SUMMARY
In October 2004, plankton samples were collected from six permanent lakes located between 4960 and 5440 m a.s.l. in the Mount Everest region (Nepal) to assess how spatial and local environmental factors affect natural bacterial community composition. Fingerprinting analysis of the bacterial 16S rRNA gene fragment was done by denaturing gradient gel electrophoresis (DGGE).
The number of DGGE bands (range: 12–23) was not correlated with lake area or remoteness, but there was a strong negative correlation with the ratio of catchment to lake area (r = −0.826, P < 0.05), suggesting that hydraulic retention time affects the establishment of the bacterial community in these seepage lakes.
Most dominant sequences belonged to Betaproteobacteria except in two lakes where members of Bacteroidetes made the largest relative contribution. Up to 81% of the phylotypes had high similarity (>98 to 100%) in partial 16S rRNA gene sequence to those reported from other alpine lakes and glaciers around the world, suggesting the presence of ‘cosmopolitan’ bacteria.
An analysis based on dissimilarity matrices and the Mantel test revealed the existence of dissimilarities in bacterial community composition related to geographical distance over a small spatial scale (<6 km), but determined by local environmental constraints.
Our results suggest that several bacterial phylotypes are ubiquitous in the freshwater aquatic realm, but taxon sorting by local environmental constraints is important.
Keywords: alpine lakes, bacteria, biogeography, denaturing gradient gel electrophoresis, spatial distribution
Introduction
Microbial ecology is undergoing a rapid process of testing hypotheses and relating results to principles that have been developed in other areas of ecology (Konopka, 2006; Prosser et al., 2007). Part of this process has been facilitated by the use of various molecular techniques that allow us to open the bacterial ‘black box’ and to assess community composition at different levels of resolution. For example, the development of fingerprint and cloning methods based on rRNA analysis to study bacterial community composition has allowed us to test ecological hypotheses derived from macroorganisms in different aquatic ecosystems (Reche et al., 2005; Horner-Devine et al., 2007; Pommier et al., 2007). Such studies are not without problems such as, for example, how to define the operational taxonomic unit to use when testing hypotheses. This is particularly difficult because there is no agreement on how to define a prokaryotic species (Cohan, 2002) and, in consequence, how to assess biodiversity. In a recent opinion article, Pedrós-Alió (2006) suggested differentiating the concepts of diversity and biodiversity in microbial studies by defining the former as being represented by that part of biodiversity that includes the abundant and active microbial taxa (so-called ‘core species’) that drive most ecosystem functions, and the latter by also including a seed bank of rare species that grow slowly or not at all. In fact, there is some evidence for bacterial community composition having few abundant and many rare taxa (Casamayor et al., 2000; Acinas et al., 2004). In any case, molecular techniques such as fingerprinting analyses of PCR-amplified rDNA, which are known to have a certain numerical detection thresholds, can be used to retrieve the diversity of the dominant bacterial taxa (Casamayor et al., 2002; Pedrós-Alió, 2006).
One of the most debated concepts in microbial ecology, particularly in the aquatic realm, is whether or not microorganisms have biogeographies. Whereas biogeographic patterns have been demonstrated among free-living microbial taxa, environmental heterogeneity has in many cases at least a partial influence on spatial variation in microbial diversity (Hughes Martiny et al., 2006; Ramette & Tiedje, 2007). The list of studies that have found nonrandom distribution of free-living microbial taxa at spatial scales ranging from 0.002 to 20 000 km is relatively large, but few studies have simultaneously tested the effects of geographic distance and local environmental factors on microbial community composition (Hughes Martiny et al., 2006). This is needed to elucidate spurious correlations between community composition and geographic distance that arise through the effects of different local environmental constraints.
An emerging result is that several groups of microbes, studied at different taxonomical resolutions, appear to follow very different biogeographic patterns (see reviews by Dolan, 2006; Hughes Martiny et al., 2006). Among bacteria, the effect of geographic and environmental factors does not appear to follow a common pattern (Hughes Martiny et al., 2006; Findlay et al., 2008; Hunt et al., 2008). However, the information available for different types of environments is still inadequate to obtain a global picture. This is the case for lakes, where relatively few studies have tested how geographic and local environmental factors affect bacterial community composition. For example, Yannarell & Triplett (2004) reported a positive effect of distance and environmental factors on bacterial community composition in northern and southern Wisconsin lakes distributed across an intermediate spatial scale (500 km). At a smaller spatial scale (<10 km), Crump et al. (2007) showed that geographic distance and lake physical connectivity (i.e. by streams) influence the distribution of bacterioplankton communities. By contrast, Reche et al. (2005) working at a similar spatial scale in high mountain lakes of Sierra Nevada, Spain, only detected a distance effect. Most of these studies used fingerprinting techniques, but relatively few studies have given details on identification of operational taxonomic units (OTUs) as obtained by partial or total 16S rRNA gene sequencing. Whereas running biogeographic analyses on OTUs is a valid procedure, the lack of information on taxonomic affiliation permits no comparison with phylotypes that might already have been described from outside the studied region.
In this study, we tested the importance of spatial and local environmental factors for bacterial community composition of remote high altitude lakes located within a small spatial scale in the Mount Everest region. We used denaturing gradient gel electrophoresis (DGGE) and sequenced the most prominent bands detected to assess their potential affiliation to other known cultured and uncultured bacteria from aquatic systems. We selected this type of ecosystem because high altitude lakes are particularly suitable for addressing biogeographic questions of natural bacterial communities. This is because these lakes are remote and lack direct anthropogenic influence but at the same time are harsh environments where bacterioplankton have to cope with low nutrient conditions, low temperatures, extreme changes in light conditions between the ice-covered and ice-free situations, and except in turbid glacier lakes, periods of strong solar UV radiation exposure (Sommaruga, 2001). These extreme environmental characteristics are probably important in causing high ‘species’ or taxon sorting at the time of colonisation. Finally, high-altitude lakes are also efficient ‘collectors’ of precipitation and direct interceptors of airborne bacteria. Consequently, they may be prone to colonisation by microorganisms transported over long distances in the troposphere.
