Abstract
Since the first demonstration of the protective effects of vaccinia inoculation, vaccination has been one of the medicine’s greatest successes. The design of vaccines against viral disease has evolved considerably over the last 50 years. Classically attenuated viruses, those created by passaging a virus in cultured cells, have proven to be an effective means for preventing many viral diseases, including smallpox, polio, measles, mumps, and yellow fever. However, empiric attenuation is not a reliable approach for all viruses and there are a number of safety issues associated with the use of live, attenuated viruses (LAVs). While inactivated viruses and subunit vaccines alleviate many of these concerns, they have generally been less efficacious than their LAV counterparts. Advances in molecular virology have provided new ways of controlling viral replication and virulence, renewing interest in LAV vaccines. These rationally attenuated viruses may lead to a new generation of safer, more widely applicable LAV vaccines. Here, we review several new approaches to viral attenuation and vaccine design, including deleterious gene mutation, altered replication fidelity, codon deoptimization, and control by microRNAs or zinc finger nucleases. While each of these approaches has garnered significant attention in recent months, they are still in their infancy and require further in vitro and animal testing before progressing to clinical trials.
Introduction
The basic goal of vaccination is to stimulate protective immunity while avoiding disease from the vaccine itself. The first generation of viral vaccines relied on empiric attenuation by repeated passage in cultured cells. Several live, attenuated viruses (LAVs) meet both criteria for vaccines; they elicit a strong and protective immune response with a low risk of disease from the vaccine itself. Despite recent successes in the development of LAV for rotavirus and several arboviruses, the classic attenuation process is somewhat unpredictable and has not always been applicable. In the present regulatory environment the use of LAVs has also been limited by safety concerns, including reversion to wild-type virulence. Because LAVs are shed from vaccinees, they sometimes present a risk to unvaccinated individuals with impaired immunity.
These safety concerns have led to a shift toward the use of inactivated viruses or viral subunits as vaccines. Despite notable successes like the inactivated poliovirus vaccine 1, inactivated viruses are generally less immunogenic than their LAV counterparts, and this strategy is limited to viruses for which there are good culture and production systems. Subunit vaccines, which use viral proteins as immunogens, have become a major focus of vaccine development and have led to several successfully licensed vaccines, including vaccines against hepatitis B, influenza viruses, and papillomaviruses 2. Production is more easily controlled and efficient than that of LAVs or inactivated viruses. However, this strategy has not achieved universal success, as many subunit vaccines have failed to elicit a protective immune response in the host. While adjuvants have increased the immunogenicity of subunit vaccines, newer methods of subunit delivery mimic a natural immune response by incorporating more viral components. There are a number of approaches to this end including liposome delivery of antigens 3, 4, virus-like particles (VLPs) 5, and virosomes, which are reconstituted viral envelopes lacking any viral genetic material 6. Another approach to increase the immunogenicity of subunit vaccines is to recombinantly encode a pathogenic antigen in a non-pathogenic, yet infectious, poxvirus or adenovirus vector 7, 8. While there have been some notable successes, the major concern with this strategy is that the vector vaccines will not induce adequate immunological responses in hosts who have pre-existing antibodies against the vector.
A less live vaccine – rational attenuation through deletion or mutation
With this shift towards multi-subunit and vector designs, the vaccine field has accepted that more viral components are required for improved efficacy. As vaccines become more complex and “virus-like,” it is not surprising that live, attenuated vaccines have received a second look. Advances in molecular virology and the advent of recombinant virus systems have led to the identification of many viral genes associated with virulence and immunogenicity. Researchers have used this information to better control the replication and pathogenesis of vaccine candidates, thereby avoiding the unpredictability of empiric attenuation.
The identification of genes essential for viral replication and assembly led to the first generation of rationally designed, live virus vaccines (Table 1). Deletion or mutation of these genes results in a “defective virus,” which cannot replicate in the host (for an excellent review of defective virus vaccines, see Dudek and Knipe 8). These defective viruses are propagated in “helper” cells that express the missing gene(s). Though the virus is unable to replicate its genome, viral genes are still expressed, which can induce a strong immune response in the inoculated host. “Single cycle viruses,” which are defective in a viral protein required for assembly or spread, are a variation on this theme. While these viruses can replicate their genome through a single cycle, they do not produce infectious virus 9.
Table 1.
Current Vaccine Strategies
| Vaccine Approach |
Construction | Safety |
|---|---|---|
| Empirically attenuated virus |
Blind passage in different cell types. By adapting to a new environment, the virus accumulates mutations that mediate attenuation. |
Host immunity is able to limit that virulence and spread of the attenuated virus |
| Inactivated virus | Virus is inactivated by chemical treatment (e.g. formaldehyde). |
Disruption of viral proteins and/or genetic material. |
| Subunit Vaccine | Recombinant expression of one or several viral proteins. |
No viral genetic material is included. |
| Viral vectors | One or several genes from a virus are inserted into the genome of a second nonpathogenic virus (the vector). Viral particles produced by the vector transduce these genes into target cells and direct their expression. |
The vector itself is attenuated (see above), but is able to express antins derived from the pathogenic virus. |
| Replication Defective Viruses |
One or several genes required for genome replication are deleted in the vaccine strain. The virus vaccine is produced in a helper cell line that expresses the missing protein(s) in trans. |
The administered virus is unable to replicate its genome. |
| Single Cycle Viruses |
One or several genes required for viral assembly and spread are deleted in the vaccine strain. Distinguished from replication defective viruses by their competence for genome replication. |
The virus is able to replicate its genome, but is defective for assembly or spread. |
The first example of a replication-defective virus used as a vaccine was an HSV-1 strain with a deletion of a gene essential for genome replication 10. This virus stimulated an immune response similar to natural infection and protected against wild-type virus challenge in a mouse model of infection 11. Replication-defective HSV-2 strains, which lack genes essential for viral DNA synthesis (UL5) and viral replication (UL29), have also been described 12. These viruses were more effective than subunit vaccines in eliciting protective immunity in mice 13 and did not establish latency 14, an important consideration in herpesviruses. HSV-1 and HSV-2 strains have also been created that lack glycoprotein H and are unable to spread from cell to cell or produce infectious progeny. These single-cycle viruses protect against wild-type challenge in rodent models 15, 16, but the block in viral spread may be leaky. Similar strategies are now being applied to viruses other than HSV. The newer smallpox vaccines are replication-defective viruses 8, and an influenza NS-2 knockout and HA cleavage site mutants were shown to provide protective immunity in mice 17, 18. Likewise, flaviviruses with a deletion in the C protein function as single-cycle viruses as they cannot spread between cells or encapsidate virus. These viruses can elicit a potent immune response and protect against wild-type challenge 19.
