Abstract
Mycobacterium tuberculosis is an obligate human pathogen capable of persisting in individual hosts for decades. To determine whether antigenic variation and immune escape contribute to the success of M. tuberculosis, we determined and analyzed 22 genome sequences representative of the global diversity of the M. tuberculosis complex (MTBC). As expected, we found that essential genes in MTBC were more evolutionarily conserved than non-essential genes. Surprisingly however, most of 491 experimentally confirmed human T cell epitopes showed little sequence variation and exhibited a lower ratio of non-synonymous to synonymous changes than essential and non-essential genes. These findings are consistent with strong purifying selection acting on these epitopes, and imply that MTBC might benefit from recognition by human T cells.
Infection with Mycobacterium tuberculosis causes enormous worldwide morbidity and mortality; there were more cases of tuberculosis in 2007 (the last year for which data are available) than at any prior point in world history1. Among the factors that contribute to the continued growth of tuberculosis as a global health problem are the efficiency of human-to-human transmission by the aerosol route, the ability of the causal agent M. tuberculosis to persist and to progress despite development of host immune responses, and the absence of a vaccine with reliable efficacy in preventing transmission of the infection. Moreover, while attempts to control tuberculosis through improved identification and treatment of infectious cases have been successful in some settings, similar approaches in other contexts have resulted in increasing rates of resistance to available anti-tuberculosis drugs2. Therefore, new approaches to controlling tuberculosis are essential and would greatly benefit from an improved understanding of the biology of the bacteria and their interactions with their human hosts. In particular, understanding the factors that drive the evolution of M. tuberculosis and allow it to evade host defences, may suggest unique opportunities to develop novel strategies against tuberculosis.
Human tuberculosis is caused by Mycobacterium tuberculosis and Mycobacterium africanum, which are members of the M. tuberculosis complex (MTBC). In addition to these human-adapted pathogens, MTBC includes various animal-adapted forms, such as Mycobacterium bovis, Mycobacterium microti, and Mycobacterium pinnipedii3. To characterize the extent and nature of the forces acting to diversify MTBC, we and others have applied several approaches to phylogenetic analysis of multiple clinical isolates from geographically diverse sources. Using single nucleotide polymorphisms (SNPs)3-6 or large sequence polymorphisms (LSPs)7-9 as genetic markers resulted in congruent groupings of human-adapted MTBC into six major lineages and consistent geographical associations for each of these lineages10. In addition, these studies found strong evidence for a clonal population structure of MTBC, without evidence of ongoing horizontal gene transfer. Analysis of SNPs in a total of 7 megabases of DNA sequence from 89 genes in 108 isolates of MTBC provided strong evidence that MTBC originated in Africa, and underwent population expansion and diversification following ancient human migrations out of Africa, followed by global spread and return to Africa of three particularly successful MTBC lineages through recent waves of travel, trade, and conquest3. Taken together, these studies have revealed that MTBC has undergone genetic diversification that corresponds to patterns of human migration, suggesting that distinct lineages have co-evolved with distinct human populations7. Moreover, they indicate that further understanding of the mechanisms and consequences of the interactions between MTBC and its human host can be obtained through comparative genomic analyses.
Host-pathogen co-evolution is characterised by reciprocal adaptive changes in interacting species11. Host immune pressure and associated parasite immune evasion are key features of this process often referred to as an ‘evolutionary arms-race’12-13. Studies in human pathogenic viruses, bacteria, and protozoa have revealed that genes encoding antigens tend to be highly variable as a consequence of diversifying selection to evade host immunity14-17. However, whether similar evolutionary mechanisms operate in MTBC, and whether the bacteria undergo antigenic variation in response to host immune pressure, is unknown.
Immunity to tuberculosis in humans, nonhuman primates, and mice depends on T lymphocytes18. Among human T lymphocyte subsets, CD4+ T cells are clearly essential for protective immunity to MTBC, as demonstrated by the observation that the incidence of active tuberculosis in people infected with HIV is inversely proportional to the number of circulating CD4+ T cells19. In addition to CD4+ T cell responses, humans infected with MTBC develop antigen-specific CD8+ T cell responses20, and MTBC antigen-specific human CD8+ T cells lyse infected cells and contribute to killing of intracellular MTBC21. Therefore, there is strong evidence that the adaptive immune system represented by CD4+ and CD8+ T cells, is an important mechanism for host recognition and control of MTBC. Recognition of foreign antigens by T lymphocytes depends on binding of short peptide fragments (termed epitopes) derived by proteolysis of foreign proteins, to MHC (major histocompatibility; in humans termed HLA (human leukocyte antigen)) proteins on the surface of macrophages and dendritic cells; CD4+ T cells recognize peptide epitopes bound to MHC/HLA class II; CD8+ T cells recognize peptide epitopes bound to MHC/HLA class I.
