Abstract
ERβ1 downregulation occurs in many breast cancers but the responsible molecular mechanisms remain unclear. Here we report that levels of ERβ1 expression are negatively regulated by the microRNA miR-92. Expression analysis in a cohort of primary breast tumours confirmed a significant negative correlation between miR-92 and both ERβ1 mRNA and protein. Inhibition of miR-92 in MCF-7 cells increased ERβ1 expression in a dose-dependent manner, whereas miR-92 overexpression led to ERβ1 downregulation. Reporter constructs containing candidate miR-92 binding sites in the 3′-UTR of ERβ1 suggested by bioinformatics analysis confirmed that miR-92 downregulated ERβ1 via direct targeting of its 3′-UTR. Our results define a potentially important mechanism for downregulation of ERβ1 expression in breast cancer.
Keywords: Breast cancer, ERβ1, 3′UTRs, miR-92
Introduction
Biological effects of 17β-estradiol are principally mediated by estrogen receptors (ER) ERα and ERβ (1). While the role of ERα in breast carcinogenesis has received much attention, our insight into ERβ function remains poor. Of the 5 known ERβ isoforms, ERβ1 is the most widely studied and consequently the best understood (2). ERβ1 is often down-regulated in cancer compared with normal cells (3), suggesting that it may function as a tumour suppressor (4,5). In support of this many studies have demonstrated ERβ1 to have anti-proliferative and pro-apoptotic properties (reviewed in 1). Mechanisms contributing to reduced ERβ1 expression in breast tumours are beginning to be elucidated and include hypermethylation of the ERβ gene (5,6) and post-transcriptional regulation via its 5′-untranslated regions (UTRs) (7).
MicroRNAs (miRs) are a class of short non-coding RNAs that regulate expression of up to one third of human genes (8). Their expression is commonly dysregulated in cancers, including those of the breast (9,10). MiRs act on target mRNAs by binding to miR recognition elements, typically within 3′-UTRs, leading to translational inhibition and/or induction of mRNA cleavage, thereby down-regulating expression of protein products (11). MiRs can function as oncogenes or tumour suppressor genes depending on their gene targets. Examples in breast cancer include the tumour suppressor function of miR-206, which targets ERα (9,10,12,13), and the oncogenic function of miR-21, which is abundant in breast tumour compared to normal breast (14). More recently, specific miR profiles have also been associated with different classes of breast cancer (15).
MiRs of the miR-17-92 cluster, also described as Oncomir-1, are known to act as oncogenes (16). There are 6 members of this cluster (miR-17, miR-18a, miR-19a, miR20a, miR-19b-1 and miR-92) and their expression has multifunctional effects, including enhanced cell proliferation and suppression of apoptosis (16). The individual function of members of this cluster are now beginning to be elucidated (17-19).The aim of this study was to establish whether one of the miRs of this cluster, miR-92, plays a role in determining ERβ1 expression in breast cancers. We examined whether ERβ1 is a functional target for miR-92 and whether there is a correlation between miR-92 and ERβ1 expression in clinical samples. Our findings are the first to demonstrate miR regulation of ERβ1 expression in cancer.
Experimental procedures
Cell lines
MCF-7, MDA-MB-453, and BT-20 breast cancer cell lines were maintained in RPMI 1640, supplemented with 5% heat-inactivated FBS (both Invitrogen, UK) and MCF10A in DMEM/F12 with 15 mM HEPES buffer, 5% horse serum, 10 μg/ml insulin, 20 ng/ml EGF, 100 ng/ml cholera toxin, 0.5 μg/ml hydrocortisone, in a 5% CO2 humidified incubator at 37°C. Bi-monthly mycoplasma checks (MycoAlert® Mycoplasma detection assay, Lonza, USA) were consistently negative and STR profiles confirmed cell identity.
