Abstract
In mammalian cells the accumulation of repair proteins to double-strand breaks is a phosphorylation and ubiquitylation regulated process. Some of the genes that encode the kinases, and ubiquitin ligases in this pathway are cancer predisposition genes, most prominently the breast cancer predisposition gene, BRCA1 which encodes a ubiquitin ligase. How BRCA1 ligase activity was regulated following DNA damage was poorly understood. In this review I summarise new data that shows a third post-translational modification, by the Small ubiquitin like modifier SUMO, is part of the same cascade, enabling and activating DNA damage-regulated processes, including the BRCA1 ligase activity.
BRCA1 in the response to DNA damage
For many years it has been clear that BRCA1 plays a central role in the response to the most genotoxic of DNA lesions, double-stranded breaks. These are formed not only on exposure to exogenous agents such as irradiation or topoisomerise inhibitors, but also in the natural life of a cell, at replication fork blocks caused by adducts generated by metabolic processes. BRCA1 in growing cells is found in punctuate sub-nuclear pattern that relocates to sites of repair following DNA damage. In the absence of BRCA1 cells accumulate chromosome rearrangements and tumours from patients (or mice) with absent BRCA1 activity show considerable genomic instability.
Two conserved regions of BRCA1 are important for its tumour suppression role. The C-terminus of BRCA1 has direct repeats of two BRCT (BRCA1 C-terminal) domains, now found in many DNA damage repair proteins and the BRCA1 N-terminus has a RING domain that interacts with E2 ubiquitin conjugating enzymes. The BRCTs create a pocket that binds phosphopeptides and patient missense variants that disrupt phosphopeptide binding or the integrity of the BRCTs are associated with disease (1, 2). BRCA1 is found in three DNA repair complexes regulated by this pocket, containing phosphorylated BRIP1/FANCJ, phosphorylated CtIP or phosphorylated Abraxas (reviewed in (3)). This latter interaction is required for BRCA1 recruitment to sites of chromatin surrounding the DNA lesion. Phospho-Abraxas is part of a multi-protein complex containing RAP80. RAP80 is recruited through the combined activities of RNF168, RNF8 and HERC2 ubiquitin ligases, which, with the ubiquitin conjugating enzyme Ubc13 generate chains of ubiquitin linked through lysine 63. These generate topologically specific structures recognised by RAP80 (4, 5). RNF8 is recruited through the phosphorylation of MCD1, co-ordinated by the ATM kinase, which also encourages MDC1 recruitment through modification of the histone H2AX within chromatin surrounding the DNA break.
The role of BRCA1 once at sites of damage repair remains unclear but what is known is that in combination with its N-terminal binding partner, BARD1, BRCA1 interacts with ubiquitin E2 conjugating enzymes and catalyses the generation of chains of ubiquitin linked through at least one lysine-6 linkage (6, 7). The ubiquitin ligase ability of BRCA1 is highly conserved, and chicken and worm versions perform the same activity at the same location (8, 9). The ligase activity is also likely to be part of BRCA1-mediated tumour suppression. The highest density of different missense substitutions in patients with a familial history of disease occurs across the N-terminal region encompassing the RING domain (10). Mutations of the Zn2+-ligating residues required to maintain the RING structure co-segregate with disease in large independent families and inhibit BRCA1 ligase activity, consistent with a requirement in tumour suppression on the activity. However as these mutations potentially affect protein stability as well (11), stronger evidence for the potential requirement of the ligase activity comes from the experimental selection from randomly generated variants of BRCA1 that inhibit ubiquitin E2 interactions. BRCA1: E2 disruptive missense changes selected in this screen were in the same amino acids as patient variants, suggesting that the E2:BRCA1 interaction plays some role in tumour suppression (12). Thus the regulation of this activity is also likely to be relevant to tumour suppression. After DNA damage chromatin-bound BRCA1 co-purifies with the ubiquitin E2, UbcH5, required for ligase activity, whereas free BRCA1, or BRCA1 in undamaged cells does not co-purify the enzyme (9), suggesting a switch between ligase-active BRCA1 present in chromatin-associated repair foci and inactive BRCA1 elsewhere.
SUMO conjugation is part of the BRCA1-response to DNA damage
There are three conjugated forms of SUMO; SUMO1, SUMO2 and SUMO3. (SUMO2 and SUMO3 differ by just 3 amino acids, are considered functionally equivalent and known as SUMO2/3). SUMOylation follows the same enzyme architecture as ubiquitin modification, requiring an E1 activating enzyme, E2 conjugating enzyme (only one of these is known, Ubc9) and E3 ligating enzymes, such as PIAS1-4, MMS21, RanBP2, Polycomb 2 or TOPORS. SUMO conjugation to target proteins tends to occur on a consensus in the target of “ψKxE′, where “ψ” is a large hydrophobic residue. SUMO2 and SUMO3 each contain a consensus site, so that these, but not SUMO1, are able to form chains of SUMO on a target protein.