To obtain a better understanding of the effects of human T cell recognition on the diversity of MTBC, and to test the hypothesis that MTBC uses antigenic variation as one mechanism of evading elimination by human immune responses, we determined the genome sequences of 21 phylogeographically diverse strains of MTBC and used those genome sequences to analyze the diversity of 491 experimentally verified human T cell epitopes. This analysis produced the unexpected finding that the known human T cell epitopes are highly conserved relative to the rest of the MTBC genome. These results provide evidence that the relationship between MTBC and its human hosts may differ from that of a classical evolutionary arms-race, and suggest that development of new approaches to control of tuberculosis must take into account the possibility that certain human immune responses may actually benefit MTBC.
Results
A Genome-wide Phylogeny of Human-adapted MTBC
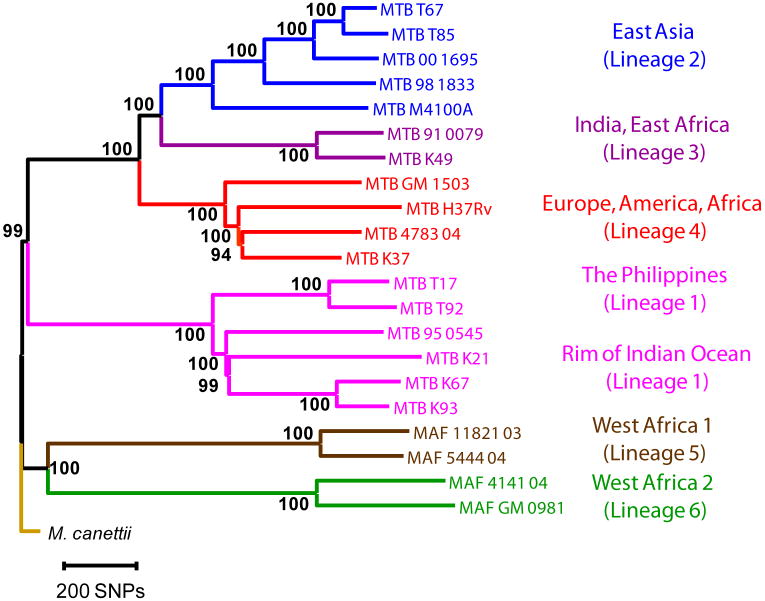
A total of 22 mycobacterial strains were included in this work. To study the sequence diversity of T cell antigens in MTBC, we used Illumina next-generation DNA sequencing to generate nearly complete genome sequences from 20 strains representative of the six main human MTBC lineages, and one strain of Mycobacterium canettii which is the closest known outgroup of MTBC3,22 (Table 1). In addition, we used the published genome sequence of the H37Rv laboratory strain of M. tuberculosis as a common reference23. For each of the 21 strains newly sequenced, a mean of 6.8 million sequence reads with a mean length of 51 base pairs were generated and mapped to the H37Rv reference genome. On average, the reads covered 98.9% of the 4.4 Mb reference genome (Table 1). The regions not covered primarily included members of the highly GC-rich and repetitive PE/PPE gene families24. A total of 32,745 SNPs were identified, corresponding to an average of 1 SNP call for every 3 kb of sequence generated. We used a total of 9,037 unique SNPs (i.e. SNPs that occurred in one or several strains) to derive a genome-wide phylogeny of 22 strains (Fig. 1, Supplementary Fig. 1). Six main lineages could be distinguished with high statistical support. These lineages were completely congruent to the strain groupings previously defined based on genomic deletion analysis and multilocus sequencing3,7,10. The perfect congruence between these different phylogenetic markers further corroborates the highly clonal population structure of MTBC and lack of ongoing horizontal gene transfer in this organism25. Because of the comprehensive nature of genome-scale data, a higher degree of phylogenetic resolution could be achieved compared to all previous studies. In this new phylogeny the brown and green lineages (also known as Mycobacterium africanum) are the most basal groups when compared to the M. canettii outgroup. M. africanum is highly restricted to West Africa for reasons that remain unclear8. However, the fact that the two M. africanum lineages represent the most ancestral forms of human MTBC reinforces the notion that human MTBC originated in Africa3,7.
Table 1.
Strains used in this study, sequencing coverage, and number of raw and filtered SNPs after comparison to the H37Rv reference genome.