Even with progress in the attenuation of viruses by deleterious gene mutation, this approach has not led to a safe and effective vaccine for human disease. While this can be attributed to the relatively short time this field has been in existence, vaccines based on deleterious gene mutation often evoke only a weak immune response because the antigen is only expressed at the site of inoculation. There are also safety concerns about the completeness of the block in viral spread in single-cycle viruses 19. Like conventional LAVs, it has proven very difficult to balance immunogenicity with safety, even with the rational design of replication-defective viruses. Over the last several years, investigators have taken advantage of recent advances in molecular virology and developed rational approaches to viral attenuation. In the following sections, we review four new methods - altered replication fidelity, codon deoptimization, and control by microRNAs or zinc finger nucleases. These novel LAV designs each allow for limited viral replication and antigen production. Because the host immune response is not required to limit viral spread, these LAVs may be safer than classical LAVs, even in immunocompromised patients.
Riboviral replication fidelity – failure then success
While LAV vaccines have been developed for many RNA viruses, the mutability of these pathogens presents unique challenges for vaccine design. The RNA-dependent RNA polymerases (RdRp) of RNA viruses exhibit characteristically low fidelity with measured mutation rates of 10−3 to 10−5 mutations per nucleotide copied per replication cycle 20. These mutation rates are orders of magnitude greater than those of nearly all DNA-based viruses and organisms. Because the genomes of RNA viruses are typically < 10,000 nucleotides, this mutation rate translates to roughly 0.1–10 mutations per genome replicated. It has been estimated that every possible point mutation and many double mutations are generated with each viral replication cycle and may be present within the population at any time 21. This impressive diversity has important biological implications. First, low frequency variants within the population may contain, or quickly acquire, mutations in key epitopes, which mediate escape from vaccine-elicited neutralizing antibody or cytotoxic T cells 22. Antigenic drift within the hemagluttinin and neuraminidase proteins of influenza virus is the best example of this process and the primary reason for annual reevaluation of vaccine strains 23. Second, many RNA viruses, including the human immunodeficiency virus (HIV) and hepatitis C virus (HCV), exhibit such dramatic intra- and interindividual genetic diversity, that it has been difficult to identify stable, conserved epitopes that provide universal protection against all strains 22. Finally, the mutability of RNA viruses has triggered real concerns about the potential reversion of live, attenuated vaccines to pathogenic strains. Both mutation and recombination are likely to play a role in this process, and the sporadic emergence of vaccine-derived polioviruses is a cautionary tale 24.
A large body of work in recent years suggests that because of their mutation rates, the evolution of RNA viruses may differ fundamentally from that of DNA-based organisms. Much of this work builds on the mathematical framework of quasispecies theory and seeks to understand the importance of genetic diversity at the population level 25. According to quasispecies theory, the mutation rates of RNA viruses place them near a critical “error threshold.” Below this threshold, the mutant spectrum within the population favors adaptability, and low fitness variants are tolerated so long as the majority remains viable. Beyond the error threshold, too many mutations accumulate, genetic information is lost, and the population becomes inviable 26. Indeed, mutagenic nucleosides increase viral mutation rate and cause population collapse, thus providing an effective treatment for several RNA viruses. However, several groups have identified mutants in poliovirus and foot and mouth disease virus (FMDV) that were resistant to nucleoside analogues 27–29. Further studies revealed that these variants replicated with higher fidelity by virtue of mutations within the viral RNA-dependent RNA polymerase. As a result, drug resistant mutants gave rise to populations with significantly less genetic diversity than wild type 30. Significantly, this decrease in diversity was responsible for attenuation in a transgenic mouse model of infection 30, 31.
We hypothesized that the observed attenuation of these high fidelity variants could be exploited for vaccine design 32. We focused on glycine 64 of the poliovirus polymerase, which regulates fidelity through a complex hydrogen bond network and mediates sensitivity to nucleoside analogues 27. Of the 19 possible amino acid substitutions at this position, only 13 gave rise to viable virus, and 8 of these were unstable. The other 5 mutants had lower mutation rates than wild type and were less adaptable in cell culture. In the transgenic mouse model, these high fidelity variants were markedly attenuated and shed less efficiently than wild type. Three of the viruses stimulated high titers of neutralizing antibody in infected mice, an order of magnitude greater than the Sabin 1 vaccine strain. They also induced long-lasting immunity. Mice vaccinated with G64S, G64A, or G64L survived a lethal challenge of wild type virus at 1 or 6 months via the intraperitoneal or intramuscular route.