Methods
Study area and sampling
From 6 to 10 October 2004, six permanent seepage lakes located between 4960 and 5440 m a.s.l. were sampled in the Himalayan Region (Khumbu Valley, Nepal) near Mount Everest using the Pyramid Laboratory (5050 m a.s.l.) as base. The lakes were located within a maximum distance of 5.9 km (flying distance) and differed in morphological and limnological characteristics, particularly in their turbidity because of the extent of glaciers in their catchments (Table 1). Thus, Lakes 5 and 6 had the highest glacier coverage in their catchments (61.0 and 43.9%, respectively) (Table 1). Lake water samples were collected from 0.5 m depth with a 3 L sampler and immediately concentrated on a single 0.2 μm pore size polycarbonate filter (type GTTP; diameter, 47 mm; Millipore, Eschborn, Germany). In all cases, filtration was done under low vacuum pressure until clogging occurred. The filters were then placed inside a liquid nitrogen dewar for transport to the laboratory at Innsbruck, Austria. Previous tests done in other alpine lakes have shown that within-lake variability in bacterial community composition assessed by DGGE, from samples collected at the same depth at three different stations, is ≤5% (R. Sommaruga & E.O. Casamayor, unpubl. data).
Table 1.
Location, morphometry and catchment characteristics of the lakes (from Tartari et al., 1998). Data on lake connectivity, pH, total phosphorus (TP), total nitrogen (TN), dissolved organic carbon (DOC), bacterial abundance and number of DGGE bands are from the present study
| Lake | Lat. (27°N) |
Long. (86°E) |
Altitude (m a.s.l.) |
Area (km2) |
Max. depth (m) |
Catchment: lake area |
Glacier cover (% catch. area) |
Connect. | pH | TP (μm) |
TN (μm) |
DOC (μm) |
Bact. abundance (cells × 106 mL−1) |
No. of bands |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 56′34″ | 47′40″ | 5160 | 0.52 | 4.8 | 63 | 38.7 | 0.63 | 6.7 | 0.02 | 2.9 | 74.2 | 0.24 | 14 |
| 13 | 56′45″ | 48′26″ | 4960 | 10.70 | 0.6 | 106 | 0.40 | 0.44 | 7.0 | 0.03 | 5.7 | 45.8 | 0.59 | 14 |
| 10 | 57′45″ | 48′56″ | 5067 | 16.70 | 14.8 | 70 | 11.5 | 0.27 | 7.1 | 0.05 | 6.4 | 25.0 | 0.92 | 16 |
| 9 | 57′54″ | 48′40″ | 5213 | 0.57 | 8.2 | 140 | 16.8 | 0.25 | 6.9 | 0.07 | 7.1 | 12.5 | 0.58 | 12 |
| 6 | 59′45″ | 49′35″ | 5440 | 3.40 | 4.0* | 87 | 43.9 | 0.33 | 6.9 | 0.13 | 5.0 | 30.0 | 1.15 | 18 |
| 5 | 59′45″ | 49′24″ | 5400 | 11.50 | 4.0* | 30 | 61.0 | 0.33 | 7.2 | 0.13 | 8.6 | 25.8 | 0.50 | 23 |
Estimated from an empirical relationship between lake area and depth for alpine lakes.
DNA extraction and PCR amplification
Polycarbonate filters were first incubated with lysozyme, proteinase K and SDS in lysis buffer (40 mm EDTA; 50 mm Tris pH = 8.3; 0.75 m sucrose) and then the DNA was extracted with phenolchloroform-isoamyl-alcohol (25 : 24 : 1, v/v/v) and chloroform : isoamyl-alcohol (24 : 1, v/v), followed by ethanol precipitation.
PCR-amplified fragments of the bacterial 16S rRNA gene suitable for subsequent genetic fingerprinting analysis were obtained with the universal bacterial primer combination BAC358FGC (5′-CCT ACG GGA GGC AGC AG-3′) with 40 nucleotide GC-rich sequence attached to the 5′ end (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG G-3′) and 907RM (5′-CCG TCA ATT CMT TTG AGT TT-3′) which amplifies a fragment approximately 585 bp long (Casamayor et al., 2000). This fragment represents the most variable region in the 16S rRNA gene (i.e. the V3 region). The amount of template DNA and the number of PCR cycles were kept at optimal conditions to minimise biases associated with PCR amplification (Casamayor, Muyzer & Pedrós-Alió, 2001). All samples were run in the same PCR run and analysed in the same DGGE gel.
Denaturing gradient gel electrophoresis
This fingerprinting technique was performed with a DGGE D-Code system (Bio-Rad, Vienna, Austria) running for 3.5 h at a constant voltage of 200 V and 60 °C in a 30–60% vertical denaturating gradient (100% denaturant agent is 7 m urea and 40% deionised formamide). After electrophoresis, the gels were stained for 30 min with SYBRGold and visualised with UV. Digitised images were obtained with a UVIDoc system (UVItec, Cambridge, UK) and saved as computer files. Files were processed with the NIH Image software (National Institutes of Health, Bethesda, MD, U.S.A.) and the relative band intensities measured. A band was defined as a stain signal with an intensity greater than 0.2% of the total intensity for each lane and was defined as an operational taxonomic unit (OTU).
rRNA sequencing and phylogenetic trees
We excised all visible bands in the DGGE gels, except that very week bands did not produce visible PCR product after reamplification and could not be sequenced. The excised bands were resuspended in 25 μL of Milli-Q water and stored overnight at 4 °C. An aliquot (2–5 μL) of supernatant was used for PCR reamplification with the original primer set, and the PCR product was purified and sequenced using external sequencing facilities (Macrogen, Seoul, South Korea). The sequence similarity search tools BLAST (Altschul et al., 1990) were used to obtain a first indication of what sequences were retrieved and the closest relative in the database. For phylogenetic analysis, sequences were aligned using the ‘ARB’ program package (http://www.arbhome.de). Partial sequences were inserted into the optimised and validated tree available in ARB (derived from complete sequence data), by using the maximum-parsimony criterion and a special ARB parsimony tool that did not affect the initial tree topology. We deposited 38 sequences for the recovered 16S rRNA genes under the accession numbers AM941673 to AM941710.