| Strain | Lineagea | Origin | Average mapped sequencing depth | Number of reads | Percent genome coverageb | Raw SNPs | Filtered SNPs |
|---|---|---|---|---|---|---|---|
| MTB_95_0545 | Lineage 1 | Laos | 77.37 | 7,621,946 | 99.75 | 3,478 | 2,017 |
| MTB_K21 | Lineage 1 | Zimbabwe | 77.99 | 7,112,888 | 99.29 | 2,853 | 2,151 |
| MTB_K67 | Lineage 1 | Comoro Islands | 78.29 | 7,097,284 | 98.95 | 2,943 | 2,070 |
| MTB_K93 | Lineage 1 | Tanzania | 65.52 | 6,017,391 | 99.22 | 2,949 | 2,041 |
| MTB_T17 | Lineage 1 | The Philippines | 72.59 | 7,130,412 | 99.36 | 3,788 | 1,988 |
| MTB_T92 | Lineage 1 | The Philippines | 46.01 | 5,068,053 | 98.85 | 4,080 | 1,994 |
| MTB_00_1695 | Lineage 2 | Japan | 77.92 | 7,394,236 | 99.02 | 2,875 | 1,351 |
| MTB_98_1833 | Lineage 2 | China | 64.49 | 6,395,114 | 99.1 | 2,962 | 1,361 |
| MTB_M4100A | Lineage 2 | South Korea | 40.47 | 4,022,290 | 98.94 | 3,316 | 1,354 |
| MTB_T67 | Lineage 2 | China | 78.77 | 7,616,603 | 98.73 | 2,820 | 1,343 |
| MTB_T85 | Lineage 2 | China | 61.65 | 6,159,284 | 99.04 | 3,046 | 1,377 |
| MTB_91_0079 | Lineage 3 | Ethiopia | 74.03 | 7,228,038 | 99.14 | 2,920 | 1,363 |
| MTB_K49 | Lineage 3 | Tanzania | 75.52 | 6,845,266 | 99.25 | 2,195 | 1,416 |
| H37Rv | Lineage 4 | USA | Reference | ||||
| MTB_4783_04 | Lineage 4 | Sierra-Leone | 78.12 | 7,466,814 | 98.78 | 1,559 | 741 |
| MTB_GM_1503 | Lineage 4 | The Gambia | 82.26 | 7,891,933 | 99.08 | 2,283 | 782 |
| MTB_K37 | Lineage 4 | Uganda | 59.86 | 5,480,451 | 98.85 | 2,496 | 822 |
| MAF_11821_03 | Lineage 5 | Sierra-Leone | 78.22 | 7,491,737 | 99.02 | 3,741 | 2,102 |
| MAF_5444_04 | Lineage 5 | Ghana | 79.75 | 7,578,690 | 98.92 | 3,686 | 2,079 |
| MAF_4141_04 | Lineage 6 | Sierra-Leone | 72.62 | 7,027,143 | 98.61 | 3,886 | 2,180 |
| MAF_GM_0981 | Lineage 6 | The Gambia | 76.39 | 7,350,873 | 99 | 4,451 | 2,213 |
| MTB_K116 | M. canettii | Somalia | 93.01 | 6,544,254 | 96.32 | 19,008 | 14,730 |
| Total MTBC | 62,327 | 32,745 |
Figure 1.
Neighbour-joining phylogeny based on 9,037 variable common nucleotide positions across 21 human M. tuberculosis complex genome sequences. The tree is rooted with M. canettii, the closest known outgroup. Node support following 1,000 bootstrap replications is indicated. Branches are coloured according to the six main phylogeographic lineages of MTBC defined previously3,7-8. Highly congruent topologies were obtained by Maximum likelihood and Bayesian inference (Supplementary Fig. 1).
Evolutionary Conservation Across Gene Categories
We used these genome sequence data and the phylogeny derived from them to compare the genetic diversity in antigens and other experimentally determined gene classes. For comparisons across different gene categories, we divided our dataset into three gene sets, including ‘essential genes’, ‘non-essential genes’, and ‘antigens’ (Supplementary Fig. 2, Supplementary Tables 1 and 2). Antigens were defined based on the presence of 491 experimentally confirmed human T cell epitopes (Supplementary Table 3), which were compiled through the Immune Epitope Database (IEDB) initiative26. The ‘essential’ gene category was defined based on genome-wide analyses of transposon insertion mutants that were defective for the ability to grow on Middlebrook 7H11 agar, or in the spleens of intravenously-infected mice, published previously27-28. We excluded from this analysis genes belonging to the PE/PPE gene family24 and those related to mobile elements as they are difficult to study using current next-generation DNA sequencing technologies (total genes excluded: 273/3,990 (6.8%) genes annotated in the H37Rv reference genome; Supplementary Table 4).