This work suggests that controlling replication fidelity is a promising approach for engineering live, attenuated vaccines. However, several important questions remain. While it is clear that such a strategy could be successful for other picornaviruses, which have structurally conserved polymerases, it may be difficult to identify the relevant residues in other viral RdRp. In these cases, selection for nucleoside analogue resistance may be an unbiased way of discovering promising mutants for further characterization. Reversion to wild type is another potential problem, since viruses containing the lower fidelity wild type polymerase appear to have a selective advantage. Although high fidelity variants would certainly revert at a lower frequency, their mutation rate is still significantly higher than that of DNA viruses 20, 30. We found no evidence for reversion of the G64S mutation after either twenty passages in HeLa cells or five mouse to mouse passages over twenty-five days 32. While encouraging, further experiments along these lines will likely be required prior to regulatory approval. Finally, the mouse model for poliovirus pathogenesis is an imperfect one and the level of attenuation observed here may not reflect the situation in human vaccinees. Nevertheless, the high fidelity variants could still be useful in the ongoing polio eradication campaign, as safer seed strains will be needed for large scale production of the inactivated polio vaccine in a post-polio world 33.
Attenuation by a thousand cuts
It is well known that many organisms exhibit a codon bias, using some synonymous codons or codon pairs more frequently than others 34. In bacteria and simple eukaryotes, codon preference is related to levels of the corresponding tRNA and affects translational efficiency 35, 36. The reasons for the observed codon bias are less clear in mammals. Because viruses rely on the host cell machinery for nearly all aspects of replication, it is not surprising that codon bias has been described in many viral genomes. In bacteriophage, codon usage closely mirrors that of the host 37. The bias is more pronounced in the highly expressed structural genes, suggesting optimization for translational efficiency 38, 39. Most mammalian viruses also have a strong preference against CpG dinucleotides, although their overall GC content is highly variable 38. Studies of HIV and influenza suggest that codons in highly variable surface proteins may be optimized for their volatility, the probability that a codon will mutate to a different amino acid class 40, 41. This would presumably facilitate immune escape and suggests that there has been selection for genetic plasticity in these highly mutable viruses.
Recent studies of poliovirus have addressed the importance of codon bias for viral replication and pathogenesis. Burns and colleagues performed large-scale mutagenesis of the Sabin 2 vaccine strain, replacing up to 50% of the capsid codons with synonymous codons that are less preferred in the human genome 42. While these codon-deoptimized viruses exhibited minimal defects in viral gene expression, they produced fewer infectious progeny and overall fitness was markedly reduced. A subsequent study found synonymous changes that increased the frequency of CpG and UpA dinucleotides had similar effects on viral fitness 43. Mueller and colleagues took a similar approach, but used gene synthesis technology to design poliovirus genomes with completely deoptimized codons in the capsid region 44. They also found a dramatic reduction in replicative fitness and a reduction in infectious progeny. However, their data strongly suggested that deoptimized viruses had reduced translational efficiency compared to wild type. They obtained similar results with viruses in which synonymous changes were determined by codon pair bias. In both cases, they found that deoptimized polioviruses were attenuated by 1000 fold on a per particle basis compared to wild type.
Because all changes are synonymous, the proteins expressed from codon-deoptimized viruses are identical to wild type and similarly immunogenic. Mueller and colleagues, therefore, hypothesized that their marked attenuation would make them ideal live vaccines 45. In their second study, they showed that deoptimized viruses provoked a robust neutralizing antibody response following three weekly intraperitoneal inoculations. All immunized mice survived subsequent lethal challenge with wild type poliovirus, demonstrating the vaccine efficacy of the engineered viruses. As a general strategy for vaccine development, codon deoptimization offers several advantages. First, attenuation does not affect antigenicity, and the immune response should closely mimic a natural infection. Second, because attenuation is systematic rather than empiric, it may be easily applied to other viruses. Finally, codon deoptimized viruses encode hundreds of point mutations, each with a fairly small individual effect on fitness. Consequently, there is little risk of reversion to virulence with even a handful of point mutations. Both Mueller and Burns found that codon deoptimized viruses are genetically stable and remain attenuated after repeated passage 42, 44. Their marked sequence divergence from circulating strains may also reduce the frequency of recombination and the risk of pathogenic, vaccine-derived variants.
Much work remains to be done before codon deoptimized viruses are employed as live, attenuated vaccines. While the results among the studies are consistent, the mechanism of attenuation is still debated. This would certainly be an issue for regulatory bodies, and the lack of clarity makes if difficult to determine whether codon-based attenuation is a unique aspect of picornaviruses, or a more generalizeable approach to vaccine design. As in the case of the high fidelity variants, the mouse model may not be the best system for assessing vaccine efficacy and safety. Nevertheless, codon deoptimization is a promising approach that has already generated significant interest in the virology community.