Physical and chemical parameters
Water samples were analysed for basic parameters such as pH, conductivity and inorganic nutrients. These data were provided by G. Tartari (Institute for Ecosystem Studies, CNR, Pallanza, Italy). Subsamples for DOC analysis were filtered immediately after sampling through a precombusted (4 h at 450 °C) GF/F filter (Whatman, Dassel, Germany) placed on a stainless steel syringe holder. The filtrate was collected in 100 mL pre-combusted glass bottles (Schott, Mainz, Germany), acidified with HCl to pH 2, and stored in the dark until further analysis within 1 month. DOC was measured by high temperature catalytic oxidation with a Shimadzu TOC analyzer Model 5000 (Shimadzu, Korneuburg, Austria). The instrument was equipped with a Shimadzu platinised-quartz catalyst for high sensitivity analysis. Three to five injections were analysed for each sample and blank (Milli-Q water). A consensus reference material (CRM) for DOC (batch 6 FS provided by RSMAS/-MAC, University of Miami) was run in parallel. Results differed from the given CRM range by 5%.
Bacterial abundance
Bacterial numbers were assessed by flow cytometry in samples preserved with formaldehyde (2% final concentration) and stored at low temperature until their analysis within 3 weeks. Subsamples of 450 μL were stained by adding 25 μL of a 50 μmol L−1 SYTO 13 solution (Molecular Probes, Paisley, UK). Fluorescent microspheres (1 mm TransFluoSpheres 488/560, Molecular Probes) were added to a final concentration of 4.7 × 105 mL−1 as a counting and internal fluorescence standard. The absolute concentration of the stock solution of the microspheres was assessed by flow cytometry combined with gravimetric volume measurement. Counts were made with a MoFlo (DakoCytomation, Glostrup, Denmark) equipped with a water-cooled argon laser tuned at 488 nm (200 mW). Bacteria were detected by their signatures in a plot of orthogonal side scatter versus green fluorescence.
Data analysis
Bands occupying the same position in the different lanes were identified, and a qualitative matrix (OTU presence/absence) was constructed. The binary data set was used to calculate a dissimilarity matrix with Jaccard’s coefficient. Lake remoteness (a measure of grouping or dispersal of a lake in geographical space) was determined using an index of habitat connectivity, modified for lakes (Reche et al., 2005):
where Γi is the connectivity of lake i with respect to 12 other nearby lakes (j ± i), dij is the Euclidean distance (in kilometers) between lake i and the remaining lakes (including those that were not sampled in the present study), and Aj is the area (in square kilometers) of each lake different from lake i (j ± i). Values for lake location and area were obtained from the lake cadastre for the Khumbu Himal region (Tartari et al., 1998). A high value of Γi indicates that lake i is surrounded by many and/or relatively large lakes, so that the likelihood of being colonised is high.
The similarity in bacterial community composition among lakes was first assessed by cluster analysis using the Jaccard index for the OTUs obtained and the Unweighted Pair Group Method with Arithmetic Mean. To determine the factors controlling bacterial composition in the lakes, standard and partial Mantel tests were performed using PASSAGE I software (Rosenberg, 2001). The standard Mantel test is used to compare two independent dissimilarity matrices describing the same set of entities and to test whether the association is stronger than expected by chance (Sokal & Rolf, 1995). The partial Mantel test is used to determine the relationship between two matrices while holding another one constant. Dissimilarity matrices were calculated using the coefficient of Euclidean distances for quantitative data. The controlling factors tested were: lake spatial distribution, physical constraints and nutrient resources. The spatial distribution matrix was obtained from the distances between pairs of lakes. The physical-constraints matrix included: the ratio of catchment to lake area, lake maximum depth, and percentage of catchment area covered by glaciers, whereas the resources matrix included: DOC, TP and TN as surrogates for bacterial resource richness.
Results
Field conditions
The lakes had characteristics typical of other high mountain lakes, including very low concentrations of total phosphorus (range: 0.02–0.13 μm), total nitrogen (2.9–8.6 μm) and DOC (12.5–74.2 μm, Table 1). The highest total phosphorus concentrations were found in the lakes most strongly influenced by glaciers (Lake 5 and Lake 6). Bacterial abundance (Table 1) ranged from 0.24 × 106 cells mL−1 (Lake 15) to 1.15 × 106 cells mL−1 (Lake 6).
Bacterial fingerprints
A total of 51 OTUs was found with the highest (23) and lowest (12) number per lake obtained in Lake 5 and Lake 9, respectively (Table 1). A significant negative correlation was found between the number of OTUs per lake and the ratio of catchment to lake area (r2 = 0.682, P < 0.05) and a positive correlation between the number of OTUs and the percentage of lake catchment covered by glaciers (r2 = 0.578, P < 0.05). No significant correlations were found with lake area, lake remoteness (connectivity) or other parameters tested.