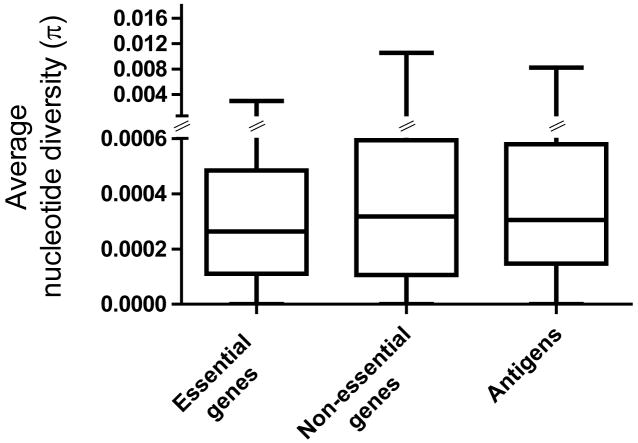
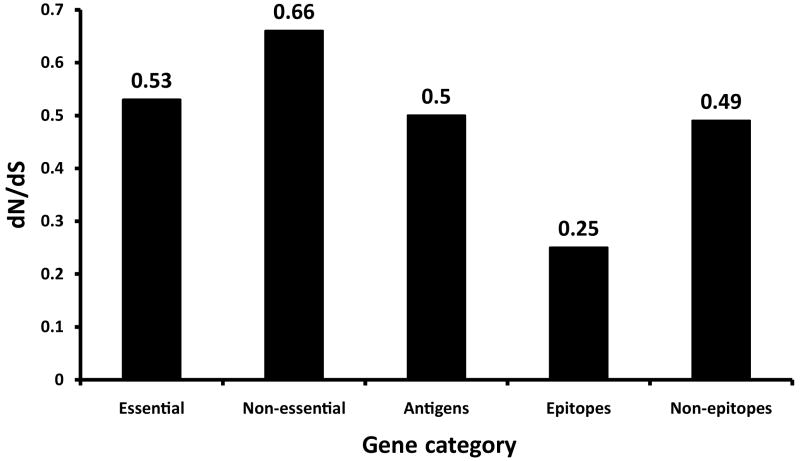
Based on evolutionary theory and findings in other bacteria29, one would expect that in contrast to non-essential genes, the essential genes in MTBC will be under stronger purifying selection and thus more evolutionary conserved. In support of this notion, we observed that on average essential genes harboured less nucleotide diversity than non-essential genes (Fig. 2; Mann-Whitney U test p<0.002). We then compared the rates of synonymous and non-synonymous SNPs in the essential and non-essential gene categories. The synonymous and non-synonymous changes were derived by comparison to the most likely recent common ancestor of MTBC, which we inferred based on our new genome-wide phylogeny (Fig. 1, Supplementary Fig. 1). Because MTBC harbours little sequence diversity, it was necessary to analyze the distribution of synonymous and non-synonymous SNPs based on gene concatenates rather than individual genes. The two measures of distribution we used were based on the number of non-redundant SNPs across all 21 MTBC strains (dN/dS based on Measure A in Table 2 and Fig. 3), and on the individual pairwise comparisons between each strain and the inferred most likely recent common ancestor (dN/dS based on Measures B in Table 2). From these analyses, we found that the dN/dS measures for essential genes were significantly lower than for non-essential genes (Measure A in Fig. 3; Measure B in Table 2, Mann-Whitney U test p<0.0001). Taken together, these data show that in MTBC essential genes are more evolutionary conserved than non-essential genes.
Figure 2.
Average gene-by-gene nucleotide diversity across three gene classes. Boxplot indicating median (horizontal line), interquartile range (box), and minimum and maximum values (whiskers).
Table 2.
Distribution of synonymous and non-synonymous SNPs in gene concatenates.
| Gene category | Length of Concatenate (base pairs) | Measure Aa | Measure Bb | |||
|---|---|---|---|---|---|---|
| Nonredundant SNPs | nonsyn/syn | dN/dS | Range | |||
| nonsyn | syn | |||||
| Essential | 907,584 | 1,124.83 | 755.17 | 1.49 | 0.53 | 0.45-0.67 |
| Non-essential | 2,674,329 | 4,392.51 | 2,338.49 | 1.88 | 0.65 | 0.78-0.56 |
| Antigens | 81,660 | 126.5 | 87.5 | 1.45 | 0.57 | 0.17-1.15 |
| Epitopes | 12,234 | 19 | 12 | 1.58 | nad | na |
| Epitopesc | 11,088 | 9 | 12 | 0.75 | na | na |
| Non-epitopesc | 68,556 | 106.5 | 75.5 | 1.41 | na | na |
Notes:
The number of non-redundant synonymous and non-synonymous SNPs after mapping the changes onto the phylogeny shown in Figure 1. An overall dN/dS was calculated based on these SNPs and is shown in Figure 3 (Measure A; see Materials and Methods).
Calculated using Measure B. The median dN/dS was calculated from the 21 strain specific dN/dS values. This measure of dN/dS could only be calculated for the essential, non-essential and antigen categories because in the epitope and non-epitope concatenates some strains had zero values for synonymous or non-synonymous changes.
After exclusion of the three outlier antigens esxH, pstS1, and Rv1986 (see main text).
not applicable.
Figure 3.
Ratio of the rates of synonymous and non-synonymous substitutions (dN/dS) in various gene classes of MTBC. Overall dN/dS was calculated based on the number of non-redundant synonymous and non-synonymous changes after comparing each of the 21 MTBC genomes to the inferred most likely recent common ancestor of MTBC. This shows that essential genes are more conserved than non-essential genes, and that antigens are as conserved as essential genes. Figures for the epitope- and non-epitope regions refer to the calculations after excluding the three outlier antigens esxH, pstS1, and Rv1986.
Because MTBC interacts with humans through antigen-specific CD4+ or CD8+ T-cells, we would expect T cell antigens to be among the most diverse genes in the genome. Particularly when invoking a co-evolutionary arms-race and associated immune evasion, we would anticipate these antigens to be under diversifying selection and to be more variable than other genes in order to escape T cell recognition. However, when we analyzed the nucleotide diversity in 78 experimentally confirmed human T cell antigens (Supplementary Table 2), we found that they were on average not more diverse than essential genes (Fig. 2, Mann-Whitney U test p=0.12). Moreover, we found that the dN/dS measures in these antigens also resembled those of essential genes (Measure A in Fig. 3; Measure B in Table 2, Mann-Whitney U test p=0.77). Thus, human T cell antigens in MTBC do not appear to be under diversifying selection. Instead, purifying selection appears to be the driving selection pressure in these genes.