MicroRNA-controlled LAVs
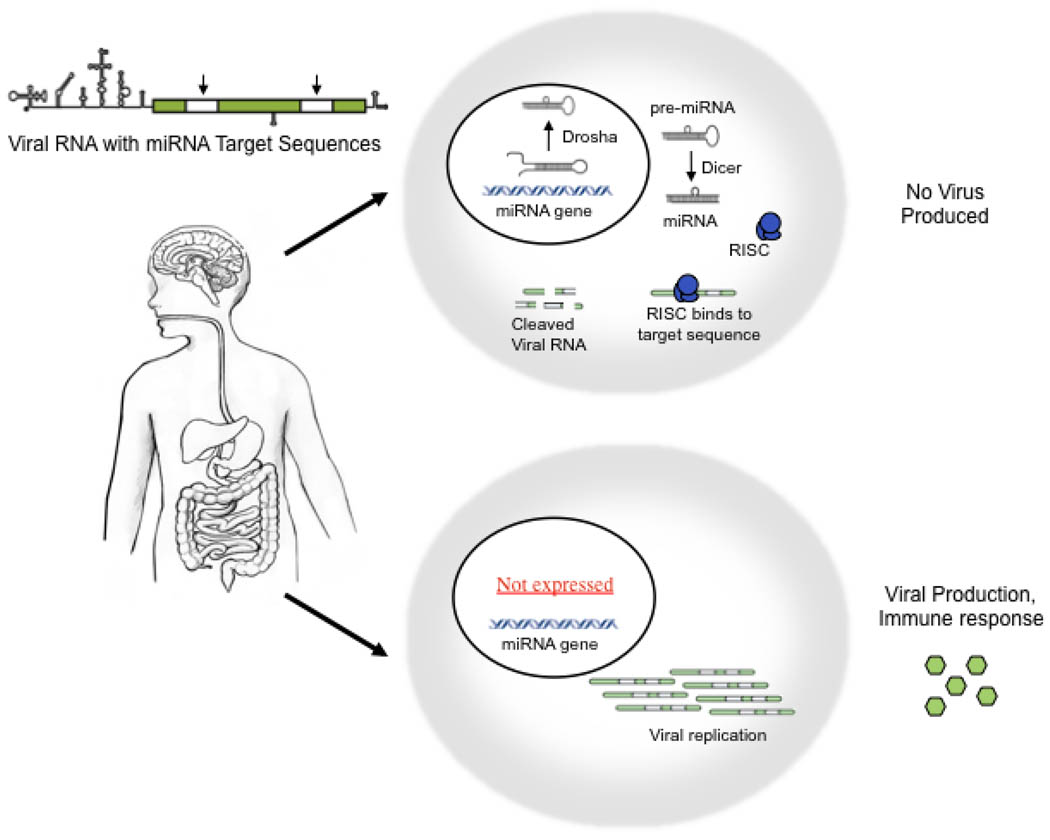
Since the discovery of RNA interference (RNAi) just a decade ago, there has been an explosion of research into this novel form of gene regulation. The two main effectors of RNAi are small interfering RNA (siRNA) and microRNA (miRNA) 46. While there has been intense interest in using siRNAs to combat mammalian RNA viruses 47, miRNAs are now also being used to limit viral pathogenesis. MicroRNA are genomically encoded and play a major role in endogenous gene regulation 48. They are transcribed as long precursor pri-miRNA, which are processed by the nuclear ribonuclease Drosha to ~ 60 nucleotide hairpin intermediates, which are then transported to the cytoplasm where they are trimmed by Dicer to roughly ~ 22 nucleotides (Figure 1). Like siRNAs, mature miRNA are loaded into the RNA-induced silencing complex (RISC), where they mediate either degradation or translational repression of target messages. The human genome encodes over 400 miRNA, many of which have tissue-specific or developmental expression patterns. Several DNA viruses also express miRNA 49. These virally derived miRNA modulate pathogenesis and host immunity through regulation of viral and cellular transcripts, respectively.
Figure 1. miRNA-virus vaccine strategy.
Genes coding for one or more microRNA are transcribed as long precursor pri-miRNA, which are processed by the nuclear ribonuclease Drosha to ~ 60 nucleotide hairpin intermediates. These small RNA are transported to the cytoplasm where they are trimmed by Dicer to roughly ~ 22 nucleotides. Mature miRNA are loaded into the RNA-induced silencing complex (RISC), where they mediate either degradation or translational repression of target messages. Viral replication can be regulated in a tissue specific manner by incorporating miRNA target sites into the viral genome. Viral RNA are cleaved in cells expressing the corresponding miRNA (e.g. brain, top cell), and viral production is restricted to cells in which the miRNA is not expressed (e.g. intestine, bottom cell). The engineered virus can therefore trigger a natural immune response in target tissues without the associated risk of dissemination and disease.
The diversity and complexity of cellular miRNA means that many cell types will have a unique miRNA profile 50. Several investigators have taken advantage of this property to better target viral gene therapy vectors 51. Silencing of specific transcripts or the entire genome can be accomplished by inclusion of miRNA binding sites in the vector sequence. In many cases, the miRNA system is used to provide a second level of control beyond receptor expression or tissue-specific promoter activity. For example, Brown and colleagues eliminated off-target expression from a hepatocyte specific promoter in antigen presenting cells by incorporating miR-142-3p binding sites in their lentiviral construct 52. In a related study, muscle-specific miRNA binding sites were used to limit secondary replication of a Coxsackie virus in a murine tumor model 53. Improved targeting of adenoviral vectors has also been achieved by the addition of miRNA binding sites to the 3’ untranslated region of the E1A transcript 54, 55.
In LAV design, empiric attenuation of viruses is often accomplished by changing the tissue tropism of a virus through repeated passage in a new cell type. We hypothesized that the same result could be achieved through miRNA restriction of poliovirus replication 56. While poliovirus replicates in many tissues, disease onset is linked to lytic infection of the central nervous system. By incorporating binding sites for either let7a (a ubiquitous miRNA) or miR124 (a CNS restricted miRNA) into the RNA genome of wild type poliovirus (Figure1), we showed that viral replication was restricted in a cell-type dependent manner and that the effect was dependent on the cellular RNAi machinery. The miRNA-targeted viruses were largely restricted from the central nervous system in a murine model of infection, and markedly attenuated as a result 56. Experiments with viruses containing mutant target sequences confirmed that the altered tropism was due to miRNA. The degree of attenuation exceeded 5 orders of magnitude and neither let7a- nor miR124-targeted viruses were pathogenic in immunocompromised mice lacking the alpha/beta interferon receptor 56. Both viruses were able to replicate in non-neuronal tissues and stimulated a strong neutralizing antibody response after a single intraperitoneal inoculation. The level of protection was impressive, as even the interferon receptor knockout mice were protected from subsequent challenge with 10,000 times the lethal dose of wild type virus 56.