Patient specimens and immunohistochemical analysis of ERβ1
Following ethical approval (06/Q1206/180), fresh frozen breast tissue samples were obtained from our Breast Tissue Bank. Formalin fixed paraffin-embedded (FFPE) tissues, matching the frozen cases, were retrieved for immunohistochemical analysis. Histological composition of frozen and FFPE tissue sections was confirmed by inspecting H&E stained images. All samples were invasive breast carcinoma. Clinicopathological data are presented in Supplementary Table 1. ERβ1 immunohistochemistry was performed as previously described (20).
RNA extraction and quantitative real-time PCR of mRNAs and miRNA
Total RNA was extracted from cell lines and tissues using the miRNeasy Mini Kit (Qiagen, Germany). For miRNA analysis, mature miRNA was reverse transcribed using a miRNA-specific stem-loop reverse transcriptase. Real-time PCR was performed using Taqman microRNA assays (PE Applied Biosystems, USA) and Sensi-Mix dT (Quantace Ltd, UK) according to the manufacturer’s instructions. RNU6B small nuclear RNA was used as an internal control to normalize all data using the Taqman RNU6B assay (PE Applied Biosystems). RNU6B was unaffected by hormone treatment, an important consideration in breast cancer studies (Supplementary Figure 1). For mRNA analysis, RNA was prepared and real-time PCR was performed in triplicates in three independent experiments using SYBR green and normalised to 36B4 as described previously (21).
Cell line transfection
MCF-7 cells were seeded in 24-well plates 24 h prior to transfection. Cells were transiently transfected with either anti-miR-92 (15, 30 and 45nM), pre-miR-92 (3nM) or negative controls (miR negative control #1 or anti-miR negative control #1), respectively (all ABI, UK) in OPTI-MEM medium using Lipofectamine 2000 (both Invitrogen) following the manufacturer’s protocol. After 48 h, cells were harvested and expression of miR-92, ERβ1 and MUC-16 were analysed by real-time PCR. Primer sequences are in Supplementary Table 2.
3′-RACE analysis of ERβ1 3′-UTR sequences in MCF-7 cells
3′RACE reactions were performed using the 3′RACE System (Invitrogen) and primers listed in Supplementary Table 2. Products were analyzed on 2.5% agarose gels and visualised by UV illumination. Products were excised from gels and cloned into pGEM-T Easy (Promega, USA); up to five clones for each were sequenced.
Green Fluorescent Protein (GFP) vector construction
A fragment of the ERβ1 3′-UTR harbouring the predicted miR-92 binding sites was PCR amplified from MCF-7 cells using ERβ1 3′-UTR primers described in Supplementary Table 2. The amplified fragment was cloned into pTH-GFPa (7) at Hind III and Bam HI sites, thereby creating pGFP-β1-UTR, to allow over-expression of transcripts coding for GFP with ERβ1 3′-UTR sequence within their 3′UTRs.
Plasmid transfection and flow cytometry
Cells were transfected with equal copy numbers of plasmids (empty pcDNA3.1(−)/myc-His A, pTH-GFPa, pGFP-β1-UTR) using Lipofectamine 2000 following the manufacturer’s protocol. Cells were trypsinised and resuspended in fresh medium containing 1% serum. GFP expression was quantified (mean fluorescent intensity of 104 events after exclusion of debris/dead cells on the basis of forward activated light scatter vs. side scatter) at 525nm (LSRII, BD Biosciences, UK). Gates were set so that <1% of untransfected cells were defined as expressing GFP.
Treatment with ER ligands
To determine whether miR-92 expression was regulated by ER ligands, MCF-7, BT-20 and MDA-MB-453 cells were seeded in 6-well plates. The cells were washed with PBS and incubated under serum-free conditions for 48 h. Cells were then treated with 10 nM 17β-estradiol (E2) or 1nM Tamoxifen (TAM), and appropriate vehicle control in phenol red-free medium containing 5% charcoal-stripped FBS for 48 h. Real-time PCR was performed to estimate the effect of ER ligands on miR-92 expression.
