron. 2007;55:69–85. doi: 10.1016/j.neuron.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY. Rules of engagement for NMDA receptor subunits. Proc Natl Acad Sci U S A. 2008;105:14163–14168. doi: 10.1073/pnas.0802075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullian EM, Barkis WB, Chen S, Diamond JS, Barres BA. Invulnerability of retinal ganglion cells to NMDA excitotoxicity. Mol Cell Neurosci. 2004;26:544–557. doi: 10.1016/j.mcn.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Wada A, Takahashi H, Lipton SA, Chen HS. NR3A modulates the outer vestibule of the "NMDA" receptor channel. J Neurosci. 2006;26:13156–13166. doi: 10.1523/JNEUROSCI.2552-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washbourne P, Liu XB, Jones EG, McAllister AK. Cycling of NMDA receptors during trafficking in neurons before synapse formation. J Neurosci. 2004;24:8253–8264. doi: 10.1523/JNEUROSCI.2555-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M. Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 1992;3:1138–1140. doi: 10.1097/00001756-199212000-00027. [DOI] [PubMed] [Google Scholar]
- Wee KS, Zhang Y, Khanna S, Low CM. Immunolocalization of NMDA receptor subunit NR3B in selected structures in the rat forebrain, cerebellum, and lumbar spinal cord. J Comp Neurol. 2008;509:118–135. doi: 10.1002/cne.21747. [DOI] [PubMed] [Google Scholar]
- Wenk GL. Neuropathologic changes in Alzheimer's disease: potential targets for treatment. J Clin Psychiatry. 2006;67 Suppl 3:3–7. quiz 23. [PubMed] [Google Scholar]
- Wong HK, Liu XB, Matos MF, Chan SF, Perez-Otano I, Boysen M, Cui J, Nakanishi N, Trimmer JS, Jones EG, Lipton SA, Sucher NJ. Temporal and regional expression of NMDA receptor subunit NR3A in the mammalian brain. J Comp Neurol. 2002;450:303–317. doi: 10.1002/cne.10314. [DOI] [PubMed] [Google Scholar]
- Wu G-Y, Malinow R, Cline HT. Maturation of a central glutamatergic synapse. Science. 1996;274:972–976. doi: 10.1126/science.274.5289.972. [DOI] [PubMed] [Google Scholar]
- Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. Embo J. 2008;27:2158–2170. doi: 10.1038/emboj.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Mayer ML. Characterization of a soluble ligand binding domain of the NMDA receptor regulatory subunit NR3A. J Neurosci. 2006;26:4559–4566. doi: 10.1523/JNEUROSCI.0560-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- Zeevalk GD, Nicklas WJ. NMDA receptors, cellular edema, and metabolic stress. Ann N Y Acad Sci. 1992;648:368–370. doi: 10.1111/j.1749-6632.1992.tb24585.x. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Liu AP, Ruan DY, Liu J. Effect of developmental lead exposure on the expression of specific NMDA receptor subunit mRNAs in the hippocampus of neonatal rats by digoxigenin-labeled in situ hybridization histochemistry. Neurotoxicol Teratol. 2002;24:149–160. doi: 10.1016/s0892-0362(01)00210-0. [DOI] [PubMed] [Google Scholar]