Bacterial composition
We used band intensity in DGGE as an index of the relative contribution of each band to the original PCR mixture of products for a sample. Only bacterial populations with abundances higher than 0.1–1% of the total bacteria were targeted (Casamayor et al., 2000, 2002), so our data represents relative contributions. The main bacterial phylogenetic groups found were Alphaproteobacteria, Betaproteobacteria, Actinobacteria, Bacteroidetes and Cyanobacteria. Consistent differences were found among lakes. For instance in Lake 5 Cyanobacteria and Actinobacteria were relatively abundant, whereas in Lake 13 and Lake 15 Bacteroidetes and Betaproteobacteria dominated.
Bacterial 16S rRNA phylogenetic affiliation
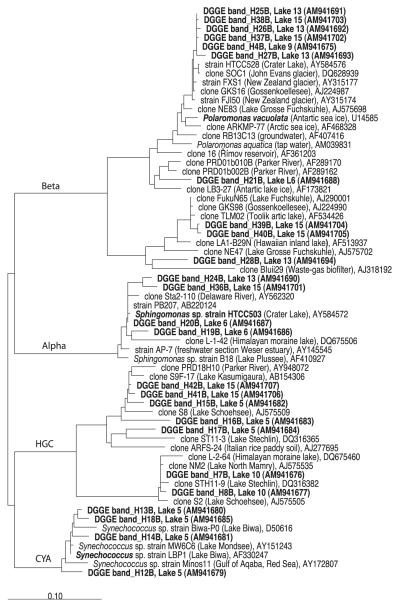
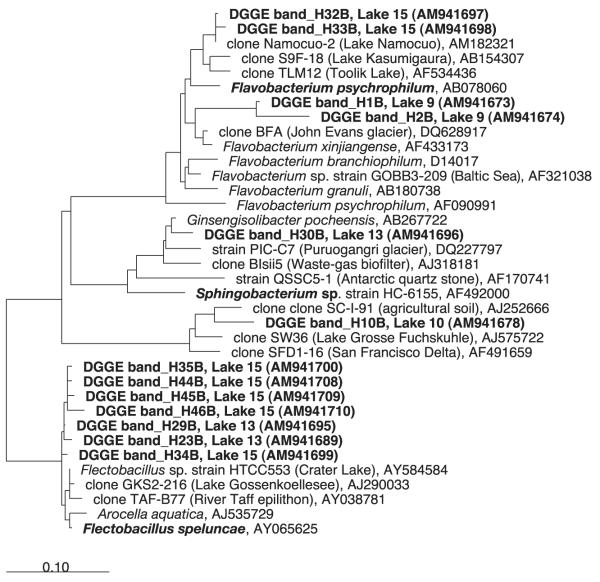
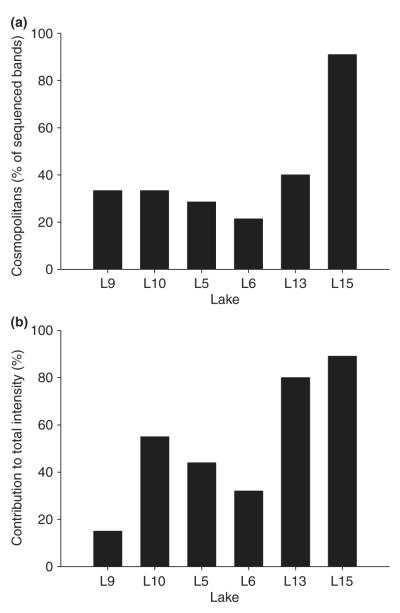
The similarities of the 16S rRNA gene sequences found with sequences available in GenBank was used as a degree of novelty. We observed a gradient of similarity for the different phylogenetic groups (Figs 1 & 2). Sequences from Betaproteobacteria affiliated with Polaromonas sp. (98% similarity) and Alcaligenes sp. (95% similarity) were found to be extremely closely related or identical (>99–100%) to previously described sequences in other high mountain and polar lakes worldwide. Next, within Alphaproteobacteria the phylotype H20B from Lake 5 was identical to Sphingomonas sp. HTCC503 recorded from Crater Lake, an alpine lake in Oregon, U.S.A. Flavobacteria (Bacteroidetes) showed 96% similarity with the closest relative in the GenBank database, but phylotype H23B from Lake 13 was 99.6% identical to Flectobacillus sp. HTCC553, also recorded from Crater Lake. Synechococcus was also very similar to other phylotypes found in Austrian alpine lakes (99% similarity). Finally, Actinobacteria were closely related (98%) to other 16S rRNA sequences recovered from pre-alpine lakes and also from rivers and estuaries. The percentage of phylotypes with ≥98% similarity in the 16S rRNA gene sequence as a consensus value ranged from 21% of sequenced bands in Lake 6 to 91% in Lake 15 (Fig. 3a). These phylotypes contributed from 15% (Lake 9) to 89% (Lake 15) of the total lane intensity (Fig. 3b).
Fig. 1.
Maximum-parsimony bacterial tree of the 16S rRNA gene sequences recovered corresponding to the phyla Beta- and Alphaproteobacteria, Actinobacteria (HGC) and Cyanobacteria (CYA). GenBank accession numbers are shown for the reference sequences. The scale bar represents 10% estimated divergence.
Fig. 2.
Maximum-parsimony bacterial tree of the 16S rRNA gene sequences recovered corresponding to the phylum Bacteroidetes. GenBank accession numbers are shown for the reference sequences. The scale bar represents 10% estimated divergence.
Fig. 3.
Relative contribution (in percentage) of ‘cosmopolitan’ phylotypes to the total number of sequenced bands (a) and to the total intensity of PCR products (b).