T Cell Epitopes are Hyperconserved
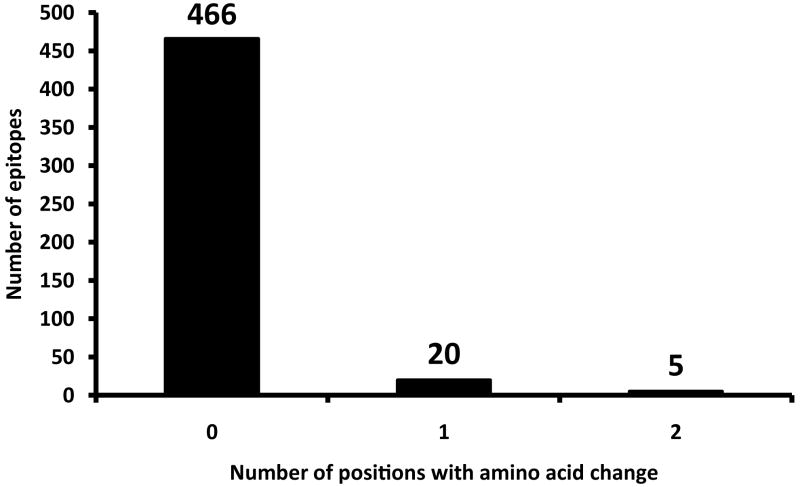
T cell antigens consist of epitope regions that interact with human T cells, and non-epitope regions which are not targets of T cell recognition. Hence, we decided to study these regions separately. To this end, we generated a separate concatenate of the epitope regions and another concatenate of all corresponding non-epitope regions. Because little data is currently available in the IEDB with respect to whether these 491 epitopes are recognized by CD4+ or CD8+ T cells, we analyzed them as one class. If immune escape was driving antigen evolution to evade T cell recognition in MTBC, we would expect non-synonymous changes to accumulate in epitope regions, leading to a high dN/dS. Contrary to this expectation however, the overall dN/dS of the epitope regions was 0.53, which was still similar to essential genes and lower than non-essential genes (Table 2, Fig. 3). Moreover, when we analyzed the distribution of amino acid replacements in individual epitopes we found that the large majority (95%) of the 491 epitopes showed no amino acid change (Fig. 4). Only five epitopes, contained in esxH, pstS1, and Rv1986, harboured more than one variable position (Supplementary Table 5). The higher number of amino acid substitutions in these five epitopes may reflect ongoing immune evasion, but further investigation is needed to determine whether the observed changes are due to immune pressure, other selection pressure(s), or mere random genetic drift3. Because these five epitopes were clear outliers compared to the large majority of T cell epitopes analyzed here, we repeated our dN/dS analysis after excluding the three antigens harbouring the five outlier epitopes. Our analysis revealed that the epitope regions had the lowest dN/dS of all gene categories (Table 2, Fig. 3). Furthermore, when we compared the proportion of non-redundant non-synonymous changes in epitope and non-epitope regions, we found that epitopes were less likely than non-epitopes to harbour changes at non-synonymous sites (Measure A in Table 2, χ2, p<0.05), whereas no difference was observed at synonymous sites (Table 2, χ2, p=0.89).
Figure 4.
Number of variable amino acid positions in 491 human T cell epitopes of MTBC. This demonstrates the remarkable lack of genetic variability among the regions of the genome that interact with the human immune system.
To further corroborate our finding of hyperconservation of human T cell epitopes in MTBC, we repeated our analysis using a data set from a previous study in which 89 individual genes were sequenced in 99 human-adapted strains representative of the six major global lineages of MTBC3. Sixteen of these 89 genes belonged to the T cell antigens analyzed here, including two of the three outlier antigens esxH and pstS13. Analysis of this additional dataset of 16 antigens in 99 MTBC strains revealed an overall dN/dS for the epitope regions of 0.74. However, after excluding the two outlier antigens, the dN/dS dropped to 0.46, which was again lower than the genome-based dN/dS values for essential and non-essential genes (Fig. 3).
Taken together, our findings strongly suggest that a large proportion of the MTBC genome known to interact with human T cells is highly conserved and under as strong, or perhaps even stronger, purifying selection than essential genes.
Discussion
In this study of 22 MTBC genomes, we demonstrate that, as expected, essential genes are more conserved than non-essential genes. These results are in agreement with a previous study which analyzed a single genome30. Surprisingly, however, we found that the large majority of the currently known T cell antigens are as conserved as essential genes. Furthermore, the epitope regions of these antigen genes are the most highly conserved regions we studied. This observation, that the regions of the genome that interact with the human adaptive immune system appear to be under even stronger purifying selection than essential genes, is inconsistent with a classical model of an evolutionary arms-race.
