Abstract
Tropomyosin is a stereotypical α-helical coiled-coil that polymerizes to form a filamentous macromolecular assembly that lies on the surface of F-actin. The interaction between the C-terminal and N-terminal segments on adjacent molecules is known as the overlap region. We report here two X-ray structures of the chicken smooth muscle tropomyosin overlap complex. A novel approach was used to stabilize the C-terminal and N-terminal fragments. Globular domains from both the human DNA ligase binding protein XRCC4 and bacteriophage φ29 scaffolding protein Gp7 were fused to 37 and 28 C-terminal amino acid residues of tropomyosin, respectively, whereas the N-terminal 29 amino acids of tropomyosin were fused to the C-terminal helix bundle of the microtubule binding protein EB1. The structures of both the XRCC4 and Gp7 fusion proteins complexed with the N-terminal EB1 fusion contain a very similar helix bundle in the overlap region which encompasses ~15 residues. The C-terminal coiled-coiled coil opens to allow formation of the helix bundle, which is stabilized by hydrophobic interactions. These structures are similar to that observed in the NMR structure of the rat skeletal overlap complex [Greenfield et al., (2006) J. Mol. Biol. 364, 80–96]. The interactions between the N- and C-terminal coiled-coils of smooth muscle tropomyosin show significant curvature, which differs somewhat between the two structures and implies flexibility in the overlap complex, at least in solution. This is likely an important attribute that allows tropomyosin to assemble around the actin filaments. These structures provide a molecular explanation for the role of N-acetylation in the assembly of native tropomyosin.
Tropomyosin was one of the first molecules to be classified as an α-helical coiled-coil and is often considered to be a stereotypical example of this structural motif (1, 2). As such, it might appear to be a simple molecule, but this belies the complexity of its biological function and the subtlety of its structure (3–5). Tropomyosin was first isolated as a component of skeletal muscle, in which it plays a key regulatory function in muscle contraction, but it is widely distributed in non-muscle eukaryotic cells (6). In non-muscle cells the different tropomyosin isoforms contribute to the modification of actin filament assembly and disassembly rates (7–9). There are more than 40 isoforms of tropomyosin found in mammals that are all associated with filamentous actin. These isoforms arise from alternative splicing of four genes (10, 11).
Tropomyosin assembles head-to-tail along the length of actin filaments following the long-pitch helical path of the F-actin subunits. Based on the length of the intact molecule in crystals or paracrystals, it was estimated that two molecules of tropomyosin overlap by approximately nine residues (2, 12). The overlap region between the C-terminus of one tropomyosin dimer and the N-terminus of another plays an important role in the function of this molecular assembly. Indeed, removal of the terminal residues eliminates polymerization (13, 14). In skeletal muscle, the N-terminal section of troponin T binds to the overlap region between adjacent tropomyosin dimers (15) where the troponin complex controls the location of tropomyosin on the actin filaments and thus serves to restrict or allow myosin access to actin depending on the calcium level in the cell (3, 16, 17). Interestingly, the amino acid sequence of the tropomyosin overlap region is one of the areas of greatest variability between tropomyosin isoforms. This variability leads to the question of whether the overlap interaction is the same for all isoforms.
Considerable effort has been devoted to determining the structure of the overlap region between tropomyosin molecules where most of the studies have focused on the striated muscle isoform (18, 19). Deriving a structure of the overlap region has been difficult task because the interactions between individual molecules are very weak. This weakness is compensated for in nature by its interactions with and cooperative assembly on the filament of actin (20, 21). In vitro studies are also complicated because N-acetylation of the N-terminal methionine is necessary for coiled-coil stability, overlap formation and actin binding. This posttranslational modification is absent in E. coli (22, 23). Originally it was proposed that the N- and C-terminal coiled-coils of adjacent dimers might overlap side-by-side (2), but as high resolution structures of these N- and C-terminal regions have become available it has become clear that a different model is more likely. The structure of the N-terminal 81 amino acids exhibits a coiled-coil throughout its length (24). In contrast, the α-helices at the C-terminus of tropomyosin separate and do not maintain a coiled-coil (25, 26). Based on these observations, it was proposed that the coiled-coil of the N-terminus is inserted between the α-helixes of the C-terminus (25).
The first structure of an overlap complex was obtained by NMR for the rat skeletal isoform of tropomyosin (27). This demonstrated that both the N-and C-terminal coiled-coils melt slightly to allow interdigitation to form a symmetrical parallel coiled-coil four helical bundle. In this structure the coiled-coils overlap by 11 residues. More recently, a model for the rat non-muscle tropomyosin isoform, which has a different N-terminal sequence than the skeletal isoform has been prepared based on the structure of the N-terminal coiled-coil (28). This isoform utilizes exons 1b and 9d instead of exons 1a and 9a, which are translated in skeletal muscle tropomyosin. The overall model is similar to the rat skeletal isoform, but the degree of overlap is greater encompassing 16 amino acid residues. In contrast to the NMR structure, the X-ray structure of the rabbit skeletal overlap complex is completely different. Here the C-terminus maintains its coiled-coil structure, but interacts obliquely with only one α-helix of the N-terminus (29). The reason for the difference between the NMR and X-ray structures is unknown, especially considering the almost identical sequences for these constructs. This might imply greater conformational variability in the overlap region than initially envisioned and that these differences would be evident in related isoforms. To address this question, we have determined the X-ray structure of the overlap region of chicken smooth muscle tropomyosin.
Chicken smooth muscle tropomyosin has the same N-terminal sequence and the same overall number of amino acids as the skeletal isoform, but differs at its C-terminus. Both skeletal and smooth isoforms of muscle tropomyosin use exon 1a for their N terminus, whereas exons 9a and 9d are used for the C-terminus of skeletal and smooth tropomyosin, respectively. This study answers the question of how the overlap region can accommodate multiple sequences. It was accomplished using a novel approach for creating the N- and C-terminal fragments.
In the previous studies of the tropomyosin overlap complex, a segment of the leucine zipper found in the yeast transcription factor GCN4 was included to stabilize the truncated coiled-coils (25, 27–32). This is the standard strategy for expressing fragments of proteins that contain a coiled-coil, however, not all coiled-coil fragments fold well as fusions with leucine zippers. In order to improve the solubility and folding characteristics, fusion proteins were prepared that included globular domains in place of the simple leucine zipper. These domains were identified in the human DNA ligase binding protein XRCC4, the bacteriophage φ29 scaffolding protein Gp7, and the C-terminal helix bundle of the microtubule binding protein EB1 (33–36) and were fused to the N- or C-terminus of tropomyosin. The fusions expressed as soluble proteins in E. coli and crystallized readily. With this approach, two similar structures of the overlap complex were obtained utilizing different combinations of globular domains. This indicates that the observed overlap is not a consequence of an individual fusion protein. The overlap region of the N and C-terminus extends over 15 residues in both structures, but with slightly different angles between the axis of the N-terminal coiled-coil and the C-terminal coiled-coil, where this suggests that the overlap region can accommodate a range of angular interactions. The parallel helix bundles are morphologically similar to those seen previously by NMR in rat skeletal and non-muscle tropomyosin (27, 28).
MATERIAL AND METHODS
Fusion Protein Design
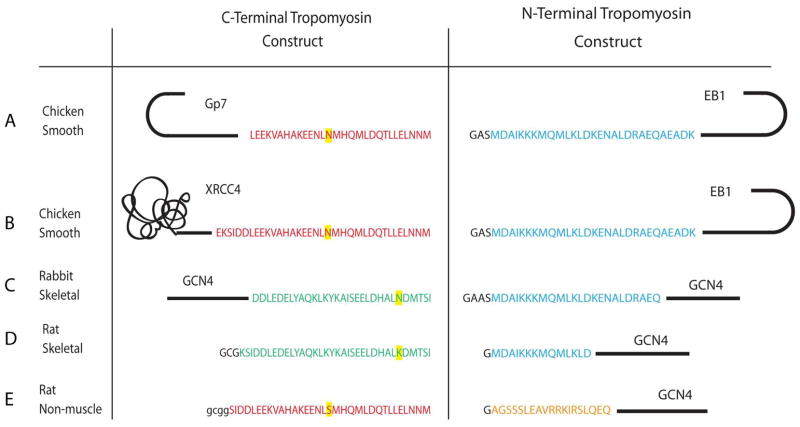
Careful consideration was given to the design of the junctions between Gp7, EB1, and XRCC4 and smooth muscle tropomyosin. First the heptad registration of the coiled-coil domains of each protein was identified with the programs Coils and Paircoil2 (37, 38). Then this information was utilized to design a series of constructs that maintained the coiled-coil motif throughout the junction between the proteins. The known structures of Gp7, EB1, XRCC4, and tropomyosin were considered to ensure that no important intra or inter-chain interactions were removed during assembly of the fusion (33, 35, 36). An alignment of the coiled-coil registration of Gp7, EB1, XRCC4, and tropomyosin is given in Supporting information Figure S1. The Gp7-TmC construct includes residues Pro2-Asp45 of Gp7 followed by residues Leu256-Met284 of tropomyosin. The XRCC4-TmC construct consists of residues Glu2-Ala135 of XRCC4 and residues Lys248-Met284 of tropomyosin. These two resulting constructs contained an additional Gly-Gly-Ser-Gly on the N-terminus after removal of the 6His-tag with TEV protease. The first glycine is the obligate P1′ residue from TEV protease cleavage site, whereas the others were included to facilitate removal of the purification tag. In the XRCC4-TmC construct Ile134 of XRCC4 was mutated to threonine and Leu249 was mutated to asparagine in tropomyosin to maintain favorable intra-chain interactions in the coiled-coil. The TmN-EB1 construct consisted of residues Met1-Lys29 of tropomyosin and residues Asp215-Asp257 of EB1. In this construct three residues, Gly-Ala-Ser, were left on the N-terminus after tag cleavage. The dipeptide Ala-Ser was included to facilitate interaction of the N- and C-termini of tropomyosin which does not occur when the native protein is expressed in bacteria due to the lack of N-acetylation (15). The glycine residue was a residual from the TEV cleavage site. A comparison of the residues of tropomyosin included in these constructs with those of earlier structural studies is shown in Figure 1.