Following the observation that c. elegans Bard interacts with the c. elegans SUMO E2 conjugating enzyme, ce-Ubc9 (13), we examined whether SUMO proteins might play a role in the way BRCA1 responds to DNA damage in mammalian cells.
We noted that SUMO isoforms, like BRCA1, relocate to sub-nuclear foci, marked by phosphorylated H2AX (γH2AX) after DNA damage, and that BRCA1 interacted with SUMO at these sites (14). The SUMO E2 conjugating enzyme, Ubc9, also accumulated at these regions, as did the SUMO-ligase enzymes PIAS1 and PIAS4. Indeed BRCA1 was conjugated to SUMO following genotoxic insult and this required the PIAS1 and PIAS4 ligases (and did not require other ligases MMS21, PIAS2 or PIAS3). The most intriguing step came when examining the ubiquitin conjugates at sites of DNA damage. When PIAS ligases where depleted K6-ubiqutin conjugates failed to locate with γH2AX labelled chromatin even when BRCA1 did so, suggesting a defect in the BRCA1 ligase activity. To establish whether this was a direct consequence of BRCA1 SUMO modification we established whether any SUMO consensus motifs might be required for BRCA1: SUMO interaction in cells. The two highest scoring motifs are located adjacent to the BRCA1 RING, and while mutation of one had no impact on the interaction, loss of the other (mutated to either ψRxE′ or ψKxA′) inhibited both the interaction of BRCA1 with SUMO and ability of exogenous BRCA1 to induce elevated ubiquitin conjugates, suggesting that the SUMO modification acts to switch on BRCA1 ligase activity. Testing these ideas in vitro we found that SUMO-modified BRCA1: BARD1 was able to generate 20-50 fold more ubiquitin-conjugates than the unmodified form, recapitulating the cellular events and showing that BRCA1 is a SUMO regulated ubiquitin ligase (SRUbL) (14).
The impact of SUMO-modification was not restricted to BRCA1. We and Galanty et al (15) also showed that events upstream of BRCA1 were effected by the depletion of the SUMO-ligases, in that PIAS1 was needed for complete accumulation of BRCA1, whereas PIAS4 was active further up the pathway and needed for proper accumulation of RNF168, and proteins subsequent to it in the cascade, (including 53BP1). The chromatin targets of RNF8/HERC2/RNF168 were no longer ubiquitylated on PIAS4 depletion, leading to the speculation that one or all of these ligases may, like BRCA1, be regulated by SUMOylation. Intriguingly Galanty et al show that PIAS4 regulated events correlate with SUMO-1 involvement and 53BP1 modification, and PIAS1 with SUMO2/3 involvement and BRCA1 modification, although how these observations translate to differential activity is not currently clear.
Galanty also made important observations regarding the requirements for PIAS recruitment to DNA damage sites. Their accumulation did not require the RNF8-repair pathway, instead the PIAS SAP domains, reported to interact with sequence or structure-specific DNA (16), were required, but not sufficient, for recruitment to DNA breaks (15). In their independent accumulation PIAS SUMO ligases perhaps provide a fail-safe for the activation of repair proteins by introducing a need for a further contact with DNA.
Significance and implications
The BRCA1 route to DNA damage repair now also includes SUMO modification events. In common with other proteins that form part of this cascade the depletion of the PIAS1 and PIAS4 SUMO ligases caused sensitivity to irradiation and cisplatin and loss of homologous recombination and non-homologous end-joining pathways of double-strand break repair (14, 15). Some genes in this pathway are mutated in the small proportion cancers that have a hereditary pattern. Might SUMO pathway components also be involved? While the SUMOs themselves and possibly the PIAS proteins are likely to be redundant, the pathway has only a single E2 enzyme, Ubc9. Rare SNPs in the UBC9 allele have recently been associated with increased risk of breast cancer and increased grade, although the causative variants remains to be discovered (17, 18). While increased Ubc9 and PIAS1 expression has been noted in Multiple Myeloma, a cancer reported to have elevated homologous recombination (19).
Other portions of the SUMO regulatory pathway not yet investigated would now be expected to play a role in the response to double-stranded breaks. Enzymes that process immature SUMO1 or SUMO2/3 and readily clip them from conjugated targets (the Sentrin specific proteases) will presumably be required, with certain SENPs required for certain portions of the pathway. Similarly the SUMO targeted ubiquitin ligase, RNF4, might be expected to play a role by targeting SUMO2-conjugated elements of the pathway for degradation, and/or by regulating the available SUMO2 levels in a cell.
Much also remains unclear about the current findings. How is the BRCA1 ligase activity regulated by SUMO, are the other ubiquitin ligases in the pathway (or elsewhere) also SRUbLs? Does SUMO-modification of 53BP1 alter its activity? What is the basis of PIAS ligase regulation?