It is possible that the known human T cell epitopes that we found to be hyperconserved represent a select subset of all of the human T cell epitopes encoded in the genome, and that certain approaches to epitope identification have favoured discovery of hyperconserved epitopes in MTBC. For example, since most, if not all of the epitope discovery efforts to date have utilized proteins and/or peptide sequences of strains from one lineage (lineage 4) and T cells from humans that are likely to have been infected by strains of other lineages, the assays used may have been especially suited to identification of hyperconserved and/or cross-reactive epitopes. While further investigation using alternative approaches to epitope discovery may reveal that variable epitopes that exhibit evidence of positive selection exist in the MTBC, it is likely that the large number of epitopes that we examined will remain a significant subset of the total, and that future vaccine development efforts will need to account for the possibility that immune recognition of certain epitopes may actually provide a net benefit to the bacteria.
Lack of antigenic variation and immune evasion has been reported for a number of other human pathogens, including RNA viruses such as measles, mumps, rubella, and influenza type C31. Theoretical studies have suggested that the absence of immune escape variants in these viruses might be due to structural constraints in viral proteins or negative mutational effects leading to reduced infectivity or transmission31. While we cannot exclude the possibility that structural and functional constraints that are independent of T cell recognition contribute to hyperconservation of the regions encoding MTBC peptides recognized by human T cells, one important characteristic of the aformentioned viral pathogens is that they spread among young and immunologically naive hosts, which might eliminate the need for immune evasion31. Moreover, infection by these viruses usually results in acute disease, followed by elimination of the infection through adaptive immunity, and acquisition of lifelong immunity against re-infection. This further indicates that these viruses are specialized pathogens of immunologically naive hosts. By contrast, MTBC causes chronic and often lifelong infections, and adaptive immunity is usually unable to completely clear the infection18. Furthermore, tuberculosis patients are prone to re-infection32, and mixed infections are also increasingly recognized33. These observations suggest that the biological basis for the lack of antigenic variation in MTBC reported here differs from what has been proposed for antigenically homogeneous RNA viruses31. In addition, we determined that the fraction of hyperconserved T cell epitopes of the MTBC that are derived from essential genes is indistinguishable from the frequency of essential genes in the MTBC genome as a whole (18% versus 21%, respectively; χ2 = 0.28, p = 0.59), indicating that our results were not skewed by over-representation of T cell epitopes in essential genes. Moreover, the T cell epitopes that we analyzed are present in genes from diverse gene ontologies, and the representation of five main gene categories (defined based on the NCBI Categories of Orthologous Groups (COG)) was no different in the T cell antigens when compared to the genome overall (χ2 with 4 degrees of freedom = 5.8, p = 0.21; Supplementary Table 6). Hence the only identifiable common property of these regions is their recognition by human T lymphocytes. These findings suggest that T lymphocyte recognition is an important factor in hyperconservation of these sequences, and that other structural or functional constraints are unlikely to fully account for the lack of sequence variation in these domains.
Our data suggest that T cell epitopes in MTBC are under strong selection pressure to be maintained, perhaps because the immune response they elicit in humans, which are essential for survival of an individual host, might partially work towards the pathogen's benefit. One potential mechanism of benefit to MTBC from human T cell recognition is that human T cell responses are essential for MTBC to establish latent infection. This notion is supported by the fact that CD4+ T cell-deficient HIV-positive individuals progress rapidly to active disease after infection, rather than to sustain prolonged periods of latent tuberculosis34. Latent infection mediated by host T cell responses, with subsequent reactivation to active disease often occurring decades after initial infection, is a key characteristic of human tuberculosis, and might have evolved as a way for MTBC to transmit to later generations of susceptible hosts35. In addition, there is evidence that T cell responses may contribute directly to human-to-human transmission of MTBC. In particular, cavitary tuberculosis, which generates secondary cases more efficiently than other disease forms36, rarely occurs in CD4+ T cell-deficient HIV-positive individuals, and the frequency of cavitary lung lesions in HIV-infected patients with tuberculosis is directly correlated with the number of peripheral CD4+ T cells37. While the mechanisms of lung cavitation in tuberculosis are poorly understood, these observations suggest that CD4+ T cells directly or indirectly mediate tissue damage in tuberculosis, and together with our finding of epitope hyperconservation indicate that certain T cell responses may be detrimental to the host and beneficial to the pathogen. Hence our findings suggest that MTBC takes advantage of host adaptive immunity to increase its likelihood of spread, and that the benefits of enhanced transmission exceed the costs of within-host cellular immune responses to these epitopes. In this manner, MTBC may resemble HIV, for which there is evidence that virulence has evolved, not to maximize replication of the virus within individual hosts, but to maximize the likelihood of its transmission38. Whether T cell responses to other epitopes, or whether specific T cell subsets (e.g. Th17 versus Th1) that benefit the host and not the bacteria can be identified will require additional studies in humans.