Figure 1.
Present and earlier constructs used in the structural studies of the tropomyosin overlap region. A) GP7 and EB1 fusions of avian smooth muscle tropomyosin. B) XRCC4 and EB1 fusions of avian smooth muscle tropomyosin. C) GCN4 fusions of rabbit skeletal muscle tropomyosin utilized in the X-ray structural determination (29). D) Rat skeletal muscle tropomyosin constructs used in the NMR structural determination (27). E) Rat non-muscle tropomyosin constructs used in the NMR structural study (28). These different tropomyosin isoforms are the consequence of different exon usage. The N-terminal sequence is identical in both the skeletal and smooth muscle isoforms (blue) and is encoded by exons 1a. The N-terminal sequence of the non-muscle isoform is encoded by exons 1b (orange). The C-terminal sequence of skeletal (green) and smooth muscle tropomyosin (red) are encoded by exons 9a and 9d respectively. Rat non-muscle tropomyosin uses the same C-terminal exon as chicken smooth muscle tropomyosin (10). Different amino acids found in the same exon from different species are highlighted in yellow.
Expression Constructs
All tropomyosin constructs were expressed in E. coli using a modified pET31b (Novagen) vector that includes an N-terminal 6His followed by the recognition site for the TEV protease (39). Cloning of the fusion constructs into expression vectors was accomplished through a combination of 1) PCR to obtain desired DNA fragments, 2) conventional restriction enzyme-based cloning, 3) QuickChange mutagenesis (essentially a home-made version of the QuikChange kit (Strategene) but using only a single primer) and 4) enzyme-independent “QuickChange cloning” technique (40, 41). Template plasmids containing cDNA were obtained from Dr. Dwight Anderson, University of Minnesota (Gp7), Open Biosystems (XRCC4 and EB1) and Geneservice (chicken alpha smooth muscle tropomyosin). All constructs were sequence-verified in both directions. The complete amino acid sequence of the expressed polypeptides can be found in the Supporting Information.
Protein Expression and Purification
The expression of all proteins was performed in Codon Plus E. coli cells (Stratagene) in 2 L flasks containing 1 L LB medium with 100 μg/ml ampicillin and 30 μg/ml chloramphenicol. Cultures were grown at 37 °C to an OD600 of ~0.6 (as measured in Beckman DU-640 spectrophotometer), chilled on ice, induced with 1 mM IPTG and allowed to grow for 14–18 hours at 16 °C. Wet cell-paste was collected by centrifugation, frozen in liquid nitrogen and stored at −80°C. Selenomethionine proteins were expressed in M9 medium utilizing a metabolic inhibition protocol (42) under otherwise identical conditions. All proteins were purified using an identical protocol. Typically, about 5 g of cells were lysed by sonication in 25 ml of buffer A (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) containing 0.5 mg/ml lysozyme, 1 mM PMSF and protease inhibitors cocktail (made from individual components to be an exact equivalent of Roche’s “Complete EDTA-free” tablets). After sonication, the lysate was supplemented with MgCl2 and DNAse (2 mM and 5 μg/ml final, respectively) and centrifuged at 45,000 g for 30 min (JA-25.5 rotor). The supernatant was loaded onto a ~4 ml Ni-NTA (Qiagen) column, washed extensively with buffer A and eluted with a 100 ml gradient from buffer A to buffer B (same as A but containing 400 mM imidazole). The purest fractions were mixed with TEV protease at ~1:40 molar ratio (TEV was prepared as in reference (43)) and dialyzed at 4 °C overnight against buffer C (20 mM Tris, 50 mM NaCl, 0.5 mM TCEP, pH8.0). Dialysate was re-purified on the same Ni-NTA column, this time collecting fractions that elute at low imidazole concentrations. The purest fractions were pooled and dialyzed against buffer C, concentrated to ~ 10 mg/ml using a Centriprep YM-3 (Millipore), clarified by centrifugation at 16,000 g, flash frozen as 30 μL droplets in liquid nitrogen, and stored at −80 °C.
Size-Exclusion Chromatography
Analytical size-exclusion chromatography was performed on a Superdex 200 HR 10/30 column (Pharmacia) at 0.5 mL/min with a running buffer composed of 25 mM MOPS, 50 mM NaCl, 0.5 mM MgCl2, 1.0 mM TCEP, 5 mM NaN3, at pH 7.0. Equimolar amounts of interacting protein partners were mixed to a final protein concentration of ~10 mg/mL, filtered through 0.22 μm filter and incubated on ice for 45 min followed by a 15 min at room temperature. Approximately 70 μl of the resulting protein complex was loaded on the column.
Crystallization, Data Collection and Structure Determination
The tropomyosin overlap complexes were prepared by mixing the individual N- and C-terminal fusion proteins in a 1:1 molar ratio based on the calculated extinction coefficients and experimental ratios determined by size exclusion chromatography. The proteins were incubated on ice for 45 min before being placed at room temperature for 15 min. The protein concentration for the components of the SeMetGp7-TmC:SeMetTmN-EB1 complex was 5.6 mg/ml for both proteins. The protein concentrations for the components of the XRCC4-TmC:TmN-EB1 complex solution were 7.4 mg/ml and 3.5 mg/ml for XRCC4-TmC and TmN-EB1 respectively.
Crystallization conditions were surveyed by vapor diffusion with a 144 conditions sparse matrix screen developed in the laboratory. The crystals for the SeMetGp7-TmC:SeMetTmN-EB1 complex were observed in three conditions from pH 5.5–7.5 with a variety of salts, but optimal crystals were obtained by mixing 1 μL of the protein with 1 μL of 16% methylether polyethylene glycol 5000, 100 mM MES, pH 6.0, 140 mM CaCl2 and 2% methanol and equilibrating against the same solution. The crystals grew as plates of dimensions of 400 × 300 × 100 μm in approximately 3 weeks at room temperature. For the X-ray diffraction studies the crystals were transferred into 1:3 solution of synthetic mother liquor:cryoprotectant where the cryoprotectant consisted of 17% methylether polyethylene glycol 5000, 100 mM MES, pH 6.0, 180 mM CaCl2, 2% methanol, and 20% ethylene glycol. This was accomplished in four stages: 25% solution for 10 min, 50% solution for 5 min, then a 75% solution for 2 min. They were then flash-frozen in liquid nitrogen.
The crystals of the SeMetGp7-TmC:SeMetTmN-EB1 complex belong to space group C222, with unit cell dimension of a = 89.1 Å, b = 285.3 Å, c = 43.5 Å. They contain two dimers of TmN-EB1 and one dimer of Gp7-TmC so that there is one complex and one free TmN-EB1 dimer in the lattice. Data were collected at the Structural Biology Center Beamline 19-BM (Advanced Photon Source, Argonne National Laboratory, Argonne, IL) using a SBC-3 CCD X-ray detector at 100 K. The structure was determined from a SeMet SAD experiment to a resolution of 2.1 Å. A total of 180 frames, each with an oscillation range of 1.0°, were collected using the peak wavelength of Se at 0.9793 Å. Integration and scaling of the data were done using HKL3000 (44). Selenium sites were located with SHELXD (45), initial phases were calculated using MLPHARE (anomalous Rcullis= 0.81) and further improved by DM (46). The experimental phases were used for an initial model building by ARP/wARP (47), which resulted in Rfree of 37.8%. This model was refined iteratively with WinCoot and PREFMAC, refining directly against SAD data. The final Rwork and Rfree values were 20.1% and 24.1%, respectively. Data collection and refinement statistics are given in Table 1.
Table 1.
X-ray data collection and refinement statistics
| GP7-TmC:TmN-EB1 | XRCC4-TmC:TmN-EB1 | |
|---|---|---|
| Space group | C222 | P41212 |
| Cell dimensions, a,b,c (Å) | 89.1, 285.3, 43.5 | 96.6, 96.6, 163.0 |
| Wavelength (Å) | 0.9795 | 0.9793 |
| Resolution (Å) | 30–2.1 (2.14–2.1) | 30–2.2 (2.24–2.2) |
| Completeness (%) | 99.8 (99.1)a | 99.3 (99.2)a |
| Redundancy | 11.8 (11.1)a | 18.6 (17.9)a |
| Unique reflections | 33097 | 39647 |
| Rmerge (%) | 10.4 (42.8)a | 5.7 (48.8)a |
| Average I/σ | 23.0 (3.6)a | 49.6 (5.8)a |
| Wilson B (Å2) | 32.1 | 51.4 |
| Rwork (%) | 20.1 (20.2)a | 20.7 (23.5)a |
| Rfree (%) | 24.1 (23.8)a | 25.0 (29.8)a |
| Protein atoms | 3361 | 3426 |
| Solvent atoms | 406 | 407 |
| Average B-factor (Å2) | ||
| Protein atoms | 42.1 | 51.2 |
| Solvent atoms | 33.9 | 32.9 |
| R.M.S. deviations | ||
| Bond lengths (Å) | 0.010 | 0.008 |
| Bond angles (°) | 1.069 | 1.016 |
| Ramachandran plot (%)b | ||
| Favored region | 98.4% | 95.8% |
| Allowed region | 1.6% | 4.2% |
Statistics for the highest resolution bins
Ramachandran analysis was performed with the program Procheck (64).