Similarity in bacterial community composition among lakes
The results of the cluster analysis using the complete OTU dataset (Fig. 4) suggested a biogeographical effect on bacterial community composition. For example, Lake 5 and Lake 6, which are lakes located close together, had the highest similarity, followed by Lake 9 and Lake 10, which were located one above the other although without apparent connection (Fig. 4). Indeed, the Mantel test indicated that the spatial distribution of the lakes had a significant effect on bacterial compositional similarity (Table 2). The physical constraints matrix but not the nutrient resource matrix had an effect on bacterial compositional similarity as confirmed by the results of the partial Mantel test (Table 2).
Fig. 4.
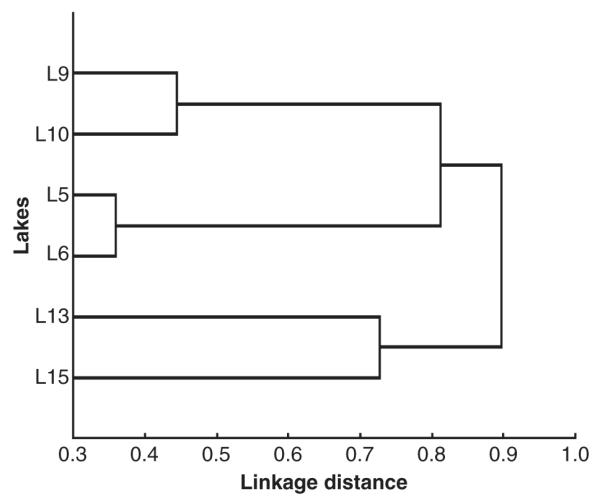
Results of the cluster analysis based on the unweighted pair group method with arithmetic mean (UPGMA) for the 51 operational taxonomic units. The similarity matrix was constructed using the Jaccard index.
Table 2.
Results of Mantel test and partial Mantel test for the comparison of the bacterial composition (OTUs) matrix with spatial distribution, the physical constraints matrix and the nutrient resources matrix
| Mantel |
Partial Mantel |
|||||
|---|---|---|---|---|---|---|
| Matrix type | Z | r | P | Z | r | P |
| Spatial distribution |
26644 | 0.555 | 0.032* | - | - | - |
| Physical constraints |
732 | 0.385 | 0.051 | 34 | 0.535 | 0.036* |
| Resources | 53 | 0.235 | 0.192 | −0.15 | −0.026 | 0.941 |
P < 0.05.
Discussion
Our results showed that the similarity in bacterial community composition among these six remote high altitude lakes was related to distance, but that local environmental factors were significant in explaining this apparent biogeographic pattern. Whether microorganisms do have biogeography is at present a highly debated topic (Hughes Martiny et al., 2006; Ramette & Tiedje, 2007). Several studies at different spatial scales have found non-random distributions of microorganisms, including examples from freshwater ecosystems although data for lakes are scarce (Hughes Martiny et al., 2006). Even less information is available about how spatial and local environmental factors affect bacterial community composition among lakes within a region or across larger space scales. However, teasing apart both types of factors in biogeographic analysis is essential because the spatial dimension may have a spurious correlation with crucial local environmental factors that affect bacterial community composition. Recently, a metacommunity study based on DGGE fingerprinting of 16S rDNA that included 98 shallow meso- to eutrophic lakes in Europe suggested that bacterial dispersal is not limited even at large spatial scales (>2500 km) and that mass effects do not affect bacterial communities even in physically connected lakes (Van der Gucht et al., 2007). In that study, though, a significant percentage of variability in bacterial community composition was not explained by the factors selected, local environmental constraints were identified as being more important than spatial distance, even if the analysis included very different geographic scales (Van der Gucht et al., 2007). Reche et al. (2005) performed the only comparable study to ours, focused on high mountain lakes, but from the Sierra Nevada in Spain. In their study, similarity in bacterial community composition among 11 lakes based on DGGE results was significantly influenced by geographical proximity. Our results have also shown that at the small geographical scale studied (ca. 6 km) there was a significant effect of the geographical proximity on bacterial community similarity. Thus, it was perhaps not surprising to find that lakes influenced by the same glacier, such as Lakes 5 and 6, had the highest similarity in bacterial community composition, even if they are isolated from each other at present. However, in contrast to the results obtained by Reche et al. (2005), we also found a significant effect of local physical constraints on bacterial community composition.
Whether the signal of local constraints on community composition can be detected may depend on the heterogeneity of ecosystems selected for analysis and the magnitude of change in environmental factors included in the analysis. In other words, by pooling ecosystems of very similar characteristics in an analysis, there is a risk in asserting only the significance of the spatial signal (Dolan, 2006), particularly at a small spatial scale. In our study, the lakes had a clear gradient in physical variables, such as lake turbidity, which was related to the extent of glaciers in the catchment, as well as in lake maximum depth, and ratio of catchment to lake area. It is difficult, however, to pinpoint how these variables directly or indirectly influence in a separate or combined way the bacterial community composition in these lakes. For example, there is evidence that in lakes with water retention times of up to 200 days, surface inflow controls bacterial community composition (Lindström et al., 2006). In our study, there was a significant negative correlation between OTU richness and the ratio of catchment to lake area (i.e. a proxy for flushing rate in seepage lakes), suggesting that bacterial community composition could be influenced too. This is feasible because many bacterial populations in surface waters of lakes with low retention times would potentially remain under the limit of detection of DGGE as a consequence of high loss rates (wash-out effect), which in turn would result in a dissimilar bacterial community composition. In fact, the period for sampling these high-altitude lakes (October) was selected among other logistic reasons to include the later period of the ice-free season and to avoid the rapid flush occurring after ice and snow melting. Nevertheless, we have no information on seasonal changes in bacterial community composition for those remote lakes to exclude the possibility of differential dynamics.