One limitation of this study was the exclusion of PE/PPE genes because of technical reasons. Some of these genes are known to vary and to be cell-surface exposed, which has lead to the hypothesis they might be involved in antigenic variation24. However, no direct evidence for this has yet been presented. Future work will need to clarify the function and evolution of PE/PPE genes. By contrast, all the T cell antigens included in this study have been experimentally confirmed26. Furthermore, some of them are being targeted by new tuberculosis diagnostics and vaccines39. Our findings thus have important implications for the development of these new tools. On the one hand, the fact that MTBC harbours little sequence diversity in T cell antigens will facilitate the development of diagnostics that are universally applicable across geographical regions where MTBC strains differ8. On the other hand, the possibility that the immune responses induced by vaccine antigens might partially benefit the pathogen suggests current efforts in vaccine research should be broadened. Most disturbing is the suggestion that vaccine induced immunity against these conserved epitopes may perversely increase transmission. In this respect, it is interesting to note that the currently available tuberculosis vaccine Bacille-Calmette-Guerin (BCG), which is a live vaccine based on an attenuated from of M. bovis, offers no protection against pulmonary tuberculosis in adults40. More importantly, some clinical trials of BCG have even reported an increased risk of tuberculosis in vaccinees compared to unvaccinated individuals41. Thus, in contrast to standard reverse vaccinology, in which the least variable antigens of a genome are targeted42, research into new tuberculosis vaccines should explore more variable regions of the MTBC genome.
While most of the T cell epitopes anlyzed here were highly conserved, five epitopes in three antigens harboured a larger number of amino acid changes. The fact that the dN/dS measure dropped sharply after excluding these outlier antigens from the analysis further supports the notion that they are indeed outliers compared to the other antigens. One of these outlier antigens, esxH (Rv0288, also known as TB10.4) is a member of a gene family known to encode a Type VII secretion system43. Importantly, this antigen is being considered as new vaccine antigen against tuberculosis39. Thus even though most of the other vaccine antigens analyzed here are conserved, our finding that this particular vaccine antigen harbours a comparatively high number of amino acid substitutions across a panel of global MTBC isolates, suggests that strain diversity should be considered during further development of the new vaccine candidates containing esxH8.
We detected significant differences in dN/dS between essential, non-essential, and antigenic genes. However, the individual dN/dS values remain high when compared to most other bacteria44. Such a high dN/dS was reported previously for MTBC, and has been linked to reduced selective constraint against slightly deleterious mutations3. It was proposed that the serial transmission bottlenecks associated with patient-to-patient transmission in MTBC could lead to an increase in random genetic drift compared to the forces of natural selection. Our new data show that even though the strength of purifying selection in MTBC might be reduced overall compared to other bacteria, it clearly is still acting on and capable of differentiating between gene categories.
In summary, we show that T cell epitopes of MTBC are highly conserved, and do not reflect any ongoing evolutionary arms-race or immune-evasion. Instead, the patterns observed might be indicative of a distinct evolutionary strategy of immune-subversion developed by this highly successful pathogen. Other intracellular bacteria such as Salmonella enterica serovar Typhi exhibit a similar lack of antigenic variation45, suggesting comparable mechanisms might exist in other pathogens with a similar lifestyle.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Supplementary Material
Acknowledgments
We thank Fernando Gonzalez-Candelas, Sonia Borrell, and Douglas Young for comments on the manuscript. This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Disease, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200400001C. J.C. is a Howard Hughes Medical Institute Research Training Fellow. J.D.E. was supported by NIH grants AI046097 and AI051242, and S.G. by the Medical Research Council, UK, the Royal Society, the Swiss National Science Foundation, and NIH grants HHSN266200700022C and AI034238.
Footnotes
Accession codes. The sequencing reads have been submitted to the NCBI Sequence Read Archive (SRA) with accession codes SRX002001-SRX002005, SRX002429, SRX003589, SRX003590, SRX005394, SRX007715, SRX007716, SRX007718-SRX007726, and SRX012272. Sequence and SNP data are also available at the Tuberculosis Database (TBDB).
Author Contribution Statements: I.C., J.D.E. and S.G. designed the study; P.M.S., S.N., K.K. and S.G. contributed sources of M. tuberculosis DNA and demographic information; I.C., J.C. and J.G. performed DNA sequencing and bioinformatics; I.C., P.M.S., J.D.E. and S.G. wrote the manuscript with comments from all authors.
Competing Interest Statement: The authors declare no competing financial interests.