The XRCC4-TmC:TmN-EB1 complex crystallized readily from 15 conditions in the screen with a range in pH of 6.0–9.0 and a variety of salts and polyethylene glycols, but the best were obtained by mixing 1 μL of the protein with 1 μL of complex with a precipitant solution that contained 18% methylether polyethylene glycol 2000, 100 mM Bis-Tris, pH 6.5, and 120 mM MgSO4 and equilibrating against the same solution. The crystals grew as rhombic plates of dimensions of 500 × 400 × 100 μm in approximately 2 weeks at room temperature. For the X-ray diffraction studies the crystals were transferred into a 1:3 solution of synthetic mother liquor:cryoprotectant where the cryoprotectant consisted of 18% methylether polyethylene glycol 2000, 140 mM MgSO4, 100 mM Bis-Tris, pH 6.5 and 20% ethylene glycol. This was accomplished in stages; 25% solution for 10 min, 50% solution for 5 min, then a 75% solution for 2 min. They were then flash-frozen in liquid nitrogen.
An initial X-ray data set for the XRCC4-TmC:TmN-EB1 complex was collected to 2.5 Å on a Bruker AXS Platinum 135 CCD detector (Bruker AXS Inc.) utilizing Cu Kα radiation generated by a Rigaku RU200 X-ray generator equipped with Montel optics and operating at 90 mA and 50 kV. The data were recorded at 100 K utilizing an Oxford Cobra cryogenic cooling system, were processed with SAINT version 7.06A (Bruker AXS Inc.), and scaled with SADABS version 2005/1 (Bruker AXS Inc.). The crystals belong to space group of P41212 with cell dimensions of a = 96.42 Å, b = 96.42 Å, c = 162.83 Å and contain one overlap complex in the asymmetric unit. A preliminary structure was determined by molecular replacement utilizing residues Glu2-Ala135 of XRCC4 (PDB accession number 1FU1) with the program Molrep (33, 48). All of the XRCC4-TmC fusion protein was visible in the initial electron density as was part of the N-terminal segment of TmN-EB1, however, the electron density for the EB1 helical bundle was ill-defined. The latter was improved with the density modification program Parrot (49) such that it was possible to autobuild parts of EB1 with the program Pirate (50). A higher resolution dataset was recorded with synchrotron radiation at the Structural Biology Center Beamline 19-BM (Advanced Photon Source, Argonne National Laboratory, Argonne, IL) using a SBC-3 CCD X-ray detector at 100 K. The data were integrated and scaled with the program HKL3000. The coordinates of the in-house structural solution for XRCC4-TmC:TmN-EB1 were used as the search model for a molecular replacement solution obtained with the program Molrep (48). Again, the EB1 region was poorly defined. TLS refinement was used to improve the electron density in this region. Manual model building and refinement were performed with the programs WinCoot and REFMAC5 (51, 52). The final Rwork and Rfree for the final 2.2 Å structure were 20.7% and 25.0% respectively. Refinement and data collection statistics are shown in Table 1. Buried surface areas were calculated with the program SurfaceRacer (53)
RESULTS AND DISCUSSION
Optimization of a Stable Overlap Complex
The N- and C-terminal regions of alpha smooth muscle tropomyosin were fused to the globular domains together with a short section of endogenous coiled-coil found in EB1 (N-terminal construct) and either Gp7 or XRCC4 (C-terminal construct). These proteins were identified as potential candidates for fusion after a search of the Protein Data Bank for structures of proteins that included a well-ordered compact globular domain that was either preceded (EB1) or succeeded (Gp7 or XRCC4) by a parallel and symmetric coiled-coil. EB1 is a microtubule binding protein that contains a small helical bundle consisting of 28 amino acids at its C-terminus (36). Gp7 is a bacteriophage φ29 scaffolding protein that carries a somewhat larger helical bundle that is composed of ~40 amino acid residues at its N-terminus (35). Lastly, XRCC4 is a DNA ligase binding protein that exhibits a mostly β-sheet domain consisting of ~120 amino acid residues at its N-terminus. The resultant fusion proteins were expressed in E. coli in a soluble form at ~25 mg of soluble protein per gram of cell paste, which highlights the utility of these constructs for expressing fragments of coiled-coils.
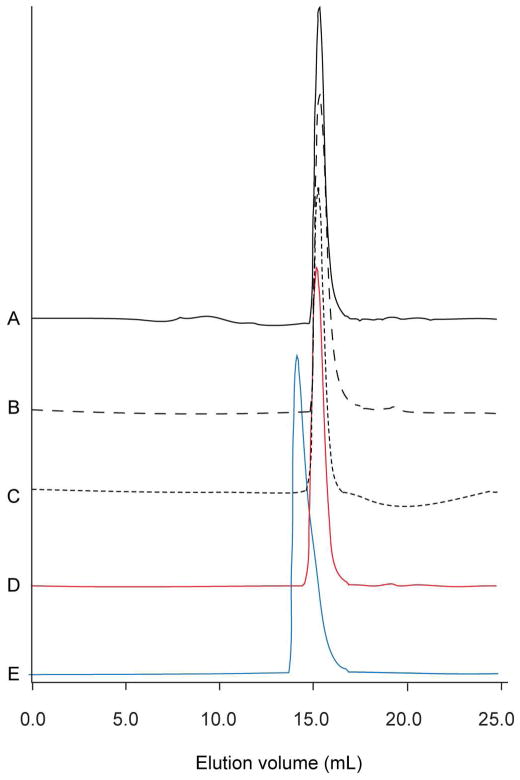
Several TmN-EB1 fusion proteins that differed at their N-termini were constructed with the goal of optimizing the stability of the overlap construct. As noted earlier, in vivo, N-acetylation of tropomyosin is required in order to generate a physiologically functional assembly on the surface of actin, where this posttranslational modification is absent in E. coli. Previously it was shown that addition of Ala-Ser to the N-terminus restores that ability of tropomyosin expressed in E. coli to polymerize (54). Later it was suggested that a single glycine at the N-terminus was a better mimic of acetylation for muscle isoforms of tropomyosin (30). In this study, constructs that carried either a single glycine or a Gly-Ala-Ser tripeptide at their N-termini were prepared. Their ability to form a stable complex with the C-terminal tropomyosin fusion proteins was tested by size-exclusion chromatography. With this assay, only the protein with Gly-Ser-Ala at its N-terminus generated a stable complex with the TmN-EB1 fusion protein (Figure 2). Consequently, this construct was utilized for the structural studies.
Figure 2.
Size exclusion chromatography demonstrates complex formation. A) TmN-EB1 construct that includes only a N-terminal Gly. B) Elution profile for the TmN-EB1 fusion with Gly-Ala-Ser added to the N-terminus. C) Elution profile for the GP7-TmC tropomyosin fusion construct. D) Elution profile for a 1:1 mixture of GP7-TmC and Gly-TmN-EB1. There is no shift in the elution time indicating the absence of a stable complex when only a single glycine is included at the N-terminus. E) Elution profile for a 1:1 mixture of GP7-TmC and Gly-Ala-Ser-TmN-EB1. The mixture elutes earlier than the individual components, consistent with formation of a stable complex when Gly-Ala-Ser is added to the N-terminus. The elution volumes for A), B), C), and D) were all similar with an average of 15.26 mL. A noticeable shift to 14.12 mL was observed on mixing GP7-TmC and Gly-Ala-Ser-TmN-EB1.
The structures reported here are for the alpha smooth muscle avian tropomyosin overlap sequence. In vivo, smooth muscle tropomyosin exists as a constitutive heterodimer of the α and β isoforms (55). The sequences of these isoforms are 85% identical, however most of the differences lie outside of the overlap region. There are two differences between the alpha and beta isoforms in the N-terminal segment and eight in the C-terminal segment for the structures reported here. Most of these differences that lie outside of the overlap region are solvent exposed and are not predicted to destabilize αα homodimers. From this it can be concluded that the determinants for heterodimer formation lie outside of the overlap region.
Structural Overview of the Overlap Complexes
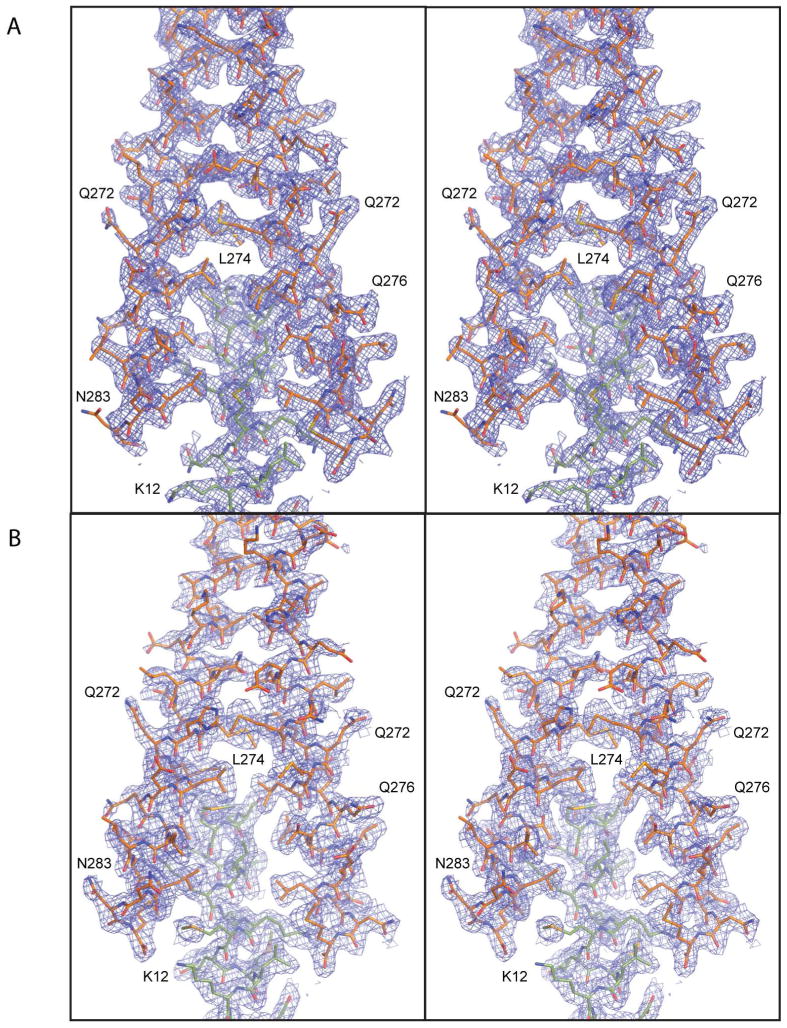
The structures of both the Gp7-TmC:TmN-EB1 and XRCC4-TmC:TmN-EB1 chicken smooth muscle tropomyosin overlap complexes have been determined to 2.1 and 2.2 Å resolution respectively. Although radically different C-terminal fusions were utilized, the overlap regions themselves are topologically similar (Figure 3). Small differences arise however from crystal packing forces. In all cases, the electron density for the overlap regions is well-defined (Figure 4), though each lattice shows some conformational disorder. Fortunately this occurs in different sections of each complex and does not involve any residues of tropomyosin.