While miRNA targeting is a promising approach to rational design of LAV, the study has several caveats worth mentioning. The let7a virus replicated poorly in most tissues, while the mir124 virus was restricted only in the central nervous system 56. As a result, the former stimulated a weaker immune response and was a less effective vaccine. On the other hand, widespread replication of the mir124 virus in non-neuronal tissues could allow the virus to accumulate mutations within the miRNA target sequence and thereby escape degradation. Indeed, several mice in the study had low titers of mir124 virus in the spinal cord, and sequence analysis showed mutations within the miRNA target sequences 56. Work from our laboratory suggests that a single let7a site can accumulate escape mutations in as little as 24–48 hours 32. The risk of miRNA-escape could be minimized by the inclusion of multiple target sequences for the same miRNA or different miRNA with the same tissue distribution. Another way of minimizing escape was highlighted in a subsequent paper on species-specific restriction of influenza virus for vaccine production 57. In this study, Perez and colleagues incorporated nonavian miRNA target sequences into a region of the viral nucleoprotein open reading frame. Because the miRNA target sequence also served as codons for conserved amino acids, escape mutations would alter protein structure and likely have a deleterious effect on viral replication. We expect that as the RNAi field matures, investigators will find other ways of controlling the replication and mutability of miRNA-targeted vaccines, though the potential for reversion to wild type will have to be mitigated to the satisfication of regulatory bodies.
Zinc finger nuclease-controlled LAVs
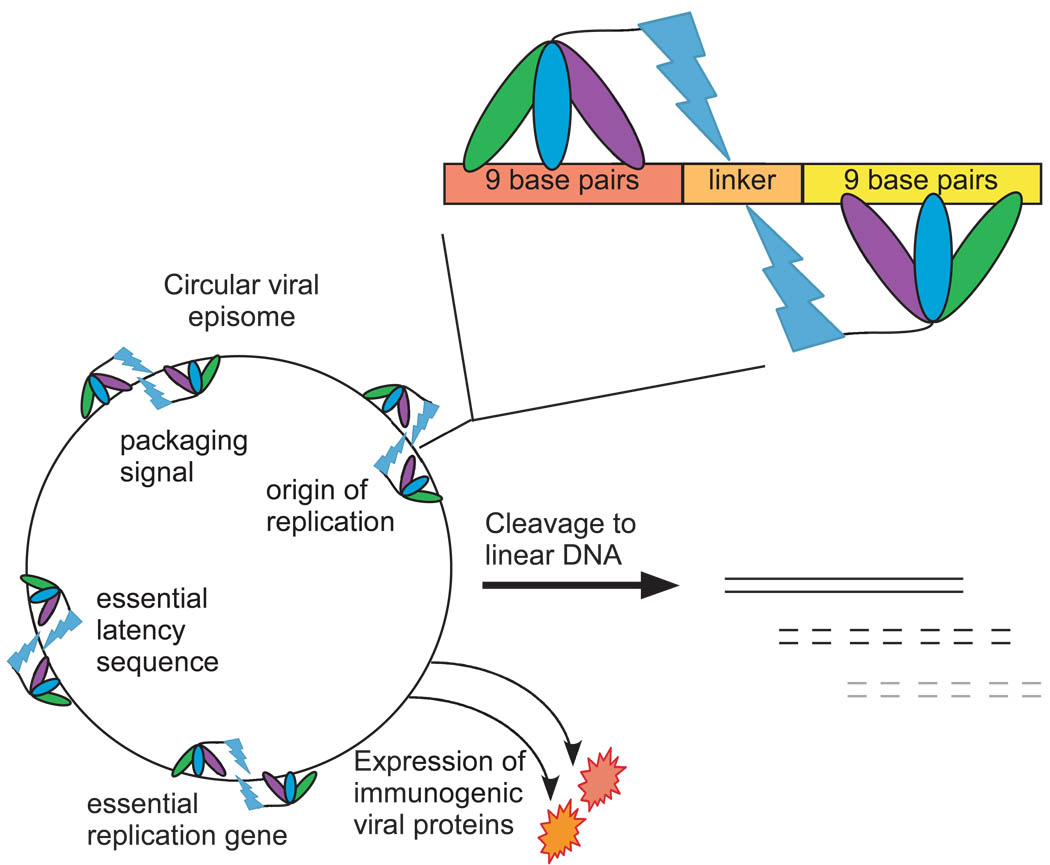
Zinc finger (ZF) domains mediate nucleotide-specific binding of proteins to DNA, a property that defines a large family of DNA binding proteins 58. Each finger makes contact with a separate DNA triplet, and natural or recombinant ZFs have been created that can recognize almost any triplet 59. The modular nature of the ZFs allows them to be joined in useful combinations. Typically, three ZFs are combined to bind to a specific 9-bp DNA sequence, and these ZFs have been coupled to various functional domains to create artificial transcription factors that can activate or repress gene transcription with remarkable promoter specificity 60. Zinc fingers have also been fused to the nuclease domain of the restriction enzyme FokI to cleave double-stranded DNA at specific sequences 61. The nuclease domain must dimerize to cleave DNA, and because the dimer interface is weak, two nuclease domains are typically brought into close proximity by pairs of ZFs binding to neighboring 9-bp sites, spaced 6-bp apart (Figure 2) 62, 63. In this configuration, the engineered ZF nuclease (ZFN) recognizes a specific 18-bp sequence, which is long enough, by a few orders of magnitude, to be unique in the human genome. Because of this specificity, this same technology could be used to distinguish between human and virus DNA.
Figure 2. ZFN-virus vaccine strategy.
Zinc finger nucleases use an array of three zinc finger (ZF) domains to recognize specific 9bp sequences in the virus genome. The ZF array is fused to DNA nuclease domain (lightning bolt) to create the zinc finger nuclease (ZFN). This nuclease is only active upon dimerization. A pair of ZFNs can be designed to bind 9bp sequences, spaced 5–6bp apart, to bring the nuclease domains close enough to dimerize, thus cleaving the double-stranded DNA sequence. ZFNs can be designed that target multiple, essential viral sequences, such as the origin of replication, the viral DNA packaging signal, sequences essential for establishment and maintenance of latency, and genes essential for viral replication. These ZFNs can be encoded in the viral genome itself using recombinant techniques. The expression of the ZFNs can be temporally controlled using viral promoters to allow a balance between expression of immunogenic viral proteins and cleavage of circular episomal DNA to linear DNA. This linear DNA is incapable of replication and establishment of latency. Thus, a ZFN-virus vaccine can elicit an immune response equal to that of the parental virus, but can limit its own replication and latency, without the need for a competent immune system.