Bioinformatic analysis
The miRGen database was used to identify potential miRNAs that may target ERβ mRNAs (http://www.diana.pcbi.upenn.edu/cgi-bin/miRGen/v3/Targets). Among the predicted miRNAs, we focused on miR-92, as it recognizes binding sites in ERβ1 transcripts. The ERβ1 3′-UTR sequence was recovered from GenBank (NM_001437) and RNA hybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html) was used to identify sites of miR-92 seed matches (binding sites) within this region.
Statistical analysis
Student’s t-test or Spearman correlation was used for statistical analyses using SPSS (SPSS, Inc., IL, USA). All tests were 2-sided. p ≤0.05 was considered significant.
Results
ERβ1 is a direct target for miR-92
Analysis of potential miR binding sites within the published 3′-UTR for ERβ1 (NM_001437) using miRGen database and RNA hybrid revealed one putative miR-92 target site (Supplementary Figure 2). The strength of the potential interaction between miR-92 and the ERβ1 target site can be estimated in terms of the minimum free energy for hybridization (ΔG); ΔG for the target site was − 20.7 kcal/mol.
MiR-92 expression is inversely correlated with ERβ1 expression in breast cancer cell lines and breast tissues
Having predicted that miR-92 may target the 3′-UTR of ERβ1 we profiled miR-92 and ERβ1 mRNA expression in 4 breast cancer cell lines. An inverse relationship was observed with cell lines which expressed high endogenous ERβ1 expressing low levels of miR-92 and vice versa (Figure 1a). We next investigated the relationship between ERβ1 and miR-92 mRNA expression in matched tumour and adjacent normal breast tissues. In all 8 pairs, we observed decreased ERβ1 mRNA expression in tumour tissue compared with matched normal tissue with miR-92 up-regulated in breast tumours (Figure 1b), and was particularly strong in cases 4 and 7. This inverse relationship was not seen between ERβ1 mRNA and another, unrelated, miR (miR-124a), suggesting that the relationship is specific (Figure 1c). In addition, using a separate cohort of 28 breast tumours we found a significant negative correlation between expression of miR-92 and ERβ1 mRNA (r = − 0.53, P = 0.001; Figure 2a). This was also seen at the protein level where immunohistochemical analysis of FFPE cases matched to frozen tumours showed a significant negative correlation between miR-92 and ERβ1 (r = −0.39, P = 0.04; Figure 2b). These points are further illustrated in Figure 2c where representative images of ERβ1 immunohistochemistry are shown alongside miR-92 expression levels. In contrast, no relationship was seen between ERα and miR-92 (r = 0.02, P = 0.9; Figure 2d). These results strongly implicate miR-92 in the negative regulation of ERβ1 expression at both mRNA and protein levels.
Figure 1. Ratios of expression of ERβ1/miR-92 in breast cell lines and clinical breast samples.
Quantitative RT-PCR analysis showed an inverse relationship between ERβ1 and miR-92in 4 breast cell lines of differing ER status (shown above each bar; a). A similar inverse correlation was observed in matched normal (N) and breast tumours (T), with high ratios in normal breast tissue and low ratios in breast tumours (b). In a subgroup of these samples this relationship was not observed with miR-124a (c), indicating specificity. Each experiment was performed in triplicate with 3 experimental replicates. Bars represent mean ± S.D.
Figure 2. miR-92 is negatively correlated with ERβ1 mRNA and protein in human breast cancer but not with ERα.
Scatterplots showing an inverse correlation between expression of miR-92 and ERβ1 mRNA (a) determined by real-time RT-PCR (P = 0.001), and protein (b) determined by immunohistochemistry and Allred scoring (P = 0.04). Examples of ERβ1 immunohistochemistry and their relationship with miR-92 expression are shown in (c). No relationship was seen with ERα (d).