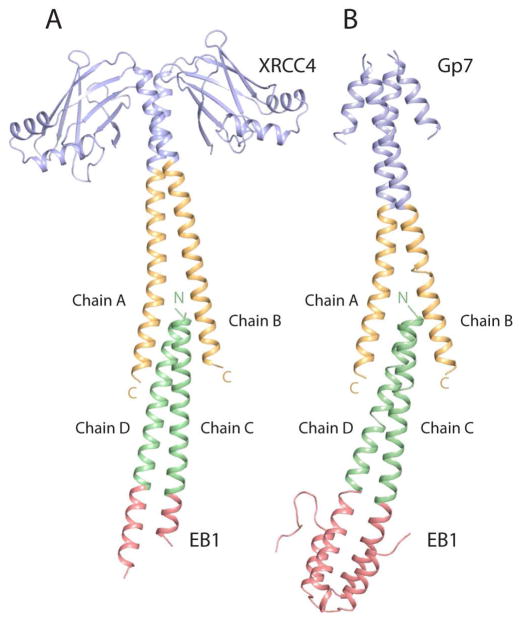
Figure 3.
Ribbon representation of the two avian smooth muscle tropomyosin overlap structures. A) The structure of the XRCC4-TmC:TmN-EB1 complex. This was obtained by fusing residues Glu2-Ala135 of XRCC4 (blue) to the C-terminal residues Lys248-Met284 of tropomyosin (orange) and fusing the N-terminal residues Met1-Lys29 of tropomyosin (green) to residues Asp215-Asp257 of the C-terminal helix bundle of EB1 (red). B) The structure of the Gp7-TmC:TmN-EB1 complex. The Gp7-TmC construct was made by fusing residues Pro2-Asp45 of Gp7 (blue) to the C-terminal residues Leu256-Met284 of tropomyosin (orange). The chain labels are included in this figure. Chains A and B belong to the C-terminal fragment of tropomyosin, whereas chains C and D belong to the N-terminal fragment. Figures 3, 4, 6, and 8 were prepared with Pymol (DeLano, 2002).
Figure 4.
Stereoelectron density maps of the overlap regions. A) The overlap observed in the XRCC4-TmC:TmN-EB1 complex. B) The overlap observed in the Gp7-TmC:TmN-EB1 complex. The electron density maps were contoured at 1.0 σ and were calculated with coefficients of the form 2Fo–Fc.
The asymmetric unit of the Gp7-TmC:TmN-EB1 tropomyosin complex contains two dimers of the TmN-EB1 fusion protein and only a single Gp7-TmC homo-dimer. As a consequence the asymmetric unit consists of one Gp7-TmC:TmN-EB1 complex and a free TmN-EB1 homodimer. Examination of the crystal lattice reveals that there is insufficient space to include a second molecule of Gp7-TmC without the introduction of serious steric clashes. In this case it would appear that the packing forces are sufficient to exclude one molecular dimer of Gp7-TmC from the lattice, even though sufficient protein was present in the crystallization mixture to form a 1:1 complex. This implies that the crystal packing interactions have a similar magnitude to those involved in complex formation and thus could influence the nature of the complex observed.
Each Gp7-TmC and TmN-EB1 chain consists of 77 and 75 amino acids, respectively. Of the two TmN-EB1 chains found within the Gp7-TmC:TmN-EB1 complex, one was completely resolved with all 75 amino acids being modeled. Chain D, however, had incomplete electron density for the last 7 amino acids of EB1 (Glu251-Asp257). In the two TmN-EB1 chains that did not participate in the complex the first two amino acids of the Gly-Ala-Ser addition to the tropomyosin were disordered. Also, the C-terminal seven and six residues of EB1 were not visible. For the Gp7-TmC fusion protein the first seven amino acids of the Gp7 helical bundle (N-terminal Gly-Gly-Ser-Gly remaining from the affinity tag and Pro2-Lys4 of Gp7) and residues Glu19-Ala21 were disordered, whereas the remainder of the chain was well-defined.
The asymmetric unit of the XRCC4-TmC:TmN-EB1 tropomyosin complex contains one dimer of the XRCC4-TmC construct and one dimer of the TmN-EB1 construct. Each chain of XRCC4-TmC consists of 175 amino acid residues. The electron density maps were well resolved throughout the structure (Figure 4) with the exception of the region corresponding to the EB1 helical bundle. Examination of the crystal lattice shows that there is a large cavity associated with where the EB1 helical bundle is located, with few intermolecular contacts. As a consequence, the electron density for the helical bundle is not well-defined. Indeed, only residues Asp215-Asn223 and Asp215-Glu230 of EB1 could be built with confidence into chains C and D respectively. Even so, the electron density for the overlap region was unequivocal (Figure 4).
The complexes are formed by the interdigitation of the N- and C-terminal coiled-coils of tropomyosin, where the overall conformation is similar to that which has been seen previously in the NMR structure of the rat skeletal and model of non-muscle tropomyosin complexes (27–29). As in those structures, the C-terminal section of tropomyosin opens to allow the insertion of the N-terminal coiled-coil. The extent of the opening of the C-terminal section of tropomyosin is approximately 2 Å larger than that observed in the structure of the free skeletal muscle fragment (25) (PDB accession number 1KQ). Thus this these complexes agree with the suggestion that the C-terminus would open on complex formation (25).
The coiled-coils of chicken smooth muscle tropomyosin overlap by 12 to 15 residues. However, the exact number for each chain varies because the overlap complex is bent as discussed below. Even with this range, more residues are implicated in the smooth muscle overlap than the eleven observed in the NMR structure of the rat skeletal tropomyosin overlap complex. Notably, the structures reported here are quite different from the crystal structure reported for the rabbit skeletal tropomyosin complex, in which the overlap involves only one of the two N-terminal α-helices of tropomyosin (29).
The N-terminal sequence of chicken smooth muscle tropomyosin is identical to that of the skeletal isoform. The structure of the first 81 amino acid residues of chicken skeletal tropomyosin has been determined previously (PDB accession 1IC2) (24). Comparison of the structure of the tropomyosin segment included in TmN-EB1 with the equivalent residues in the skeletal isoform shows a rms difference between equivalent α-carbons of only 0.40 Å after exclusion of two N-terminal residues which adopt different conformations due to the addition of Gly-Ala-Ser at the N-terminus of the EB1 construct (see below). Similarly, the rms difference between equivalent α-carbons of the TmN-EB1 non-crystallographically related dimers in this crystal form (that in the complex and the free dimer) is only 0.29 Å suggesting that complex formation has little impact on the structure of the N-terminal segment of tropomyosin. Likewise, the rms difference between the tropomyosin component of TmN-EB1 dimer in the complexes Gp7-TmC:TmN-EB1 and XRCC4-TmC:TmN-EB1 is only 0.63 Å. The similarity of the N-terminal segment of tropomyosin across a broad range of constructs and complexes confirms, as suggested before, that the conformational changes needed to form a complex occur primarily in the C-terminal coiled-coil of tropomyosin (27).
The interactions between the N- and C-terminal coiled-coils of smooth muscle tropomyosin show significant curvature (Figure 3). This differs somewhat in the two structures and probably reflects the inherent flexibility associated with the overlap region and its response to crystal packing forces. Deviation from linearity was also seen in the model for non-muscle tropomyosin (28). This flexibility is likely an important attribute that allows tropomyosin to wrap around the actin filaments. The nature of the contacts observed in the smooth muscle tropomyosin overlap regions is discussed below, starting with the interactions seen in the XRCC4-TmC:TmN-EB1 complex, and then followed by those seen in the structure of Gp7-TmC:TmN-EB1.