Several groups have used recombinant ZF proteins to control aspects of the viral life cycle. ZF proteins fused to the KOX-1 repression domain were created that targeted the HSV-1 ICP4 promoter 64. These proteins bound the promoter with nanomolar affinity, and one was able to significantly repress VP16-activated transcription in vitro. This ZF-KOX-1 fusion, when delivered in trans into HSV-1 infected cells, was able to limit HSV-1 replication and reduced viral titer by 90%. In a similar strategy, recombinant ZF proteins were designed to recognize the HPV-18 replication origin 65. When expressed in vitro, these ZFs were able to compete with the replication protein E2 for binding to viral DNA. This competitive antagonism led to reduced HPV replication in transient replication assays in mammalian cells. By fusing the origin-targeted ZF protein to a nuclease domain this ZFN was able to cleave viral DNA and reduce viral replication in cultured cells 66. These experiments demonstrate that ZFN can effectively target and eliminate viral DNA in mammalian cells.
It may be feasible to deliver a therapeutic virus-specific ZFN in trans to eradicate latent viral DNA. However, delivery of the ZFNs to all latently infected cells is technically challenging. Alternatively, virus-specific ZFNs could be delivered using the viral genome itself and serve as a vaccine. In the ZFN-vaccine strategy, ZFNs targeting sequences for viral replication and other essential viral processes would be introduced into the viral genome (Figure 2). Following inoculation, immunogenic viral genes and virus-specific ZFNs would be expressed. While the viral proteins would stimulate a natural immune response, the ZFNs would cleave viral DNA, and limit replication. ZFN-LAVs have potential both as prophylactic vaccines, protecting against wild-type challenge, as well as therapeutic vaccines, delivering ZFNs to cells already harboring latent viral DNA.
The immunogenicity of ZFN vaccines can be controlled by temporal and spatial regulation of ZF expression to balance viral protein expression with the ability of the ZFNs to eliminate all replication-competent viral DNA. This could best be accomplished using promoters that are temporally controlled by the virus itself. For instance, herpesvirus gene transcription occurs in at least three distinct stages; immediate-early (before most of viral protein synthesis), early (before viral replication), and late (after viral replication begins) 67. Other DNA viruses for which ZFNs would be useful are similarly regulated. There is also the potential to encode ZFNs behind inducible promoters, so that ZFN expression would commence upon the administration of a small molecule 68. Nuclease activity can also be controlled directly by addition of small molecule-sensitive residues to the ZFN 69. These strategies would provide an ideal way to optimize the balance between ZFN-virus replication and nuclease activity.
The ability to create a ZFN-vaccine that can prevent and eliminate persistent viral infections is a long way from being realized. As with any LAV, safety issues are always a concern. The ZFN vaccine approach would likely be limited to non-integrating, DNA viruses, as random breaks in host chromosomal DNA caused by ZFN-cleavage of integrated viral DNA could be catastrophic. There are many non-integrating human viruses, includes the herpes-, polyoma-, adeno-, and papillomaviruses, that establish a persistent infection and provide particularly difficult challenges for the treatment of their respective diseases. ZFN-based vaccines may offer a way to prevent or eliminate these hard-to-treat latent infections. Reversion to wild type is another concern, but the risk can be reduced by including ZFNs against multiple, essential viral sequences to ensure that the intrinsic mutation rate of the virus will not allow the mutation of every ZFN target site. It is also possible that DNA cleaved by ZFNs could be repaired via homologous recombination using uncleaved viral genomes. However, if the sequence were repaired accurately, it would be subject to repeated cleavage; if it is repaired inaccurately, the virus should not be viable due to mutation of an essential sequence. Ideally, we will arrive at a live virus strain that will have limited replication, not establish latency, and elicit a protective immune response. In essence, we would turn an otherwise detrimental latent infection into an asymptomatic, acute infection.
Conclusions
LAV vaccines have provided ideal protection from several major diseases, but have not lived up to their potential due to limited applicability and safety concerns. Advances in molecular biology have opened the door to novel approaches to viral attenuation and may lead to a new generation of safer LAVs (Table 2). Though replication-defective LAVs have encountered some problems, this approach to attenuation is on the cusp of providing safe, effective vaccines for several diseases. Several other approaches to attenuation are poised to overcome other problems specifically associated with vaccine design for RNA and DNA viruses. For many RNA viruses where high mutation rates limit the efficacy of vaccines, altering the replication fidelity can attenuate the entire virus population, leading to population collapse without mutation of key immunogenic epitopes. Codon deoptimization provides a systematic means by which to attenuate any virus. By substituting synonymous codons throughout a viral genome, there is no loss of immunogenicity and little risk of reversion to wild type. Zinc finger nucleases and miRNAs can be used to control the replication of DNA and RNA viruses, respectively. By controlling viral replication temporally or spatially, a strong, natural immune response can be elicited before the virus is eliminated. These may be particularly useful approaches for designing vaccines against persistent or latent viruses, as ZFNs and miRNAs lead to the elimination of all viral DNA or RNA, thus preventing chronic infection.
Table 2.