Manipulation of miR-92 expression in vitro modulates ERβ1 expression and other known miR-92 targets genes
To determine the functional effect of miR-92 on ERβ1, we aimed to manipulate miR-92 expression in cultured cells and to examine the influence on expression of ERβ1. We used MCF-7 cells; as shown above they express easily detectable levels of both miR-92 and ERβ1 and are a well recognised breast cancer model. First, we confirmed that the potential miR-92 target sequences are present within the ERβ1 3′-UTR in these cells by sequencing the ERβ1 3′-UTR. The ERβ1 3′-UTR was amplified using Rapid Amplification of cDNA Ends reactions; a single 3′-UTR species, of 242 nucleotides containing the potential miR-92 target site was identified. Expression of endogenous miR-92 was reduced in a dose-dependent fashion by transfection with anti-miR-92 resulting in a corresponding significant up-regulation of endogenous ERβ1 expression (Figure 3a). In the reverse experiment, transfection of pre-miR-92 into MCF-7 cells resulted in over-expression of miR-92 with a concomitant significant down-regulation of ERβ1 mRNA expression (Figure 3b). To further validate the anti-miR-92 effect, another putative miR-92 target, MUC16, was measured following miR-92 knockdown. MiR-92 silencing significantly restored MUC16 expression (Figure 3c).
Figure 3. Effects of miR-92 manipulation on expression of ERβ1 and other target genes.
Using quantitative RT-PCR, suppression of miR-92 inhibits miR-92 gene expression in MCF-7 cells in a dose-dependent manner and after 48 hours resulted in upregulation of ERβ1 mRNA expression relative to negative controls (a) while overexpression of miR-92 led to downregulation of ERβ1 mRNA expression (b). MiR-92 silencing restored MUC16 expression (c). Each experiment was performed in triplicate with 3 experimental replicates. Each data point is the mean ± S.D. *P < 0.05, **P <0.001, ***P < 0.0001.
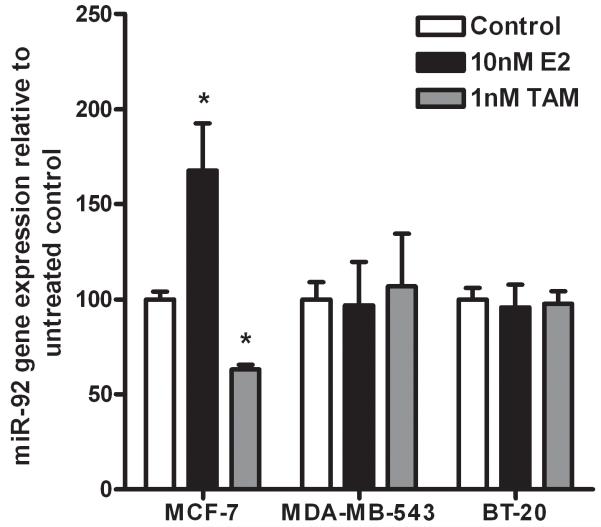
E2 and Tamoxifen regulate miR-92 expression in MCF-7 breast cancer cells
To determine whether miR-92 was hormonally regulated, MCF-7, BT-20 and MDA-MB-453 cells growing in estrogen-depleted conditions were treated with either 10nM E2 or 1nM Tamoxifen. E2 significantly induced an increase in miR-92 expression and TAM induced a reduction in miR-92 expression in ER-positive MCF-7 cells, while no effect was seen in the ER-negative BT-20 and MDA-MB453 cells (Figure 4). We did not observe parallel decreases in ERβ1 expression (data not shown).
Figure 4. Hormonal regulation of miR-92 expression.
Quantitative RT-PCR analysis showed inhibition of miR-92 expression by TAM and its upregulation by E2 in MCF-7 cells but not in BT-20 and MDA-MB-453. Values are fold expression compared to vehicle control (EtOH) for miR-92. Each experiment was performed in triplicate with 3 experimental replicates. Each data point is the mean ± S.D. *P < 0.05.
