Abstract
A novel angucycline metabolite, 2,3-dehydro-UWM6, was identified in a jadH mutant of Streptomyces venezuelae ISP5230. Both UWM6 and 2,3-dehydro-UWM6 could be converted to jadomycin A or B by a ketosynthase a (jadA) mutant of S. venezuelae. These angucycline intermediates were also converted to jadomycin A by transformant of the heterologous host Streptomyces lividans expressing the jadFGH oxygenases in vivo and by its cell-free extracts in vitro; thus the three gene products JadFGH are implicated in catalysis of the post-polyketide synthase biosynthetic reactions converting UWM6 to jadomycin aglycone. Genetic and biochemical analyses indicate that JadH possesses dehydrase activity, not previously associated with polyketide-modifying oxygenase. Since the formation of aromatic polyketides often requires multiple dehydration steps, bifunctionality of oxygenases modifying aromatic polyketides may be a general phenomenon.
Polyketides, a large structurally diverse group of secondary metabolites, are produced in bacteria, fungi and plants by polyketide synthase (PKS)1 complexes (1, 2). The remarkable diversity of polyketides is to a great extent due to post-PKS modifications that determine the ultimate structures of polyketide metabolites formed. Prominent among the tailoring enzymes responsible for post-PKS modifications are oxidoreductases and group transferases (3–5).
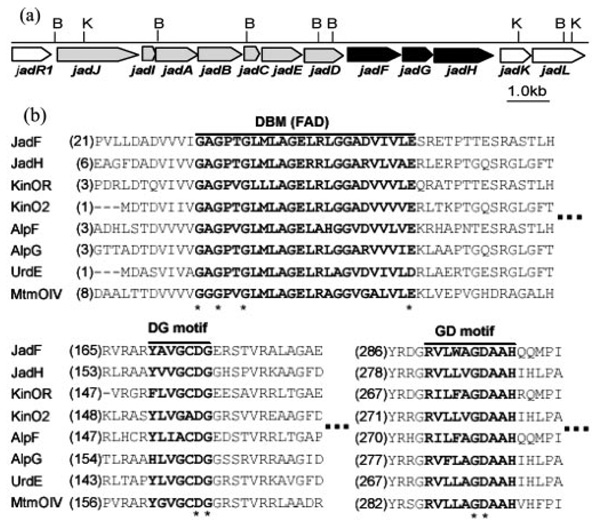
The atypical angucycline antibiotic jadomycin B (1) and its aglycone jadomycin A (2) are produced in cultures of Streptomyces venezuelae ISP5230 grown under stress conditions in a medium containing isoleucine (6, 7). The angucycline origin of jadomycins was verified by accumulation of a well known antibiotic rabelomycin (3) in the jadF disruption mutant VS655 (7) and by production of UWM6 (4) in Streptomyces lividans transformed with the jad PKS gene cluster (i.e. jadABCDEIJ) (8). One unique feature of the jadomycin family is its nitrogen-containing pentacyclic benz[b]oxazolophenanthridine ring, formed via a hitherto uncharacterized oxidative cleavage between C-5 and C-6 of ring B in an angucyclic polyketide intermediate. The phenanthridine ring system is plausibly derived by condensation of an amino acid with the postulated acid/aldehyde intermediate (5), formed during angucyclic ring opening (7, 9). Strengthening this possibility is the formation of jadomycin analogues when different amino acids are supplied as precursors (10, 11). Until now, the only oxygenase gene identified in the jad biosynthetic cluster (Fig. 1a) is jadF, which is implicated in oxidative ring cleavage and displays strong sequence homologies with FAD- and NADPH-dependent mono-oxygenases (7). Sequence analyses suggest that the two genes, jadG and jadH, immediately downstream of jadF also encode oxygenases. JadG resembles anthrone oxygenases such as ActVA-orf6 and TcmH, while JadH strongly resembles JadF in its amino acid sequence. Consequently, JadF and JadH are potential candidates for participation in oxidative ring B cleavage (7, 12).
FIG. 1.
a, partial gene organization map of jadomycin biosynthetic gene cluster. Genes responsible for UWM6 biosynthesis are indicated in gray, and genes studied in this paper are indicated in black. B, BamHI; K, KpnI. b, conserved motifs in JadF, JadH, and related proteins. Three conserved motifs are indicated in bold and marked on top. The highly conserved residues in motifs are marked with asterisk.
In this work, we further investigated the functions of JadF, JadG and JadH by product profile analyses of their mutants, enzymatic assay and in vivo/in vitro bioconversion experiments. We demonstrate the requirement for the co-presence of JadF, JadG and JadH to completely convert UWM6 to jadomycin A and established JadH as a bifunctional oxygenase/dehydrase.
EXPERIMENTAL PROCEDURES
Materials
S. venezuelae ISP5230 and the derived strains VS655 (jadF mutant) and VS662a (jadR2 mutant) have been described previously (7, 13). Escherichia coli ET12567 has been described by MacNeil et al. (14); other E. coli strains were from commercial sources; S. lividans TK24 was described by Hopwood et al. (15). Plasmid pWHM1238, described in Kulowski et al. (1999), was kindly provided by Dr. Ben Shen (8); plasmid pUWL201, described in Doumith et al. (2000), was kindly provided by Dr. Udo Wehmeier (16). Ultrafiltration centrifugation tubes (Centriplus YM series) were purchased from Millipore. Restriction enzymes, T4 DNA ligase, and Pfu DNA polymerase were purchased from Promega or Takara.