References
- 1.WHO. Global tuberculosis control - surveillance, planning, financing. WHO; Geneva, Switzerland: 2009. [Google Scholar]
- 2.Shenoi S, Friedland G. Extensively drug-resistant tuberculosis: a new face to an old pathogen. Annu Rev Med. 2009;60:307–20. doi: 10.1146/annurev.med.60.053107.103955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershberg R, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker L, Brown T, Maiden MC, Drobniewski F. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg Infect Dis. 2004;10:1568–77. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filliol I, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188:759–72. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutacker MM, et al. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J Infect Dis. 2006;193:121–128. doi: 10.1086/498574. [DOI] [PubMed] [Google Scholar]
- 7.Gagneux S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–37. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 9.Reed MB, et al. Major Mycobacterium tuberculosis lineages associate with patient country of origin. J Clin Microbiol. 2009;47:1119–28. doi: 10.1128/JCM.02142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comas I, Gagneux S. The past and future of tuberculosis research. PLoS Pathog. 2009;5:e1000600. doi: 10.1371/journal.ppat.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–77. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 12.Brunham RC, Plummer FA, Stephens RS. Bacterial antigenic variation, host immune response, and pathogen-host coevolution. Infect Immun. 1993;61:2273–6. doi: 10.1128/iai.61.6.2273-2276.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc Lond B Biol Sci. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 14.Kawashima Y, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–5. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farci P, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–44. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 16.Jeffares DC, et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–5. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urwin R, et al. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect Immun. 2004;72:5955–62. doi: 10.1128/IAI.72.10.5955-5962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.North RJ, Jung YJ. Immunity to tuberculosis. Annu Rev Immunol. 2004;22:599–623. doi: 10.1146/annurev.immunol.22.012703.104635. [DOI] [PubMed] [Google Scholar]
- 19.Shafer RW, Edlin BR. Tuberculosis in patients infected with human immunodeficiency virus: perspective on the past decade. Clin Infect Dis. 1996;22:683–704. doi: 10.1093/clinids/22.4.683. [DOI] [PubMed] [Google Scholar]
- 20.Lewinsohn DA, et al. Immunodominant tuberculosis CD8 antigens preferentially restricted by HLA-B. PLoS Pathog. 2007;3:1240–9. doi: 10.1371/journal.ppat.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruns H, et al. Anti-TNF immunotherapy reduces CD8+ T cell-mediated antimicrobial activity against Mycobacterium tuberculosis in humans. J Clin Invest. 2009;119:1167–77. doi: 10.1172/JCI38482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez C, et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathogens. 2005;1:1–7. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 24.Brennan MJ, Delogu G. The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol. 2002;10:246–9. doi: 10.1016/s0966-842x(02)02335-1. [DOI] [PubMed] [Google Scholar]
- 25.Supply P, et al. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol Microbiol. 2003;47:529–38. doi: 10.1046/j.1365-2958.2003.03315.x. [DOI] [PubMed] [Google Scholar]
- 26.Ernst JD, et al. Meeting Report: NIH Workshop on the Tuberculosis Immune Epitope Database. Tuberculosis (Edinb) 2008;88:366–70. doi: 10.1016/j.tube.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 28.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–94. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12:962–8. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plotkin JB, Dushoff J, Fraser HB. Detecting selection using a single genome sequence of M. tuberculosis and P. falciparum. Nature. 2004;428:942–5. doi: 10.1038/nature02458. [DOI] [PubMed] [Google Scholar]
- 31.Frank SA, Bush RM. Barriers to antigenic escape by pathogens: trade-off between reproductive rate and antigenic mutability. BMC Evol Biol. 2007;7:229. doi: 10.1186/1471-2148-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small PM, et al. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis in patients with advanced HIV infection. N Engl J Med. 1993;328:1137–44. doi: 10.1056/NEJM199304223281601. [DOI] [PubMed] [Google Scholar]
- 33.Warren RM, et al. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med. 2004;169:610–4. doi: 10.1164/rccm.200305-714OC. [DOI] [PubMed] [Google Scholar]
- 34.Daley CL, et al. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus. An analysis using restriction-fragment-length polymorphisms. N Engl J Med. 1992;326:231–5. doi: 10.1056/NEJM199201233260404. [DOI] [PubMed] [Google Scholar]
- 35.Blaser MJ, Kirschner D. The equilibria that allow bacterial persistence in human hosts. Nature. 2007;449:843–9. doi: 10.1038/nature06198. [DOI] [PubMed] [Google Scholar]
- 36.Rodrigo T, et al. Characteristics of tuberculosis patients who generate secondary cases. Int J Tuberc Lung Dis. 1997;1:352–7. [PubMed] [Google Scholar]
- 37.Mukadi Y, et al. Spectrum of immunodeficiency in HIV-1-infected patients with pulmonary tuberculosis in Zaire. Lancet. 1993;342:143–6. doi: 10.1016/0140-6736(93)91346-n. [DOI] [PubMed] [Google Scholar]
- 38.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc Natl Acad Sci U S A. 2007;104:17441–6. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young DB, Perkins MD, Duncan K, Barry CE. Confronting the scientific obstacles to global control of tuberculosis. J Clin Invest. 2008;118:1255–1265. doi: 10.1172/JCI34614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen P, Doherty TM. Opinion: The success and failure of BCG - implications for a novel tuberculosis vaccine. Nat Rev Microbiol. 2005;3:656–62. doi: 10.1038/nrmicro1211. [DOI] [PubMed] [Google Scholar]
- 41.Colditz GA, et al. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. Jama. 1994;271:698–702. [PubMed] [Google Scholar]
- 42.Pizza M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–20. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 43.Abdallah AM, et al. Type VII secretion--mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–91. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 44.Rocha EP, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239:226–35. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 45.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–93. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 48.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 49.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 50.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 51.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 52.Vilella AJ, Blanco-Garcia A, Hutter S, Rozas J. VariScan: Analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics. 2005;21:2791–3. doi: 10.1093/bioinformatics/bti403. [DOI] [PubMed] [Google Scholar]
- 53.Korber B. Computational analysis of HIV molecular sequences. Kluwer Academic Publishers; Dordrecht, Netherlands: 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.