Approaches to viral attenuation for vaccine design
| Vaccine Approach |
Advantages | Disadvantages | Examples |
|---|---|---|---|
| Empirically attenuated virus |
Excellent immunogenicity, few doses required |
Limited applicability, reversion to wild type, breakthrough disease |
Measles, mumps, rubella (MMR); Oral polioirus vaccine (OPV), Influenza, Rotavirus, Yellow Fever, Varicella |
| Subunit Vaccine | Widely applicable, very safe |
Poor immunogenicity, multiple doses usually required |
Hepatitis B virus, Human papilloma virus |
| Viral vectors | Good immunogenicity, delivery of multiple antigens |
Neutralizing antibodies to vector, possible safety issues |
Many examples (experimental) |
| Defective Viruses |
Good immunogenicity, known mechanism of attenuation |
Limited to inoculation site, possible safety issues |
HSV-1, HSV-2. Influenza (experimental) |
| Replication Fidelity |
Strong immunogenicity, known mechanism of attenuation, not susceptible to antigenic shift/drift |
RNA viruses only, possible reversion to wild type |
Poliovirus (experimental) |
| Codon deoptimization |
Strong immunogenicity, no reversion to wild type, possibly applicable to many viruses |
Possible safety concerns | Poliovirus (experimental) |
| miRNA- controlled virus |
Strong immunogenicity, known mechanism of attenuation, prevent latent infection |
Limited to some RNA viruses |
Poliovirus, adenovirus, coxsackievirus, influenza (experimental) |
| ZFN-controlled virus |
Strong immunogenicity, known mechanism of attenuation, prevent latent infection |
Limited to non-integrating DNA viruses |
Each of these approaches is aimed to address long-standing problems with LAV vaccine design. While they could potentially change the way we think about attenuation, significant hurdles lie ahead. Live vaccines present an inherent trade-off between safety and efficacy, and regulatory bodies are right to be concerned about viral escape or reversion to wild type. The studies described here have largely been carried out in murine models with relatively short-term measures of immunogenicity and limited characterization of viral genetic stability. Much more work is needed in relevant animal models before contemplating an initial dosing and safety trial in humans. We expect that each strategy will need to be modified to optimize its safety and efficacy profile. Nevertheless, the efficacy demonstrated by available LAV, particularly the recent success in developing safe and effective live-attenuated rotavirus, influenza, and varicella zoster vaccines is a strong incentive to redouble efforts to improve the safety characteristics of this type of vaccine. Rational attenuation may also facilitate the development of inactivated vaccines for high-risk agents by providing safer seed stocks for large-scale production. The next several years will clearly be an exciting time in vaccine research as advances in molecular biology are further translated into preventive strategies for viral disease.
Acknowledgements
This work was supported by grants from the NIAID to RA (R01 AI36178 and R01 AI40085) and ASL (K08 AI081754-01).
References
- 1.Salk J. Polio vaccines and polioviruses. Br Med J. 1977;2:765. doi: 10.1136/bmj.2.6089.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvin AM, Greenberg HB. New viral vaccines. Virology. 2006;344:240–249. doi: 10.1016/j.virol.2005.09.057. [DOI] [PubMed] [Google Scholar]
- 3.Rogan D, Babiuk LA. Novel vaccines from biotechnology. Rev Sci Tech. 2005;24:159–174. [PubMed] [Google Scholar]
- 4.Kersten GF, Crommelin DJ. Liposomes and ISCOMs. Vaccine. 2003;21:915–920. doi: 10.1016/s0264-410x(02)00540-6. [DOI] [PubMed] [Google Scholar]
- 5.Jennings GT, Bachmann MF. The coming of age of virus-like particle vaccines. Biol Chem. 2008;389:521–536. doi: 10.1515/bc.2008.064. [DOI] [PubMed] [Google Scholar]
- 6.Felnerova D, Viret JF, Gluck R, Moser C. Liposomes and virosomes as delivery systems for antigens, nucleic acids and drugs. Curr Opin Biotechnol. 2004;15:518–529. doi: 10.1016/j.copbio.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Panicali D, Paoletti E. Construction of poxviruses as cloning vectors: insertion of the thymidine kinase gene from herpes simplex virus into the DNA of infectious vaccinia virus. Proc Natl Acad Sci U S A. 1982;79:4927–4931. doi: 10.1073/pnas.79.16.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudek T, Knipe DM. Replication-defective viruses as vaccines and vaccine vectors. Virology. 2006;344:230–239. doi: 10.1016/j.virol.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 9.Loudon PT, et al. Preclinical safety testing of DISC-hGMCSF to support phase I clinical trials in cancer patients. J Gene Med. 2001;3:458–467. doi: 10.1002/jgm.206. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen LH, Knipe DM, Finberg RW. Replication-defective mutants of herpes simplex virus (HSV) induce cellular immunity and protect against lethal HSV infection. J Virol. 1992;66:7067–7072. doi: 10.1128/jvi.66.12.7067-7072.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison LA, Knipe DM. Mechanisms of immunization with a replication-defective mutant of herpes simplex virus 1. Virology. 1996;220:402–413. doi: 10.1006/viro.1996.0328. [DOI] [PubMed] [Google Scholar]
- 12.Da Costa X, Kramer MF, Zhu J, Brockman MA, Knipe DM. Construction, phenotypic analysis, and immunogenicity of a UL5/UL29 double deletion mutant of herpes simplex virus 2. J Virol. 2000;74:7963–7971. doi: 10.1128/jvi.74.17.7963-7971.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshino Y, et al. Comparative efficacy and immunogenicity of replication-defective, recombinant glycoprotein, and DNA vaccines for herpes simplex virus 2 infections in mice and guinea pigs. J Virol. 2005;79:410–418. doi: 10.1128/JVI.79.1.410-418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Da Costa XJ, Jones CA, Knipe DM. Immunization against genital herpes with a vaccine virus that has defects in productive and latent infection. Proc Natl Acad Sci U S A. 1999;96:6994–6998. doi: 10.1073/pnas.96.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLean CS, et al. Protective vaccination against primary and recurrent disease caused by herpes simplex virus (HSV) type 2 using a genetically disabled HSV-1. J Infect Dis. 1994;170:1100–1109. doi: 10.1093/infdis/170.5.1100. [DOI] [PubMed] [Google Scholar]