XRCC4-TmC:TmN-EB1 Overlap Complex
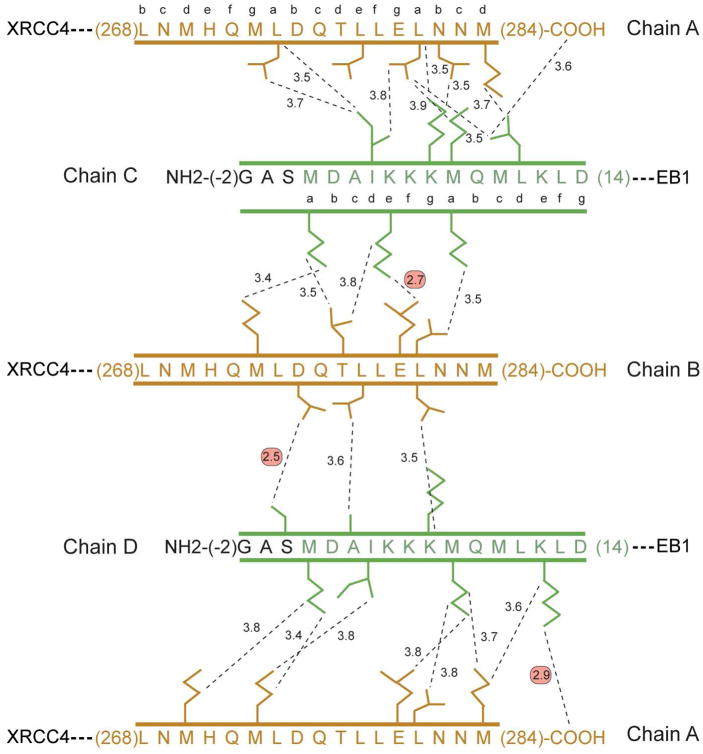
The curvature of the XRCC4-TmC:TmN-EB1 tropomyosin overlap region results in a shorter overlap of the α-helices on the outside of the curve and a deeper overlap of the α-helices on the inside. Since the structure of the N-terminal section of tropomyosin is approximately invariant in all of the structures it is convenient to describe the complexes in terms of the position of the C-terminal helices relative to the fixed N-terminal coiled-coils within context of the curvature of the complex. Helix A of XRCC4-TmC lies on the inside of the curvature whereas helix B of XRCC4-TmC lies on the outside (Figure 3). Because of the curvature, the helix A of XRCC4-TmC and helix D of TmN-EB1 are approximately parallel to each other. This requires that all other helices are not parallel to each other because of the intrinsic angle of ~−20° between α-helices in a coiled-coil. Thus the interaction angle between B and D of TmC and TmN, and the angle between A and C of TmC and TmN are both ~45°.
Each of the inside and outside helices of XRCC4-TmC has a substantial number of pair-wise interactions with both helices of the opposing TmN-EB1 dimer. These are shown schematically in Figure 5. The outside helix B of XRCC4-TmC overlaps with at most 12 residues of the N-terminal coiled-coil which corresponds to ~3.3 α-helical turns. Most of the interactions are non-polar except for salt bridge between the carboxylate oxygen of Glu280B and Nζ of Lys5C, and a hydrogen bond between a carboxylate oxygen of Asp275B and the hydroxyl of Ser0D. Both of these interactions provide insight into the structure of the overlap complex. The first interaction between Glu280 and Lys5 would be expected to occur in the native tropomyosin and may be functioning as a positioning element for the α-helical interaction in a similar manner to the lone hydrogen bonding event that occurs with Asn16 in the trigger sequence of GCN4 (56, 57). The difference here is that the polar interaction in the overlap complex lies on the periphery of the helical bundle, whereas it is buried in GCN4.
Figure 5.
Schematic representation of the pair-wise intermolecular contacts in the XRCC4-TmC:TmN-EB1 overlap complex. Chains A and B correspond to XRCC4-TmC, whereas chains C and D correspond to TmN-EB1. The hydrophobic interactions were selected based on a distance cutoff of 3.9 Å using Contact. The predicted coiled-coil heptad registration is shown for both the N- and C-terminal sequences of smooth muscle tropomyosin (37, 38). The hydrogen bonding and ionic interactions are highlighted by red circles, whereas all other interactions are non-polar interactions. The color scheme for this figure is the same as that defined in the ribbon representation shown in Figure 3.
The interaction between the carboxylate oxygen of Asp275B and the side chain hydroxyl of Ser0D would not occur in native tropomyosin because it involves the serine of the Gly-Ala-Ser peptide that was added to the N-terminus to compensate for the absence of N-acetylation, where either acetylation or the inclusion of a few extra residues is required in order to form a stable filament of tropomyosin. Thus at first sight, this interaction might be ignored, but the resulting conformation of the N-terminus provides insight into the molecular role of N-acetylation in native tropomyosin.
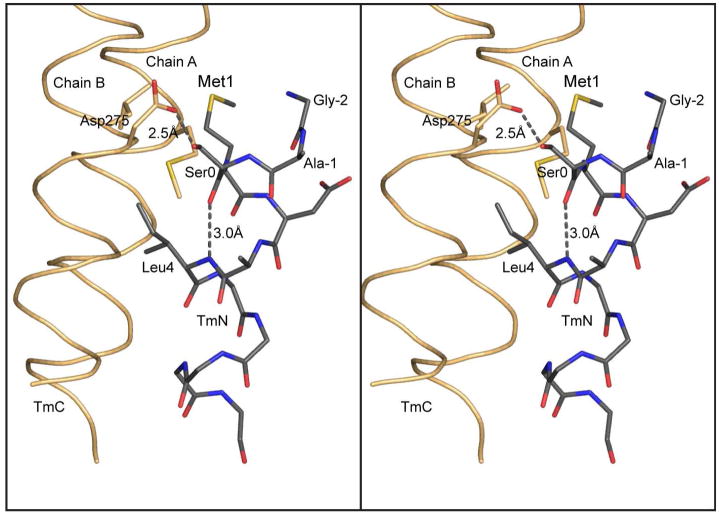
Originally it was proposed that acetylation of the N-terminus eliminated the destabilizing effect of a positively charge amino group in the “a” position of the coiled-coil (54), since this position is normally occupied by a hydrophobic side chain. It was proposed that introduction of Ala-Ser to the N-terminus of bacterially expressed tropomyosin would move the N-terminus to a surface location more forgiving of a positive charge. Examination of the structure here suggests that the function of Ala-Ser is primarily to maintain Met1 in an α-helical conformation. The additional serine introduces a peptide carbonyl oxygen that forms an intrahelical hydrogen bond to the amide hydrogen of Leu4 thus maintaining Met1 in a helical conformation (Figure 6). The presence of a hydrogen bond between the carboxylate oxygen of Asp275B and the Ser0D hydroxyl is probably secondary to this effect, since the same intrahelical backbone hydrogen bond is seen in chain C TmN even though the hydrogen bond between the carboxylate oxygen of Asp275A and the hydroxyl of Ser0C is not observed. In both chains C and D of TmN-EB1 Met1 lies in a hydrophobic pocket which should lend to considerable stabilization. This stabilization would be lost if Met1 were to adopt a non-helical conformation. In nature, acetylation of the N-terminus could perform the same function as the additional serine residue by providing a terminal carbonyl oxygen that would be able to hydrogen bond to the backbone amide hydrogen four residues away and thus ensure that Met1 is maintained within a α-helical conformation.
Figure 6.
Hydrogen bonding pattern at the N-terminus of tropomyosin. This figure shows a stereo close-up of the hydrogen bonding pattern associated with the N-terminal helix of tropomyosin and the Gly-Ala-Ser addition that is required to allow bacterially expressed tropomyosin to assemble into filaments. As seen here, the additional serine residue maintains the N-terminus of tropomyosin in an α-helical conformation. This is accomplished through an intrahelical hydrogen bond between the carbonyl oxygen of Ser0 and the amide hydrogen of Leu4, together with a hydrogen bond between Oδ1 of Asp275 and Oγ Ser0. In this conformation, Met1 lies in a hydrophobic pocket which will contribute to the hydrophobic stabilization of the complex. In nature, acetylation of the N-terminus would provide a terminal carbonyl oxygen that would be able to hydrogen bond back to the backbone amide hydrogen four residues away and thus allow Met 1 to remain in an α-helical conformation.
The inside helix of XRCC4-TmC, chain A, overlaps the N-terminal coiled-coil of TmN-EB1 by 15 residues corresponding to ~4.2 α-helical turns. There are considerably more interactions between chain A of XRCC4-TmC and helices C and D of TmN-EB1 than formed by the outside helix which reflects the effect of the curvature of the complex. A total of sixteen residues are involved the interactions between chain A of XRCC4-TmC and helices C and D of TmN-EB1, compared with twelve residues in the interaction of outer helix B with C and D. Again, almost all of the interactions are hydrophobic. There is only a single hydrogen bond between chains A and D between the C-terminal oxygen of Met284A and Nζ of Lys12D (Figure 5). A similar overall group of interactions is seen in the Gp7-TmC:TmN-EB1 overlap complex.
Gp7-TmC:TmN-EB1 Overlap Complex
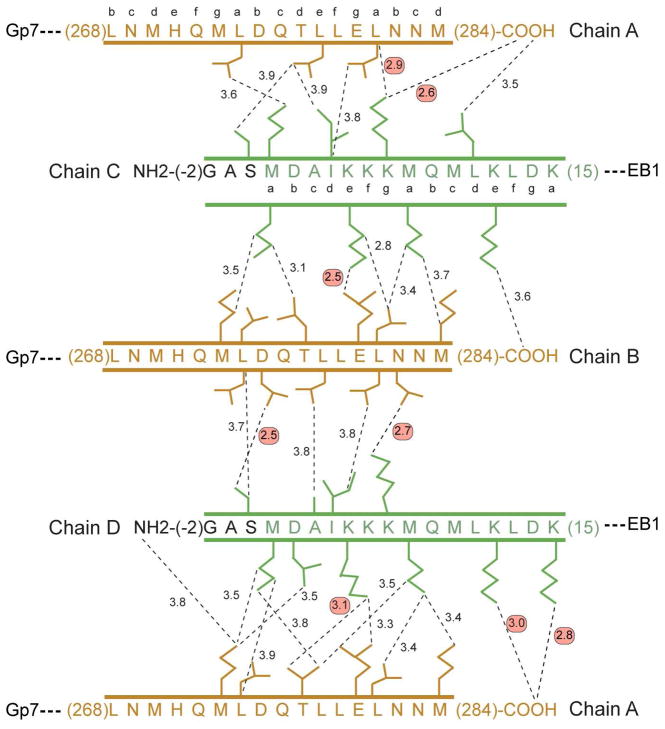
In the Gp7-TmC:TmN-EB1 complex the curvature is more pronounced by ~5 ° which results in further differences in the interactions between the helices (Figure 7). However the extent of the overlap is similar. The outside helical segment of the TmC overlaps the N-terminal coiled-coil by ~12 residues whereas the inside helix of TmC overlaps by 15 residues. Because of the additional curvature, the outer helix of TmC translates ~0.5 Å over the TmN coiled-coil relative to the position adopted in the XRCC4-TmC:TmN-EB1 overlap complex. Interestingly, the changes in the interactions are largely compensatory, however, there are more polar interactions in the Gp7-TmC:TmN-EB1 than in the complex with XRCC4-TmC.