Another factor that potentially influences bacterial community composition in surface waters of high altitude mountain lakes is the strong incident solar UV radiation and high water transparency for these wavelengths (Sommaruga, 2001). Alpine lakes, especially those located at high altitudes such as those in the Himalayas, receive high instantaneous incident solar UV fluxes due to the natural increase of UV irradiance levels with elevation. Moreover, with the exception of those in glaciated catchments, alpine lakes are among the most UV transparent aquatic ecosystems (Sommaruga & Augustin, 2006). Solar UV radiation is known to cause several different types of negative effects on phototrophic and heterotrophic aquatic organisms including photosynthesis inhibition, DNA damage and reduction in cell growth (Sommaruga, 2003). Although organisms in alpine lakes have various strategies to minimise damage caused by UV radiation (Sommaruga, 2001), bacteria are relatively sensitive to this damaging radiation particularly in surface waters (Sommaruga et al., 1997). Thus, differences in bacterial community composition might be expected between transparent and turbid lakes resulting from differential tolerances to this environmental factor (Sommaruga et al., 2005; Alonso-Sáez et al., 2006). In our lake data set, the percentage of the catchment covered by glaciers varied from 0.4 to 61%, corresponding to a large range in lake turbidity. Interestingly, Lakes 5 and 6 were the systems with the highest minerogenic turbidity and at the same time not only had the highest OTU richness among the lakes studied (Table 1) but also unique phylotypes corresponding to Cyanobacteria (Synechoccocus-like, Fig. 1). In Lake 5, for example, these phylotypes accounted for 46% of the total PCR products detected in the DGGE, suggesting they were numerically important. Furthermore, in Lake 6 we recorded the highest bacterial density (Table 1), which further suggests that conditions for bacteria were more favourable in glacier lakes than in transparent ones. Glacier lakes also had the highest concentration of total phosphorus, an element limiting production in alpine lakes.
Interestingly, Bertoni, Callieri & Contesini (1998), using epifluorescence microscopy, reported that Synechoccocus-like cells were present in densities greater than 500 cells mL−1 in surface waters of the transparent Lakes 9 and 10 in September–October. Thus, it is apparent that picoplanktonic cyanobacteria in transparent lakes were below the detection limit of DGGE. Though marine counterparts of Synechococcus are relatively tolerant to high UV radiation (Sommaruga et al., 2005), the extreme PAR and UV radiation at the surface of transparent lakes at this altitude may suppress their growth. The contrasting results between DGGE and direct counts for cyanobacteria not only illustrate the threshold limit of this fingerprinting method but also suggest the important influence of crucial environmental factors such as UV radiation on bacterial community composition, as detected by DGGE. In fact, by highlighting differences in these environmental local factors, we do not imply that other variables known to affect bacteria, such as grazing by bacterivores or mortality caused by viruses, are unimportant, but we did not have data to test for their roles.
An interesting result from our study was the apparent occurrence of ubiquitous phylotypes, which judging from their relative contribution to the PCR products detected by DGGE, were also numerically important. Previous studies have shown the existence of freshwater bacterial clusters among the Alpha- and Betaproteobacteria that seem to be globally distributed, based on the criterion of 97–100% sequence similarity (Zwart et al., 1998). Certainly, the definition of ‘cosmopolitan’ bacteria as phylotypes having ≥98% partial sequence similarity in the 16S RNA gene is debatable. Even identical 16S rRNA gene sequences may not necessarily correspond to identical genotypes and even less so to ecotypes (Jaspers & Overmann, 2004; Hahn & Pöckl, 2005). However, in our study, partial sequences of several phylotypes (Figs 1 & 2) were identical to others exclusively found until now in alpine lakes or glaciers distributed around the world. Although we cannot exclude the possibility that these phylotypes are also present in other types of freshwater systems (perhaps in low abundance), a substantial number of sequences deposited in GenBank suggest that these phylotypes may be characteristic of alpine lakes. Thus, several sequences of Betaproteobacteria retrieved from Lakes 9, 13 and 15 were, for example, identical in their partial sequence to the bacterium HTCC528 (sequence AY584576) described from the high mountain Crater Lake in Oregon, U.S.A. (Page, Connon & Giovannoni, 2004), or to the culturable bacterium FXS1 (sequence AY315177) isolated from a glacier in the Southern Hemisphere (Foght et al., 2004). Those ecosystems can be generally characterised as cold and oligotrophic and, in the case of lakes, affected by strong UV radiation. One may ask whether this is just a coincidence resulting from a biased comparison of genetical identity or the result of bacterial dispersal and successful colonisation of a similar suitable environment in different parts of the world. We cannot give a definitive answer, but there are several potential mechanisms of bacterial dispersal that may explain a global or cosmopolitan distribution of certain bacterial taxa. One mechanism that seems particularly relevant to lakes located at high altitude is the inclusion of microorganisms into aerosols that can be transported for long distances through the atmosphere (Bovallius, Roffey & Henningson, 1980). There is evidence that bacterial endospores and fungal spores can be transported even over an inter-hemispheric scale (Griffin et al., 2002; Prospero et al., 2005) and recent estimates suggest that globally up to 1018 bacterial cells per year are transported by aerosols (Griffin et al., 2002). In particular, annual emissions of dust to the global atmosphere are high and range from 500 to more than 5000 × 106 t (Goudie & Middleton, 2006). Moreover, several recent studies also report on the importance of soils and the sea surface microlayer as sources of microorganisms entering the atmosphere (Aller et al., 2005; Schlesinger, Mamane & Grishkan, 2006). For example, a recent study showed that culturable bacteria deposited over a Himalayan glacier at 6518 m a.s.l., relatively close to our study area, were associated with the pre-monsoon and monsoon long-range transportation of continental dust and marine aerosols (Zhang et al., 2007).