- 16.Farrell HE, et al. Vaccine potential of a herpes simplex virus type 1 mutant with an essential glycoprotein deleted. J Virol. 1994;68:927–932. doi: 10.1128/jvi.68.2.927-932.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe T, Watanabe S, Neumann G, Kida H, Kawaoka Y. Immunogenicity and protective efficacy of replication-incompetent influenza virus-like particles. J Virol. 2002;76:767–773. doi: 10.1128/JVI.76.2.767-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stech J. Attenuated influenza A viruses with modified cleavage sites in hemagglutinin as live vaccines. Expert Rev Vaccines. 2008;7:739–743. doi: 10.1586/14760584.7.6.739. [DOI] [PubMed] [Google Scholar]
- 19.Widman DG, Frolov I, Mason PW. Third-generation flavivirus vaccines based on single-cycle, encapsidation-defective viruses. Adv Virus Res. 2008;72:77–126. doi: 10.1016/S0065-3527(08)00402-8. [DOI] [PubMed] [Google Scholar]
- 20.Holland J, et al. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 21.Vignuzzi M, Stone JK, Andino R. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 2005;107:173–181. doi: 10.1016/j.virusres.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Walker B, Burton D. Toward an AIDS Vaccine. Science. 2008;320:760–764. doi: 10.1126/science.1152622. [DOI] [PubMed] [Google Scholar]
- 23.Couch RB. Seasonal inactivated influenza virus vaccines. Vaccine. 2008;26(Suppl 4):D5–D9. doi: 10.1016/j.vaccine.2008.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minor P. Vaccine-derived poliovirus (VDPV): Impact on poliomyelitis eradication. Vaccine. 2009;27:2649–2652. doi: 10.1016/j.vaccine.2009.02.071. [DOI] [PubMed] [Google Scholar]
- 25.Domingo E, et al. Viruses as quasispecies: biological implications. Curr Top Microbiol Immunol. 2006;299:51–82. doi: 10.1007/3-540-26397-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biebricher CK, Eigen M. The error threshold. Virus Res. 2005;107:117–127. doi: 10.1016/j.virusres.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Arnold JJ, Vignuzzi M, Stone JK, Andino R, Cameron CE. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J Biol Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci USA. 2003;100:7289–7294. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sierra M, et al. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J Virol. 2007;81:2012–2024. doi: 10.1128/JVI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- 33.Chumakov K, Ehrenfeld E. New generation of inactivated poliovirus vaccines for universal immunization after eradication of poliomyelitis. Clin Infect Dis. 2008;47:1587–1592. doi: 10.1086/593310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carbone A. Codon bias is a major factor explaining phage evolution in translationally biased hosts. J Mol Evol. 2008;66:210–223. doi: 10.1007/s00239-008-9068-6. [DOI] [PubMed] [Google Scholar]
- 38.Jenkins GM, Holmes EC. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/s0168-1702(02)00309-x. [DOI] [PubMed] [Google Scholar]
- 39.Shackelton LA, Parrish CR, Holmes EC. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J Mol Evol. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 40.Plotkin JB, Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc Natl Acad Sci USA. 2003;100:7152–7157. doi: 10.1073/pnas.1132114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens CR, Waelbroeck H. Codon bias and mutability in HIV sequences. J Mol Evol. 1999;48:390–397. doi: 10.1007/pl00006483. [DOI] [PubMed] [Google Scholar]
- 42.Burns CC, et al. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J Virol. 2006;80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burns CC, et al. Genetic Inactivation of Poliovirus Infectivity by Increasing the Frequencies of CpG and UpA Dinucleotides Within and Across Synonymous Capsid Region Codons. J Virol. 2009 doi: 10.1128/JVI.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2030;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 51.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 52.Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 53.Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 54.Cawood R, et al. Use of tissue-specific microRNA to control pathology of wild-type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog. 2009;5:e1000440. doi: 10.1371/journal.ppat.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ylösmäki E, et al. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific MicroRNA. J Virol. 2008;82:11009–11015. doi: 10.1128/JVI.01608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–248. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez JT, et al. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat Biotechnol. 2009;27:572–576. doi: 10.1038/nbt.1542. [DOI] [PubMed] [Google Scholar]
- 58.Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright DA, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 60.Dhanasekaran M, Negi S, Sugiura Y. Designer zinc finger proteins: tools for creating artificial DNA-binding functional proteins. Acc Chem Res. 2006;39:45–52. doi: 10.1021/ar050158u. [DOI] [PubMed] [Google Scholar]
- 61.Porteus M. Design and testing of zinc finger nucleases for use in mammalian cells. Methods Mol Biol. 2008;435:47–61. doi: 10.1007/978-1-59745-232-8_4. [DOI] [PubMed] [Google Scholar]
- 62.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mani M, Smith J, Kandavelou K, Berg JM, Chandrasegaran S. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem Biophys Res Commun. 2005;334:1191–1197. doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Papworth M, et al. Inhibition of herpes simplex virus 1 gene expression by designer zinc-finger transcription factors. Proc Natl Acad Sci U S A. 2003;100:1621–1626. doi: 10.1073/pnas.252773399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mino T, et al. Inhibition of DNA replication of human papillomavirus by artificial zinc finger proteins. J Virol. 2006;80:5405–5412. doi: 10.1128/JVI.01795-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mino T, Mori T, Aoyama Y, Sera T. Inhibition of human papillomavirus replication by using artificial zinc-finger nucleases. Nucleic Acids Symp Ser (Oxf) 2008:185–186. doi: 10.1093/nass/nrn094. [DOI] [PubMed] [Google Scholar]
- 67.Fields BN, Knipe DM, Howley PM, Griffin DE. Fields virology. Edn. 4th. Philadelphia: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 68.Zhu Z, Zheng T, Lee CG, Homer RJ, Elias JA. Tetracycline-controlled transcriptional regulation systems: advances and application in transgenic animal modeling. Semin Cell Dev Biol. 2002;13:121–128. doi: 10.1016/s1084-9521(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 69.Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS Genet. 2009;5:e1000376. doi: 10.1371/journal.pgen.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]