Figure 7.
Schematic representation of the pair-wise intermolecular contacts in the Gp7-TmC:TmN-EB1 chicken smooth muscle tropomyosin overlap complex. Chains A and B correspond to Gp7-TmC, whereas chains C and D correspond to TmN-EB1. The hydrophobic interactions were selected based on a distance cutoff of 3.9 Å. The predicted coiled-coil heptad registration is shown for both the N- and C-terminal residues of smooth muscle tropomyosin. Red circles highlight the hydrogen bonding and ionic interactions, whereas all other interactions are non-polar. The color scheme for this figure is the same as that defined in the ribbon representation shown in Figure 3.
As in the XRCC4-TmC complex, there are somewhat fewer interactions between the outside helix (chain B) and TmN-EB1 than between the inside helix (A) and the N-terminal coiled-coil. Seventeen residues participate in interactions with contact distances less than 3.9 Å between the C-terminal chain B and the N-terminal coiled-coil. Of these, only three are polar. The interactions between the carboxylate of Glu280B and Nζ of Lys5C, and the carboxylate of Asp275B and side-chain hydroxyl of Ser0D are identical to those seen in the XRCC4-TmC complex discussed above. The intramolecular hydrogen bond that serves to maintain Met1 of chain D in an α-helical conformation is also present, demonstrating that this is not a consequence of crystal packing. An additional hydrogen bond is formed between Asn282B and Lys7D.
The inside helix makes more contacts with the N-terminal coiled-coil. Nineteen residues in chains A, C, and D are involved in contacts shorter than 3.9 Å, of which eight are polar. The constellation of interactions is similar to those seen in the XRCC4-TmC:TmN-EB1 complex, but there are differences, which demonstrate the plasticity of the interactions between these chains. For example, the C-terminal carboxylate moiety of Met284A of the inside helix forms three salt-bridge interactions with Lys7C, Lys12D, and Lys15D. This may keep the side chain of Met284A in a hydrophobic environment. In the XRCC4-TmC complex, Met284A still lies in a hydrophobic environment, though the salt bridges are mostly lacking. Although salt bridges are unlikely to contribute significantly to the overall stability, they are expected to influence the specificity of the interactions between the chains. These subtle differences indicate that a variety of interaction angles can be achieved with little energy difference.
The curvature of the XRCC4-TmC:TmN-EB1 and Gp7-TmC:TmN-EB1 overlap complexes raises the question whether the overlap region is constitutively bent. That is to imply that the most stable conformation is non-linear. It is not possible to ascertain whether this is the case on the basis of the structures described here because of the potential effect of crystal packing effects which will statistically select against a linear arrangement. But, a non-linear interaction would be consistent with the assessment that the conformation of the tropomyosin molecule as a whole adopts a structure that follows the helical path dictated by F-actin (58, 59). However, the degree of curvature necessary to fit the overall helical path of tropomyosin need not necessarily be that great. Simple calculations, based on the diameter of actin and the helical pitch of tropomyosin, suggest that an angular change of 4–5 degrees is all that is needed to encompass the overlap region. In which case the complexes seen here are over-bent and thus reflect the small energies needed to distort the overlap region.
The extent and character of the buried surface area are similar for both complexes. XRCC4-TmC:TmN-EB1 and GP7-TmC:TmN-EB1 bury a total of 2114 Å2 and 2054 Å2 respectively on complex formation (53). Within this area, 72% of the surface is non-polar in the XRCC4-TmC:TmN-EB1 complex and 68% is non-polar in the GP7-TmC:TmN-EB1 complex. This further strengthens the claim that the four helical bundle is stabilized by the hydrophobic effect.
As noted earlier, smooth muscle tropomyosin exists as an αβ heterodimer in vivo (55). The structure reported here is for an αα homodimer. There are only differences between the α- and β- isoforms in the N-terminal segment included in these structures (Asp2Glu and Leu19Ile) where neither of these is predicted to favor the heterodimer or destabilize the overlap complex. In contrast there are eight differences in the C-terminal segment. Of these Asn269Gly, Met270Ile, Met 273Val, and Met284leu lie within the overlap complex and may influence its stability. In particular, the Asn269Gly isoform difference may allow the C-terminal coiled-coils to separate more easily to allow formation of the complex. However, none of the differences in the C-terminal segment are predicted to stabilize the αβ heterodimer over the αα homodimer, thus the determinants for heterodimer formation must lie elsewhere in the protein.
Comparison to Previous Overlap Structures
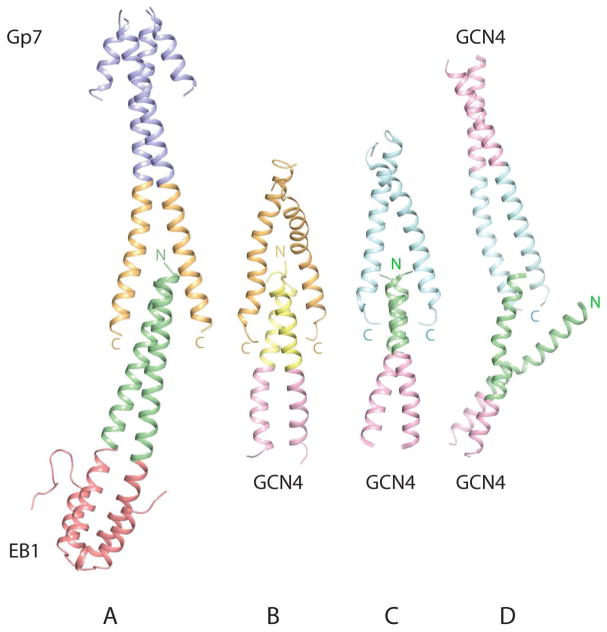
The XRCC4-TmC:TmN-EB1 and GP7-TmC:TmN-EB1 overlap structures most closely match the structures determined previously by NMR (Figure 8) (27, 28). In all of these structures the C-terminal coiled-coils separate to allow interdigitation of the N- and C-termini of tropomyosin. Furthermore the structures of the chicken smooth muscle tropomyosin overlap complex exhibit similar degrees of curvature to that seen in the non-muscle NMR model (28). The four-helix bundle observed in the structure of skeletal tropomyosin as determined by the NMR structure is more symmetric in the overlap region, although the complex as a whole is somewhat bent. These comparisons are important because of the high conservation of the amino acid sequence of tropomyosin in these regions (Figure 1). The sequence of the rat skeletal N-terminal coiled-coil is identical to that of chicken smooth muscle tropomyosin studied here although different amino acids were included to stabilize the N-terminus. Likewise, the sequence of the C-terminal coiled-coil is identical to that used in the study of non-muscle tropomyosin overlap complex except that Asn269 in chicken smooth muscle is replaced by Ser in the non-muscle isoform. However, this position is not involved in any interactions within the different overlap complexes. Thus it is possible to compare the overlaps accomplished by identical sequences in different combinations.
Figure 8.
Comparison of current and previous structures of the tropomyosin overlap complex. A) The structure of chicken smooth muscle GP7-TmC:TmN-EB1 determined here. B) Rat non-muscle tropomyosin complex model determined by NMR. C) Rat skeletal tropomyosin complex solved by NMR. D) X-ray crystal structure of rabbit skeletal tropomyosin. Panel B was prepared from the coordinates supplied in the supporting information of reference (28). Panels C and D were prepared from PDB files with the accession numbers 2G9J and 2Z5I (27–29). Note; the free N-terminal chain in the X-ray structure of rabbit skeletal tropomyosin interacts with a crystallographically related C-terminal coiled coil, which doubtless stabilizes this arrangement.
The primary difference between the NMR structure and model is the extent of the overlap. In the non-muscle model the overlap extends over 18 residues or 5 helical turns, whereas the skeletal complex covers a maximum of 13 residues or 3.6 helical turns. This corresponds to a buried surface area of 2302 Å2 and 1835 Å2 for the non-muscle and skeletal tropomyosin overlap complex, where 66.2% and 66.8% are non-polar respectively. The buried surface areas for the chicken smooth muscle overlap complexes lie within the range seen for the non-muscle and skeletal complexes and exhibit a similar proportion of buried non-polar surface. It is expected that the stability of the complex will be proportional to the buried hydrophobic surface area, which would imply that the non-muscle complex should be somewhat more stable than the smooth muscle complex based on the area. Conversely, the skeletal complex would appear to be the least stable. The differences in buried surface area correlate with the observation that gizzard smooth muscle tropomyosin polymerizes more readily than the striated muscle isoform (60). This might indicate that other elements are required to stabilize the striated muscle complex in vivo, where the obvious candidate is troponin which binds to the overlap region in skeletal tropomyosin.
A alternative explanation for the stronger complex might be the net charge on the N-terminal and C-terminal coiled-coils. It has been shown that a stronger complex is formed when the negative charge on the C-terminus is increased (61). Indeed, the overlap region of the N-terminus contains more positively charged residues whereas the C-terminal coiled-coil is more negatively charged. The net charge on the C-terminal 11 residues for chicken smooth muscle tropomyosin is −3 (including the C-terminal carboxyl group) compared to −1 for rat skeletal. Ironically, the skeletal muscle C-terminus contains the same number of negatively charged residues as the smooth muscle isoform, but the overall negative charge is lower due to the inclusion of two additional positively charged residues. Examination of the structures reveals that it is the position of the charges rather than the simple number that might be important. In the case of the smooth muscle complex the C-terminal carboxyl group interacts with a cluster of lysine residues (Lys7C, Lys12D, and Lys15D) as mentioned above. The rat skeletal NMR structure exhibits only one salt bridge between Lys7 and the carboxyl terminus that suggests that it is the degree of interdigitation of the coiled-coils rather than the total number of charges that is important in the stability.