Bacteria known to produce endospores that are resistant to desiccation and can survive extreme conditions during atmospheric transport are typical members of low GC gram-positive Firmicutes (Reisenman & Nicholson, 2000). Members of this bacterial division are usually not found in the bacterioplankton of freshwater and marine ecosystems. Thus, the question remains of how other bacteria manage to survive dispersal through the atmosphere. Aeolian dust aerosols may be an important vehicle for dispersal of bacteria unable to produce resistant spores as these aerosols have characteristic particle sizes in the μm range (see Goudie & Middleton, 2006). Furthermore, these particles could provide some direct protection against high UV radiation exposure during transport in the troposphere. In addition, typical dust aerosols include clay minerals containing quantitatively important amounts of water and organic matter (Goudie & Middleton, 2006), which may provide conditions to maintain basal metabolism of bacteria until they eventually find a suitable habitat to colonise. High altitude lakes are probably very efficient ‘collectors’ of airborne bacteria transported in the atmosphere because orographic uplifting of air masses causes precipitation to increase with elevation (Lovett & Kinsman, 1990). Other mechanisms of dispersal are possible but, for example, zoochory by birds is probably less important at high elevations.
In conclusion, our results from remote high altitude freshwater lakes provide evidence for the existence of several phylotypes that are found everywhere on earth, but also for the selective effect or ‘species’ sorting of the environment. A lot remains to be done to investigate more exhaustively whether the putatively cosmopolitan bacteria do play the same function in different alpine lakes.
Acknowledgments
We thank M. T. Pérez for bacterial counts, G. Tartari for providing data on water chemistry, and A. Lami and S. Musazzi for helping during field work. A. Alfreider and R. Psenner provided useful comments to an earlier version. The logistics for the field work and work at the Pyramid Research Laboratory was supported by a project granted to RS by the Committee on High Altitude Scientific and Technological Research (Ev-K2-CNR) in collaboration with the Nepal Academy of Science and Technology, and thanks to contributions from the Italian National Research Council and the Italian Ministry of Foreign Affairs. Funds for the analyses were provided by the Austrian Science Fund (Project Nr. 19245-BO3 to RS), the University of Innsbruck through the Daniel Swarovski Fund (to RS), the Austrian Academic Exchange Service with Spain (ÖAD project, Nr. 16-2006 and MEC project HU2005-0032 granted to both authors), the Spanish project granted to EOC CGL2006-12058-CO2-02/BOS, and ECOSENSOR BIOCON04/009 from the BBVA Foundation.
References
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- Aller JY, Kuznetsova MR, Jahns CJ, Kemp PF. The sea surface microlayer as a source of viral and bacterial enrichment in marine aerosols. Journal of Aerosol Science. 2005;36:801–812. [Google Scholar]
- Alonso-Sáez L, Gasol JM, Lefort T, Hofer J, Sommaruga R. Effect of natural sunlight on bacterial activity and differential sensitivity of natural bacterioplankton groups in NW Mediterranean coastal waters. Applied and Environmental Microbiology. 2006;72:5806–5813. doi: 10.1128/AEM.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bertoni R, Callieri C, Contesini M. Organic carbon and microorganisms in two Nepalese lakes. Memorie dell’ Istituto Italiano di Idrobiologia. 1998;7:99–106. [Google Scholar]
- Bovallius A, Roffey R, Henningson E. Long range air transmission of bacteria. Annals of the New York Academy of Sciences. 1980;353:186–200. doi: 10.1111/j.1749-6632.1980.tb18922.x. [DOI] [PubMed] [Google Scholar]
- Casamayor EO, Schäfer H, Bañeras L, Pedrós-Alió C, Muyzer G. Identification of and spatiotemporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Applied and Environmental Microbiology. 2000;66:499–508. doi: 10.1128/aem.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor EO, Muyzer G, Pedrós-Alió C. Composition and temporal dynamics of planktonic archaeal assemblages from anaerobic sulfurous environments studied by 16S rDNA denaturing gradient gel electrophoresis and sequencing. Aquatic Microbial Ecology. 2001;25:237–246. [Google Scholar]
- Casamayor EO, Pedrós-Alió C, Muyzer G, Amann R. Microheterogeneity in 16S rDNA-defined bacterial populations from a stratified planktonic environment is related to temporal succession and to ecological adaptations. Applied and Environmental Microbiology. 2002;68:1706–1714. doi: 10.1128/AEM.68.4.1706-1714.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. What are bacterial species? Annual Review of Microbiology. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- Crump BC, Adams HE, Hobbie JE, Kling GW. Biogeography of bacterioplankton in lakes and streams of an Arctic tundra catchment. Ecology. 2007;88:1365–1378. doi: 10.1890/06-0387. [DOI] [PubMed] [Google Scholar]
- Dolan J. Microbial biogeography? Journal of Biogeography. 2006;33:199–200. [Google Scholar]
- Findlay RH, Yeates C, Hullar MAJ, Stahl DA, Kaplan LA. Biome-level biogeography of streambed microbiota. Applied and Environmental Microbiology. 2008;74:3014–3021. doi: 10.1128/AEM.01809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foght J, Aislabie J, Turner S, Brown CE, Ryburn J, Saul DJ, Lawson W. Culturable bacteria in subglacial sediments and ice from two Southern Hemisphere glaciers. Microbial Ecology. 2004;47:329–340. doi: 10.1007/s00248-003-1036-5. [DOI] [PubMed] [Google Scholar]
- Goudie AS, Middleton NJ. Desert Dust in the Global System. Springer; Berlin: 2006. p. 287. [Google Scholar]
- Griffin DW, Kellogg CA, Garrison VH, Shinn EA. The global transport of dust. American Scientist. 2002;90:230–237. [Google Scholar]