ERβ1 is targeted by miR-92 via its 3′-UTR
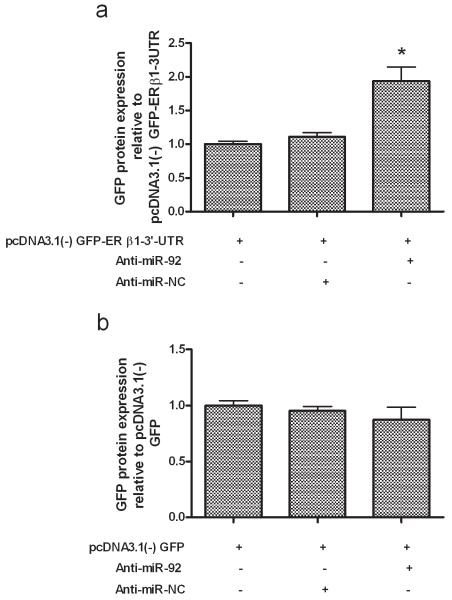
Next we examined whether miR-92 is capable of influencing ERβ1 expression via the ERβ1 3′-UTR. A fragment of the ERβ1 3′-UTR harbouring the miR-92 binding site was cloned downstream of the GFP reading frame in a mammalian expression vector, thereby creating a fluorescent reporter for the function of the 3′-UTR region (Supplementary Figure 3). MCF-7 cells were either transfected with this reporter, or transfected to over-express transcripts coding for GFP lacking a specialised 3′-UTR, along with either non-targeting control anti-miRNAs or with anti-miR-92. GFP protein expression was measured by flow-cytometry. In the presence of anti-miR-92 a significant increase in GFP protein expression from the reporter containing the ERβ1 3′-UTR (P<0.05; Figure 5a) was seen demonstrating that endogenous miR-92 can directly target the 3′-UTR of ERβ1 mRNA. This increase was not seen with the GFP reporter that lacked a specialised 3′-UTR (Figure 5b).
Figure 5. ERβ1 is targeted by miR-92 via its 3′UTR in MCF-7 cells.
MCF-7 cells were transiently transfected with plasmids to allow over-expression of GFP transcripts with either 3′-UTRs containing sequence from the ERβ1 3′-UTR including the potential miR-92 sites, or with unspecialised 3′-UTRs. Transfections also included either anti-miR-92 or a non-targeting control (NC). Reduction of endogenous miR-92 by anti-miR-92 led to an increase in GFP protein expression specified by the ERβ1 3′-UTR (a) and was not observed with the plasmid containing the unspecialised 3′-UTRs (b). Each experiment was performed in triplicate with 3 experimental replicates. Bars represent mean ± S.D. *P < 0.05.
Discussion
It is well recognised that ERβ1 is frequently downregulated in breast cancer compared to normal mammary gland where it is constitutively expressed (1,3,5). However, little is known about the mechanisms responsible for its reduction in some breast tumours. Here we present novel evidence that ERβ1 expression is deregulated in breast cancer cells by miR-92.
In silico analysis using miRGen and RNA hybrid revealed a putative miR-92 target site within the ERβ1 transcript. MiR-92 is a component of the miR-17–92 cluster containing 6 miRs; miR-17, miR-18a, miR-19a, miR20a, miR-19b-1 and miR-92, which appear to play a role in cell proliferation and have been shown to be potential oncogenes in several tumour types (16,22). MiR-92 was the only miR of this cluster predicated to target the 3′-UTR of ERβ1and functional in vitro data demonstrated for the first time the potential role of this miR in the regulation of ERβ1 expression in breast cancer cells.