Examination of the detailed interactions and conformations of the identical sequences and their opposing partners in the complexes reveals that for the most part the same amino acids participate in interactions in all complexes, though the exact nature of the interactions are different. This reflects that in both the NMR structures and X-ray structures reported here the C-terminal coiled-coils separate somewhat to allow the interdigitation, but do not fundamentally change their structure. Thus the same amino acid side chains will be present in the overlap region, but those that they contact will be different. A schematic representation of the intermolecular contacts in the NMR non-muscle and skeletal tropomyosin overlap complexes is shown in Supporting information Figures S2 and S3. A few features are worthy of note.
The hydrogen bond between Lys5 and Glu280 seen in the crystal structures reported here is replaced in the NMR rat skeletal structure by a hydrogen bond between Lys5 and the backbone carbonyl oxygen of Ala277. Likewise, Lys7 participates in polar interactions in both the Gp7-TmC:TmN-EB1 and the NMR skeletal structure. In the smooth muscle tropomyosin complex the interaction occurs between side chains, whereas in the skeletal muscle isoform the lysine interacts with a main-chain carbonyl oxygen atom. Apparently, lysine residues can accommodate different binding partners because of the flexibility of the side chain. Lastly, there are differences in the structural orientation of the last few residues of the tropomyosin depending on the structural technique. In the X-ray structures reported here the α-helical motif is maintained all the way to the N and C-terminus. However, in the skeletal NMR structure the last three residues of the C-terminus lose their α-helical nature. Likewise, the first few residues at the N- terminus in the rat non-muscle NMR model adopt non-helical conformations.
In contrast to the comparisons with the NMR structures, the overlap region of chicken smooth muscle tropomyosin determined here is markedly different from the X-ray structure reported for rabbit skeletal tropomyosin (Figure 8D) (29). The primary disparity is that the N-terminal coiled-coil in the rabbit skeletal muscle tropomyosin overlap complex opens and separates so that only a single α-helix interacts with the C-terminal coiled-coil. In this latter structure, as in the structure of Gp7-TmC:TmN-XRCC4 presented here, there is also a C-terminal tropomyosin coiled-coil that does not participate in any interactions. The difference is that the N-terminal coiled-coil separates to interact with two symmetry-related molecules of the C-terminal fragment (The coordinates are reported in the lower space group P21 rather than the crystallographic symmetry C2221 to which the crystals belong in order to generate a complete molecule of the N-terminal coiled-coil in the final coordinates). Furthermore, in the rabbit skeletal X-ray structure the C-terminal coiled-coil does not separate to allow interdigitation by the N-terminal coiled-coil, as observed in all other structures, but remains a coiled-coil throughout its entirety. The resultant complex generates a trimeric coiled-coil instead of the four-helix bundle observed in all other structures of the overlap complex. This results in a relatively small buried surface area of 1045 Å2 for the complex, where 65% is non-polar. This is below the magnitude of surface area expected for a stable protein-protein interaction (62). The small buried surface area is reflected in the intermolecular contacts. There are very few interactions between the single helix and the C-terminal coiled-coil in the X-ray structure of skeletal tropomyosin (Figure 9) compared to the other structures of the overlap complex where the interactions are evenly distributed across the C-terminal coiled-coil. Indeed, there are only two interactions within 3.9Å with one C-terminal helix and three interactions with the other C-terminal helix. Interestingly, the same type of arrangement is observed in the low resolution structure of the troponin-T overlap complex (29). There is no simple explanation for the difference between this structure and all others, but it certainly reflects the flexibility of the α-helices at the termini of tropomyosin and the plasticity of their interactions. However, based on energetic considerations, one would expect that the helix-bundle overlap complex would be more physiologically relevant.
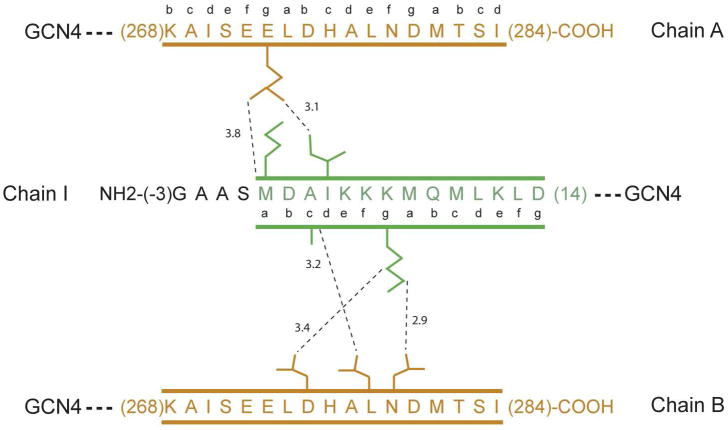
Figure 9.
Schematic representation of the pair-wise intermolecular contacts in the rabbit skeletal tropomyosin overlap obtained by X-ray diffraction. Chains A and B are the α-helices of the tropomyosin C-terminus, which remain as a coiled-coil throughout their length. Chain I is the single α-helix of the N-terminal tropomyosin segment that interacts in an oblique manner with the C-terminal coiled-coil. The intermolecular contacts shown in this figure were abstracted from coordinates obtained from the RCSB (accession number 2Z5I) (29).
CONCLUSIONS
The structures of the chicken smooth muscle tropomyosin overlap complex determined here reveal some important features of the tropomyosin filament. First, the small differences in the overlap region found in the two structures determined here suggest that the interaction between the N- and C-terminal ends of tropomyosin is intrinsically flexible, at least in solution. Second, the complex shows an inherent curvature. Both of these observations are consistent with the need for the tropomyosin filament to follow the helical path of the actin subunits, especially since the observed curvature than that necessary to fit around the filament. The hydrophobic effect is without question the major stabilizing factor the four-helix bundle that constitutes the overlap region, where the specificity for the interaction is provided by a few polar interactions at the periphery of the assembly. Interestingly, lysine residues contribute disproportionately to the polar interactions. The longer side chains appear to provide specificity and yet also accommodate conformational flexibility due to the length of the side chain.
The structure of the smooth muscle tropomyosin overlap complex is morphologically similar to that of skeletal tropomyosin complex obtained by NMR, but differs from the X-ray crystal structure of the skeletal tropomyosin complex. It is difficult to reconcile the dramatic differences between the three-helical bundle observed in the X-ray skeletal structure and the four-helix bundle seen in all other complexes. This may reflect the sensitivity of the weak overlap interaction in skeletal tropomyosin to outside forces such as crystal packing interactions, where the solution NMR approach is less susceptible to these issues.
Finally, the overlap structures presented here were obtained by creating a novel series of fusion proteins to stabilize the coiled-coil regions of tropomyosin and to modify the shape of the resulting construct. Inclusion of compact globular domains to the tropomyosin fragments greatly facilitated protein expression and allowed for multiple structural determinations of the same overlap complex. The fact that a very similar complex was formed from two completely different fusion constructs suggests that the complex observed is not an artifact of crystallization. Based on the success achieved here, it seems likely that XRCC4, Gp7, and EB1 could prove useful as general tools for expressing domains or regions of proteins that contain coiled-coils. Indeed, the structure of the C-terminal domain of Cnm67 from Saccharomyces cerevisiae was recently determined with this strategy (Frye et al., (2010) manuscript in preparation). In this case, the fusion protein with Gp7 was expressed readily in E. coli, whereas the equivalent construct with GCN4 failed to produce any soluble protein. Given that about ~6% of all eukaryotic proteins are annotated to contain coiled-coils (63), these additions to the repertoire of expression strategies should prove generally useful.
Supplementary Material
Acknowledgments
We thank Dr. Dwight Anderson (University of Minnesota) for the expression vector encoding the scaffolding protein Gp7 from bacteriophage φ29. We also thank Kirsten Dennison for help in constructing the Gp7 expression vector and Robert Smith for assistance in protein purification. I.R. thanks Dr. David B. Niemann and his team (University of Wisconsin) for facilitating the completion of this work.
ABBREVIATIONS
- Tm
tropomyosin
- TmC
C-terminal overlap region of tropomyosin
- TmN
N-terminal overlap region of tropomyosin
- SeMet
selenomethionine
- PEG
polyethylene glycol
- Gp7
scaffolding protein from bacteriophage φ29
- EB1
C-terminal helical bundle observed in the microtubule binding protein EB1
- XRCC4
human DNA ligase binding protein
- Gp7-TmC
a fusion protein that includes residues Pro2-Asp45 of Gp7 followed by residues Leu256-Met284 of chicken smooth muscle tropomyosin
- XRCC4-TmC
a fusion protein that includes residues Glu2-Ala135 of XRCC4 and chicken smooth muscle tropomyosin residues Lys248-Met284
- TmN-EB1
a fusion protein that includes residues Met1-Lys29 of chicken smooth muscle tropomyosin fused to residues Asp215-Asp257 of EB1
- rms
root mean square
Footnotes
This work was supported in part by NIH grants GM083987 and GM086351 to I.R. Use of the SBC BM19 beamline at the Argonne National Laboratory Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research, under Contract No. W-31-109-ENG-38.