- Hahn MW, Pöckl M. Ecotypes of planktonic Actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Applied and Environmental Microbiology. 2005;71:766–773. doi: 10.1128/AEM.71.2.766-773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Silver JM, Leibold MA, et al. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology. 2007;88:1345–1353. doi: 10.1890/06-0286. [DOI] [PubMed] [Google Scholar]
- Hughes Martiny JB, Bohannan BJ, Brown JH, et al. Microbial biogeography: putting microorganisms on the map. Nature Review Microbiology. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- Jaspers E, Overmann J. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Applied and Environmental Microbiology. 2004;70:4831–4839. doi: 10.1128/AEM.70.8.4831-4839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka A. Microbial ecology: searching for principles. Microbe. 2006;1:175–179. [Google Scholar]
- Lindström ES, Forslund M, Algesten G, Bergström A-K. External control of bacterial community structure in lakes. Limnology and Oceanography. 2006;51:339–342. [Google Scholar]
- Lovett GM, Kinsman JD. Atmospheric pollution deposition to high elevation ecosystems. Atmospheric Environment. 1990;24:2767–2786. [Google Scholar]
- Page KA, Connon SA, Giovannoni SJ. Representative Freshwater Bacterioplankton isolated from Crater Lake, Oregon. Applied and Environmental Microbiology. 2004;70:6542–6550. doi: 10.1128/AEM.70.11.6542-6550.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrós-Alió C. Marine microbial diversity: can it be determined? Trends in Microbiology. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pommier T, Canbäck B, Riemann L, Boström KH, Simu K, Lundberg P, Tunlid A, Hagström A. Global patterns of diversity and community structure in marine bacterioplankton. Molecular Ecology. 2007;16:867–880. doi: 10.1111/j.1365-294X.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- Prospero JM, Blades E, Mathison G, Naidu R. Interhemispheric transport of viable fungi and bacteria from Africa to the Caribbean with soil dust. Aerobiologia. 2005;21:1–19. [Google Scholar]
- Prosser JI, Bohannan BJ, Curtis TP, et al. The role of ecological theory in microbial ecology. Nature Review Microbiology. 2007;5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- Ramette A, Tiedje JM. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2761–2766. doi: 10.1073/pnas.0610671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche I, Pulido-Villena E, Morales-Baquero R, Casamayor EO. Does ecosystem size determine aquatic bacterial richness? Ecology. 2005;86:1715–1722. [Google Scholar]
- Reisenman PJ, Nicholson WL. Role of the spore coat layers in Bacillus subtilis spore resistance to hydrogen peroxide, artificial UV-C, UV-B, and solar UV radiation. Applied and Environmental Microbiology. 2000;66:620–626. doi: 10.1128/aem.66.2.620-626.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MS. PASSAGE. Pattern Analysis, Spatial Statistics, and Geographic Exegis. Department of Biology, Arizona State University; Tempe, AZ: 2001. [Google Scholar]
- Schlesinger P, Mamane Y, Grishkan I. Transport of microorganisms to Israel during Saharan dust events. Aerobiologia. 2006;22:259–273. [Google Scholar]
- Sokal RR, Rolf FJ. Biometry. 3rd edn W. H. Freeman and Company; New York, NY: 1995. [Google Scholar]
- Sommaruga R. The role of UV radiation in the ecology of alpine lakes. Journal of Photochemistry and Photobiology B: Biology. 2001;62:35–42. doi: 10.1016/s1011-1344(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Sommaruga R. Ultraviolet radiation and its effects on species interactions. In: Helbling W, Zagarese H, editors. UV Effects in Aquatic Organisms and Ecosystems. ESP, Royal Society of Chemistry; London: 2003. pp. 485–508. (Comprehensive Series in Photosciences). [Google Scholar]
- Sommaruga R, Obernosterer I, Herndl GJ, Psenner R. Inhibitory effect of solar radiation on thymidine and leucine incorporation by freshwater and marine bacterioplankton. Applied and Environmental Microbiology. 1997;63:4178–4184. doi: 10.1128/aem.63.11.4178-4184.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommaruga R, Augustin G. Seasonality in UV transparency of an alpine lake is associated to changes in phytoplankton biomass. Aquatic Sciences. 2006;68:129–141. [Google Scholar]
- Sommaruga R, Hofer J, Alonso-Sáez L, Gasol JM. Differential sensitivity to sunlight of picophytoplankton from surface Mediterranean coastal waters. Applied and Environmental Microbiology. 2005;71:2154–2157. doi: 10.1128/AEM.71.4.2154-2157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartari GA, Panzani P, Adreani L, Ferrero A, De Vito C. Lake cadastre of Khumbu Himal Region: geographical-geological-limnological data base. Memorie dell’ Istituto Italiano di Idrobiologia. 1998;7:151–235. [Google Scholar]
- Van der Gucht K, Cottenie K, Muylaert K, et al. The power of species sorting: Local factors drive bacterial community composition over a wide range of spatial scales. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20404–20409. doi: 10.1073/pnas.0707200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannarell AC, Triplett EW. Within- and between-lake variability in the composition of bacterioplankton communities: Investigations using multiple spatial scales. Applied and Environmental Microbiology. 2004;70:214–223. doi: 10.1128/AEM.70.1.214-223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Hon S, Ma X, Qin M, Chen T. Culturable bacteria in Himalayan glacial ice in response to atmospheric circulation. Biogeosciences. 2007;4:1–9. [Google Scholar]
- Zwart G, Hiorns WD, Methé BA, van Agterveld MP, Huismans R, Nold SC, Zehr JP, Laanbroek HJ. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Systematic and Applied Microbiology. 1998;21:546–556. doi: 10.1016/S0723-2020(98)80067-2. [DOI] [PubMed] [Google Scholar]