Our results showed that miR-92 levels were upregulated in breast tumours as compared with matched normal tissues, something which has been suggested in earlier work using northern blotting, but not followed up in detail (23) We further demonstrated that miR-92 expression was negatively associated with ERβ1 but not with ERα in breast cancer tissues and also in cell lines. We also showed that reduction of endogenous miR-92 expression was associated with upregulation of ERβ1 expression. Moreover, treatment of MCF-7 cells with anti-miR-92 increased expression of MUC-16, a predicted miR-92 target by bioinformatics, providing further evidence for target specificity and adding to the growing body of data demonstrating the importance of members of the miR-17-92 cluster in cancer (17-19).
Upregulation of miR-92 expression was also associated with E2 sensitivity. This finding is in line with recent reports showing increased miR-92 expression during E2-induced rat mammary carcinogenesis (24) and in E2-treated MCF-7 cells (25). This is at odds with a previous in vitro study which found no change in miR-92 expression following E2 treatment of MCF-7 cells (26). However this group also failed to demonstrate E2-dependent regulation of miR-206 in these cells which has been consistently shown by several other independent groups (12,13,27). To add further complexity, a detailed miR analysis across a wide range of tissues and cell lines found no miR-206 expression in MCF-7 cells, although they did detect miR-92 (28). Inter-laboratory variation in MCF-7 cells is well recognised (29) and could explain these discrepancies.
We also observed that a reduction in endogenous miR-92 suppressed cell growth (Supplementary Figure 4); given the well documented anti-proliferative effects of ERβ1 (4,30-31) mechanistically, this may be associated with miR-92 regulation of ERβ1, although this clearly requires further experimental validation. Furthermore, we confirmed that the predicted miR-92 binding site within the ERβ1 3′-UTR represents a true target using reporter assays. Our data are in accordance with much published work demonstrating that this miR cluster has a critical regulatory role in the expression of many oncogenes and tumour suppressor genes thereby influencing cell proliferation and apoptosis (16,32).
Taken together, these results suggest that one likely oncogenic role for miR-92 in breast cancer is the inhibition of ERβ1 expression, although the likelihood of miR-92 influencing other genes cannot be excluded. Our data complements other recent observations showing the growing importance of miRs in defining breast carcinogenesis (10, 12-15, 33). These findings could provide the basis of potential therapeutic strategies for breast cancer aimed at reactivating expression of ERβ1 through manipulation of miR-92 expression.
Supplementary Material
Acknowledgments
Financial support: Government of Saudi Arabia (HAN), Breast Cancer Campaign (VS, TAH, AMH, LS), Cancer Research UK (VS, MC), US Department of Defence (SS, TAH).
References
- 1.Fox EM, Davis RJ, Shupnik MA. ERβ in breast cancer-onlooker, passive player, or active protector? Steroids. 2008;73:1039–51. doi: 10.1016/j.steroids.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung YK, Mak P, Hassan S, Ho SM. Estrogen receptor (ER)-β isoforms: a key to understanding ERβ signaling. Proc Natl Acad Sci U S A. 2006;103:13162–7. doi: 10.1073/pnas.0605676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speirs V, Skliris GP, Burdall SE, et al. Distinct expression patterns of ERα and ERβ in normal human mammary gland. J Clin Pathol. 2002;55:371–4. doi: 10.1136/jcp.55.5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Treeck O, Lattrich C, Springwald A, et al. Estrogen receptor β exerts growth inhibitory effects on human mammary epithelial cells. Breast Cancer Res Treat. 2009 May 12; doi: 10.1007/s10549-009-0413-2. DOI 10.1007/s10549–009-0413–2. [DOI] [PubMed] [Google Scholar]
- 5.Skliris GP, Munot K, Bell SM, et al. Reduced expression of oestrogen receptor beta in invasive breast cancer and its re-expression using DNA methyl transferase inhibitors in a cell line model. J Pathol. 2003;201:213–20. doi: 10.1002/path.1436. [DOI] [PubMed] [Google Scholar]