SUMO modification plays a role in various forms of DNA repair (20) as well as many other cellular pathways and now in BRCA1 accumulation and activity. These new data suggest that successful hits from drug discovery programmes targeting the SUMO pathway are likely to disrupt BRCA1 activity, presenting both a hazard to their use and suggesting potential efficacy in cancer treatment.
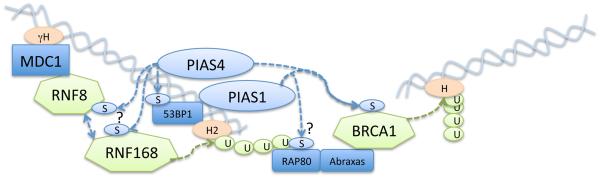
Figure 1. SUMO conjugation components regulate ubiquitin processes at chromatin around double-stranded DNA breaks.
The PIAS4 SUMO ligase is required for the accumulation of RNF168 and subsequent activity of RNF8:RNF168 ubiquitin ligases possibly through direct SUMOylation. 53BP1 SUMO conjugation depends on PIAS4.
The PIAS1 SUMO ligase is required for complete accumulation of BRCA1, possibly through RAP80, and PIAS1 and PIAS4 are both required for SUMOylation of BRCA1 which in turn increases its ubiquitin ligase activity.
Ubiquitin ligases are shown as green diamonds, and ubiquitin as green circles labelled “U”. SUMO ligases are shown as light blue ovals and SUMO as small light blue ellipses labelled “S”. Chromatin components are in orange.
References
- 1.Drikos I, Nounesis G, Vorgias CE. Characterization of cancer-linked BRCA1-BRCT missense variants and their interaction with phosphoprotein targets. Proteins. 2009;77:464–76. doi: 10.1002/prot.22460. [DOI] [PubMed] [Google Scholar]
- 2.Williams RS, Chasman DI, Hau DD, Hui B, Lau AY, Glover JN. Detection of protein folding defects caused by BRCA1-BRCT truncation and missense mutations. J Biol Chem. 2003;278:53007–16. doi: 10.1074/jbc.M310182200. [DOI] [PubMed] [Google Scholar]
- 3.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–48. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan J, Jetten AM. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett. 2008 doi: 10.1016/j.canlet.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Bartek J, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol. 2010;12:80–6. doi: 10.1038/ncb2008. sup pp 1-12. [DOI] [PubMed] [Google Scholar]
- 6.Xia Y, Pao GM, Chen HW, Verma IM, Hunter T. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J Biol Chem. 2003;278:5255–63. doi: 10.1074/jbc.M204591200. [DOI] [PubMed] [Google Scholar]
- 7.Morris JR, Solomon E. BRCA1 : BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet. 2004;13:807–17. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- 8.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–75. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Polanowska J, Martin JS, Garcia-Muse T, Petalcorin MI, Boulton SJ. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. Embo J. 2006;25:2178–88. doi: 10.1038/sj.emboj.7601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solomon E, Morris JR. Recent advances in understanding the cellular functions of BRCA1. In: Welcsh PL, editor. The Role of Genetics in Breast and Reproductive Cancers. Springer Springer Science + Business Media and Humana Press; New York, NY, USA: 2009. Chapter 4. [Google Scholar]
- 11.Joukov V, Chen J, Fox EA, Green JB, Livingston DM. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc Natl Acad Sci U S A. 2001;98:12078–83. doi: 10.1073/pnas.211427098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris JR, Pangon L, Boutell C, Katagiri T, Keep NH, Solomon E. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum Mol Genet. 2006;15:599–606. doi: 10.1093/hmg/ddi476. [DOI] [PubMed] [Google Scholar]
- 13.Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, Vidal M. BRCA1/BARD1 Orthologs Required for DNA Repair in Caenorhabditis elegans. Curr Biol. 2004;14:33–9. doi: 10.1016/j.cub.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 14.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–90. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 15.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–9. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–4. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 17.Dunnebier T, Bermejo JL, Haas S, Fischer HP, Pierl CB, Justenhoven C, et al. Common variants in the UBC9 gene encoding the SUMO-conjugating enzyme are associated with breast tumor grade. Int J Cancer. 2009;125:596–602. doi: 10.1002/ijc.24286. [DOI] [PubMed] [Google Scholar]
- 18.Dunnebier T, Bermejo JL, Haas S, Fischer HP, Pierl CB, Justenhoven C, et al. Polymorphisms in the UBC9 and PIAS3 genes of the SUMO-conjugating system and breast cancer risk. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0530-y. [DOI] [PubMed] [Google Scholar]
- 19.Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR, et al. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood. 2009 doi: 10.1182/blood-2009-03-211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris JR. SUMO in the mammalian response to DNA damage. Biochem Soc Trans. 2010;38:92–7. doi: 10.1042/BST0380092. [DOI] [PubMed] [Google Scholar]