DNA Manipulation and Transformation
Competent E. coli cells were prepared and transformed by standard procedures (17). Plasmid DNA was isolated from E. coli by the alkaline method (17). Cultures of S. venezuelae strains used for DNA extraction were grown in MYME medium (13) at 30 °C for 36 h; genomic DNA was isolated as described by Kieser et al. (18). DNA was manipulated by standard procedures (17). Protoplasts of S. venezuelae were prepared and transformed as described previously (7).
Sequencing of jadFGH and Sequence Analysis
pJV69A was constructed by inserting a 7.2-kb XhoI fragment (with intact jadDFGHK and partial jadL) into pBluescript II KS(+). The jadFGH portion of pJV69A was re-sequenced; its revised sequence was deposited in GenBank™ (accession number AY773079). Related proteins were searched with BLASTP (www.ncbi.nlm.nih.gov), and selected sequences were aligned with ClustalX (ftp-igbmc.u-strasbg.fr/pub/ClustalX/) (19).
Gene Inactivations
Disruption of jadH
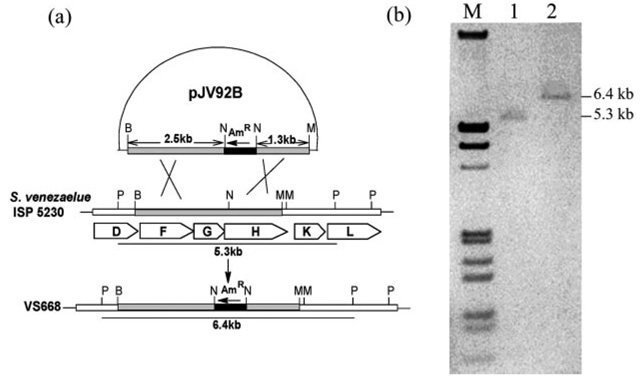
To facilitate inactivation of this gene, a 6.0-kb BamHI fragment of S. venezuelae DNA containing jadFGHK was cloned in pHJL400, furnishing pJV77A. Construction of the jadH disruption plasmid involved removing an EcoRI/MluI fragment from pJV77A. The remaining fragment, consisting of the vector plus a 3.8-kb BamHI/MluI insert, was blunt-ended by treatment with the Klenow fragment of DNA polymerase I and religated to generate pJV91. Digestion of pJV91 with NcoI (internal to jadH) and ligation of the linear product to an apramycin resistance gene with NcoI ends yielded the two recombinant plasmids, pJV92A and pJV92B, which carried the apramycin resistance gene in opposite orientations. When both plasmids were introduced into S. venezuelae, only pJV92B gave transformants (VS667). Selection for an apramycin-resistant and thiostrepton-sensitive phenotype gave the jadH mutant VS668 (Fig. 2).
FIG. 2.
a, diagram illustrating the construction of VS668 by disrupting jadH with an apramycin-resistance gene (AmR). B, BamHI; N, NcoI, M, MluI; P, PstI. b, Southern blotting of S. venezuelae ISP5230 and VS668 genomic DNA digested with PstI. Lane M, digoxigeninlabeled λ DNA (HindIII- and EcoRI-digested); lane 1, S. venezuelae ISP5230 genomic DNA; lane 2, VS668 genomic DNA.
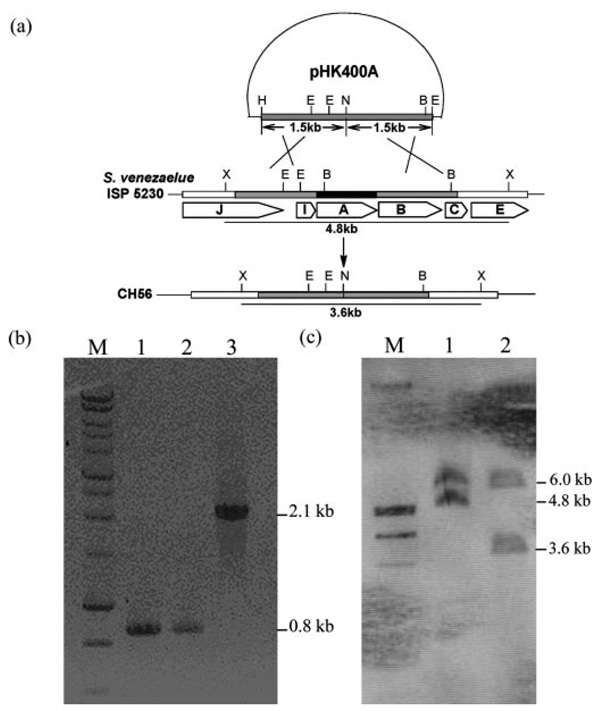
In-frame Deletion of jadA
Two 1.5-kb fragments flankingjadA were obtained by PCR using primer pairs P1 and P2. For the fragment upstream of jadA, P1F (5′-CCCAAGCTTGCAGTGCCTGGCCGACCA-3′, HindIII) and P1R (5′-GGAATTCCATATGTCACGCGTTCGCCTC-CCA-3′, NdeI) were used; for the fragment downstream of jadA, P2F (5′-GGAATTCCATATGAGCGCGTCCGTGGTG-3′, NdeI) and P2R (5′-CGGAATTCAGGCGGCGGCGACGGC-3′, EcoRI) were used. After digestion with appropriate enzymes, the two fragments were inserted into HindIII/EcoRI-digested pHJL400 to generate pHK400A. The latter was used to transform protoplasts of VS662a. Transformants were propagated on MYM (maltase, yeast extract, and malt extract; Ref. 6) agar without thiostrepton selection for three generations; colonies from spores collected after non-selective propagation were picked for sensitivity to thiostrepton and examined for loss of jadomycin B production in cultures grown under conditions supporting biosynthesis of the antibiotic. The non-producing mutant was designated CH56. Evidence that the loss of jadomycin production in CH56 correlated with deletion of jadA was investigated by PCR using primers P3F (5′-GGCCACCCGCTTCTACAAC-3′) and P3R (5′-CGAAGGTGGAGCCGTATCC-3′) (Fig. 3).