- 6.Zhao C, Lam EW, Sunters A, et al. Expression of estrogen receptor β isoforms in normal breast epithelial cells and breast cancer: regulation by methylation. Oncogene. 2003;22:7600–6. doi: 10.1038/sj.onc.1207100. [DOI] [PubMed] [Google Scholar]
- 7.Smith L, Brannan RA, Hanby AM, et al. Differential regulation of estrogen receptor β isoforms by 5′ untranslated regions in cancer. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00867.x. DOI 10.1111/j.1582-4934.2009.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.09.016. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 9.Verghese ET, Hanby AM, Speirs V, et al. Small is beautiful: microRNAs and breast cancer-where are we now? J Pathol. 2008;215:214–21. doi: 10.1002/path.2359. [DOI] [PubMed] [Google Scholar]
- 10.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondo N, Toyama T, Sugiura H, et al. miR-206 Expression is down-regulated in estrogen receptor α-positive human breast cancer. Cancer Res. 2008;68:5004–8. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 13.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERα) and represses ERα messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–47. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 14.Si ML, Zhu S, Wu H, et al. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 15.Lowery AJ, Miller N, Devaney A, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. doi: 10.1186/bcr2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, et al. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707–14. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–11. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–49. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaaban AM, Green AR, Karthik S, et al. Nuclear and cytoplasmic expression of ERβ1, ERβ2, and ERβ5 identifies distinct prognostic outcome for breast cancer patients. Clin Cancer Res. 2008;14:5228–35. doi: 10.1158/1078-0432.CCR-07-4528. [DOI] [PubMed] [Google Scholar]
- 21.Maraqa L, Cummings M, Peter MB, et al. Carcinoembryonic antigen cell adhesion molecule 6 predicts breast cancer recurrence following adjuvant tamoxifen. Clin Cancer Res. 2008;14:405–11. doi: 10.1158/1078-0432.CCR-07-1363. [DOI] [PubMed] [Google Scholar]
- 22.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sempere LF, Christensen M, Silahtaroglu A, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 24.Kovalchuk O, Tryndak VP, Montgomery B, et al. Estrogen-induced rat breast carcinogenesis is characterised by alterations in DNA methylation, histone modifications and aberrant microRNA expression. Cell Cycle. 2007;6:2010–18. doi: 10.4161/cc.6.16.4549. [DOI] [PubMed] [Google Scholar]
- 25.Klinge CM. Estrogen regulation of microRNA expression. Curr Genomics. 2009;10:169–83. doi: 10.2174/138920209788185289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maillot G, Lacroix-Triki M, Pierredon S, et al. Widespread estrogen-dependent repression of microRNAs involved in breast tumor cell growth. Cancer Res. 2009;69:8332–40. doi: 10.1158/0008-5472.CAN-09-2206. [DOI] [PubMed] [Google Scholar]
- 27.Leivonen SK, Mäkelä R, Ostling P, et al. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926–36. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- 28.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burdall SE, Hanby AM, Lansdown MR, Speirs V. Breast cancer cell lines: friend or foe? Breast Cancer Res. 2003;5:89–95. doi: 10.1186/bcr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartman J, Lindberg K, Morani A, et al. Estrogen receptor β inhibits angiogenesis and growth of T47D breast cancer xenografts. Cancer Res. 2006;66:11207–13. doi: 10.1158/0008-5472.CAN-06-0017. [DOI] [PubMed] [Google Scholar]
- 31.Hartman J, Edvardsson K, Lindberg K, et al. Tumor repressive functions of estrogen receptor β in SW480 colon cancer cells. Cancer Res. 2009;69:6100–6. doi: 10.1158/0008-5472.CAN-09-0506. [DOI] [PubMed] [Google Scholar]
- 32.Castellano L, Giamas G, Jacob J, et al. The estrogen receptor-α-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci U S A. 2009;106:15732–7. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maillot G, Lacroix-Triki M, Pierredon S, et al. Widespread estrogen-depdent repression of microRNAs involved in breast tumour cell growth. Cancer Res. 2009;69:8332–40. doi: 10.1158/0008-5472.CAN-09-2206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.