X-ray coordinates for the XRCC4-TmC:TmN-EB1 and the Gp7-TmC:TmN-EB1 complexes have been deposited in the Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, N. J. (Protein Data Bank entries 3MUD, and 3MTU respectively)
Supporting Information Available
The alignment of the coiled-coil registration of Gp7, EB1, XRCC4, and tropomyosin together with the sequence of the final fusion proteins is shown in Figure S1. A schematic representation of the pair-wise intermolecular contacts in the rat skeletal tropomyosin NMR overlap complex is shown in Figure S2 in the same format used in Figures 5 and 7. Figure S3 shows a schematic representation of the pair-wise intermolecular contacts in the model for the rat non-muscle tropomyosin NMR overlap complex. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Crick FHC. The Packing of a-Helices: simple coiled coils: Simple Coiled-Coils. Acta Cryst. 1953:689–697. [Google Scholar]
- 2.McLachlan AD, Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- 3.Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv Protein Chem. 2005;71:121–159. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- 4.Gunning P. Introduction and historical perspective. Adv Exp Med Biol. 2008;644:1–5. doi: 10.1007/978-0-387-85766-4_1. [DOI] [PubMed] [Google Scholar]
- 5.Hitchcock-DeGregori SE. Tropomyosin: function follows structure. Adv Exp Med Biol. 2008;644:60–72. doi: 10.1007/978-0-387-85766-4_5. [DOI] [PubMed] [Google Scholar]
- 6.Gunning P, O’Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- 8.Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem. 1989;264:7490–7497. [PubMed] [Google Scholar]
- 10.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Vrhovski B, Theze N, Thiebaud P. Structure and evolution of tropomyosin genes. Adv Exp Med Biol. 2008;644:6–26. doi: 10.1007/978-0-387-85766-4_2. [DOI] [PubMed] [Google Scholar]
- 12.Phillips GN, Jr, Lattman EE, Cummins P, Lee KY, Cohen C. Crystal structure and molecular interactions of tropomyosin. Nature. 1979;278:413–417. doi: 10.1038/278413a0. [DOI] [PubMed] [Google Scholar]
- 13.Johnson P, Smillie LB. Polymerizability of rabbit skeletal tropomyosin: effects of enzymic and chemical modifications. Biochemistry. 1977;16:2264–2269. doi: 10.1021/bi00629a035. [DOI] [PubMed] [Google Scholar]
- 14.Cho YJ, Liu J, Hitchcock-DeGregori SE. The amino terminus of muscle tropomyosin is a major determinant for function. J Biol Chem. 1990;265:538–545. [PubMed] [Google Scholar]
- 15.Palm T, Greenfield NJ, Hitchcock-DeGregori SE. Tropomyosin ends determine the stability and functionality of overlap and troponin T complexes. Biophys J. 2003;84:3181–3189. doi: 10.1016/S0006-3495(03)70042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galinska-Rakoczy A, Engel P, Xu C, Jung H, Craig R, Tobacman LS, Lehman W. Structural basis for the regulation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2008;379:929–935. doi: 10.1016/j.jmb.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul DM, Morris EP, Kensler RW, Squire JM. Structure and orientation of troponin in the thin filament. J Biol Chem. 2009;284:15007–15015. doi: 10.1074/jbc.M808615200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitby FG, Kent H, Stewart F, Stewart M, Xie X, Hatch V, Cohen C, Phillips GN., Jr Structure of tropomyosin at 9 angstroms resolution. J Mol Biol. 1992;227:441–452. doi: 10.1016/0022-2836(92)90899-u. [DOI] [PubMed] [Google Scholar]
- 19.Phillips GN, Jr, Fillers JP, Cohen C. Tropomyosin crystal structure and muscle regulation. J Mol Biol. 1986;192:111–131. doi: 10.1016/0022-2836(86)90468-7. [DOI] [PubMed] [Google Scholar]
- 20.Tobacman LS. Cooperative binding of tropomyosin to actin. Adv Exp Med Biol. 2008;644:85–94. doi: 10.1007/978-0-387-85766-4_7. [DOI] [PubMed] [Google Scholar]
- 21.Hitchcock-DeGregori SE, Singh A. What makes tropomyosin an actin binding protein? A perspective. J Struct Biol. 2010;170:319–324. doi: 10.1016/j.jsb.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hitchcock-DeGregori SE, Heald RW. Altered actin and troponin binding of amino-terminal variants of chicken striated muscle alpha-tropomyosin expressed in Escherichia coli. J Biol Chem. 1987;262:9730–9735. [PubMed] [Google Scholar]
- 23.Greenfield NJ, Stafford WF, Hitchcock-DeGregori SE. The effect of N-terminal acetylation on the structure of an N-terminal tropomyosin peptide and alpha alpha-tropomyosin. Protein Sci. 1994;3:402–410. doi: 10.1002/pro.5560030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock-DeGregori SE, Cohen C. Deciphering the design of the tropomyosin molecule. Proc Natl Acad Sci U S A. 2001;98:8496–8501. doi: 10.1073/pnas.131219198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Mui S, Brown JH, Strand J, Reshetnikova L, Tobacman LS, Cohen C. The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site. Proc Natl Acad Sci U S A. 2002;99:7378–7383. doi: 10.1073/pnas.102179999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenfield NJ, Swapna GV, Huang Y, Palm T, Graboski S, Montelione GT, Hitchcock-DeGregori SE. The structure of the carboxyl terminus of striated alpha-tropomyosin in solution reveals an unusual parallel arrangement of interacting alpha-helices. Biochemistry. 2003;42:614–619. doi: 10.1021/bi026989e. [DOI] [PubMed] [Google Scholar]
- 27.Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J Mol Biol. 2006;364:80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 28.Greenfield NJ, Kotlyanskaya L, Hitchcock-DeGregori SE. Structure of the N terminus of a nonmuscle alpha-tropomyosin in complex with the C terminus: implications for actin binding. Biochemistry. 2009;48:1272–1283. doi: 10.1021/bi801861k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami K, Stewart M, Nozawa K, Tomii K, Kudou N, Igarashi N, Shirakihara Y, Wakatsuki S, Yasunaga T, Wakabayashi T. Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T. Proc Natl Acad Sci U S A. 2008;105:7200–7205. doi: 10.1073/pnas.0801950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenfield NJ, Huang YJ, Palm T, Swapna GV, Monleon D, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein. J Mol Biol. 2001;312:833–847. doi: 10.1006/jmbi.2001.4982. [DOI] [PubMed] [Google Scholar]
- 31.Minakata S, Maeda K, Oda N, Wakabayashi K, Nitanai Y, Maeda Y. Two-crystal structures of tropomyosin C-terminal fragment 176–273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule. Biophys J. 2008;95:710–719. doi: 10.1529/biophysj.107.126144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown JH, Zhou Z, Reshetnikova L, Robinson H, Yammani RD, Tobacman LS, Cohen C. Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc Natl Acad Sci U S A. 2005;102:18878–18883. doi: 10.1073/pnas.0509269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J. 2000;19:5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- 35.Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- 36.Slep KC, Rogers SL, Elliott SL, Ohkura H, Kolodziej PA, Vale RD. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J Cell Biol. 2005;168:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 38.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 39.Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen GJ, Qiu N, Karrer C, Caspers P, Page MG. Restriction site-free insertion of PCR products directionally into vectors. Biotechniques. 2000;28:498–500. 504–495. doi: 10.2144/00283st08. [DOI] [PubMed] [Google Scholar]
- 41.van den Ent F, Lowe J. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods. 2006;67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 43.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 46.Cowtan K. ‘dm’: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 47.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 48.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 49.Zhang KY, Cowtan K, Main P. Combining constraints for electron-density modification. Methods Enzymol. 1997;277:53–64. doi: 10.1016/s0076-6879(97)77006-x. [DOI] [PubMed] [Google Scholar]
- 50.Cowtan K. General quadratic functions in real and reciprocal space and their application to likelihood phasing. Acta Crystallogr D Biol Crystallogr. 2000;56:1612–1621. doi: 10.1107/s0907444900013263. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Skubak P, Murshudov GN, Pannu NS. Direct incorporation of experimental phase information in model refinement. Acta Crystallogr D Biol Crystallogr. 2004;60:2196–2201. doi: 10.1107/S0907444904019079. [DOI] [PubMed] [Google Scholar]
- 53.Tsodikov OV, Record MT, Jr, Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J Comput Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 54.Monteiro PB, Lataro RC, Ferro JA, Reinach FC. Functional alpha-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. J Biol Chem. 1994;269:10461–10466. [PubMed] [Google Scholar]
- 55.Lehrer SS, Stafford WF., 3rd Preferential assembly of the tropomyosin heterodimer: equilibrium studies. Biochemistry. 1991;30:5682–5688. doi: 10.1021/bi00237a007. [DOI] [PubMed] [Google Scholar]
- 56.Oakley MG, Kim PS. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry. 1998;37:12603–12610. doi: 10.1021/bi981269m. [DOI] [PubMed] [Google Scholar]
- 57.Akey DL, Malashkevich VN, Kim PS. Buried polar residues in coiled-coil interfaces. Biochemistry. 2001;40:6352–6360. doi: 10.1021/bi002829w. [DOI] [PubMed] [Google Scholar]
- 58.Holmes KC, Lehman W. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil. 2008;29:213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- 59.Li XE, Lehman W, Fischer S, Holmes KC. Curvature variation along the tropomyosin molecule. J Struct Biol. 2009 doi: 10.1016/j.jsb.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanders C, Smillie LB. Chicken gizzard tropomyosin: head-to-tail assembly and interaction with F-actin and troponin. Can J Biochem Cell Biol. 1984;62:443–448. doi: 10.1139/o84-060. [DOI] [PubMed] [Google Scholar]
- 61.Paulucci AA, Katsuyama AM, Sousa AD, Farah CS. A specific C-terminal deletion in tropomyosin results in a stronger head-to-tail interaction and increased polymerization. Eur J Biochem. 2004;271:589–600. doi: 10.1111/j.1432-1033.2003.03961.x. [DOI] [PubMed] [Google Scholar]
- 62.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q Rev Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 63.Rose A, Schraegle SJ, Stahlberg EA, Meier I. Coiled-coil protein composition of 22 proteomes--differences and common themes in subcellular infrastructure and traffic control. BMC evolutionary biology. 2005;5:66. doi: 10.1186/1471-2148-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.