FIG. 3.
a, diagram illustrating the construction of CH56 by in-frame deleting of jadA. H, HindIII; E, EcoRI; N, NdeI; B, BamHI; X, XhoI. b, PCR detection of jadA deletion. Lane M is DNA ladder marker, and lanes 1,2, and 3 are fragments obtained by PCR with pHK400A, CH56, and wild-type genomic DNA as template, respectively. c, Southern blotting of S. venezuelae ISP5230 and CH56 genomic DNA digested with XhoI. Lane M, digoxigenin-labeled λ DNA (HindIII- and EcoRI-digested); lane 1, S. venezuelae ISP5230 genomic DNA; lane 2, CH56 genomic DNA.
Southern Hybridization
For mutants with disruptions in jadH, the genomic DNA was completely digested with PstI, fractionated by agarose gel electrophoresis, and transferred to a positively charged nylon membranes. The probe was a 552-bp KpnI/PshAI fragment from pJV69A labeled with digoxigenin-dUTP by the random priming method (Fig. 2).
The genomic DNA from jadA deletion mutants was digested with XhoI and a 561-bp PstI/EcoRI fragment from pWHM1238 was labeled as a probe. Hybridization of probes with DNA fragments on the nylon membrane was detected by the chromogenic method using procedures described by Roche Diagnostics (Fig. 3).
Isolation and Structural Characterization of the Product Accumulated by VS668
Filtered cultures of VS668 grown in d-galactose-l-isoleucine liquid medium containing 25 µg/ml apramycin as described by Doull et al. (10) were extracted with ethyl acetate. After fractionation of the crude extract by semipreparative high performance liquid chromatography (HPLC), the main product was isolated and its structure was elucidated by NMR, as described elsewhere (20).
Expression of jadFGH in S. lividans
The 2.3-kb insert of pJV60 (7) was excised by SacI digestion, and the purified fragment was ligated into the SacI site of pUC19 to yield plasmid pUC19-jadF, in which the transcriptional direction of jadF is opposite to that of lacZ. pUC19-jadF was digested with EcoRI and XbaI to re-excise the insert, which was then cloned into EcoRI/XbaI-digested expression vector pUWL201 (16) to create pUWL201+jadF. Two fragments, 6.3 kb (KpnI) and 0.5 kb (PstI/KpnI) from pUWL201+jadF were ligated with a 4.9-kb (PstI/KpnI) fragment of pJV69A to generate pUWL2014, in which jadFGH is positioned downstream of the “ermE up” promoter of pUWL201. Introduction of pUWL2014 into S. lividans TK24 by standard procedures gave the transformant LC2014 (18).
Preparation of Substrates for Bioconversion Assays
Rabelomycin was isolated from a culture of VS655 as described previously (7). 2,3-Dehydro-UWM6 was isolated from S. venezuelae mutant VS668 as described above. To obtain UWM6, S. lividans TK24 was transformed with pUWL1238. From cultures of the transformant (LC1238) grown as described earlier, UWM6 was obtained (8); however, UWM6 was unstable and was partially converted to rabelomycin. LC1238 culture extracts used here as the substrate for bioconversion and enzyme assays were shown by HPLC to consist mainly of UWM6.
In Vivo Bioconversion Assays
CH56
The in-frame deletion jadA mutant was cultured for jadomycin production by the procedure described by Doull et al. (10) except that the culture medium was supplemented with 25 µg/ml apramycin. Putative substrates (UWM6, 2,3-dehydro-UWM6, and rabelomycin) were added to the cultures at the time (6 h after inoculation) they were stressed with 6% (v/v) ethanol. After another 24 h, the cultures were extracted with ethyl acetate and the extracts were evaporated in vacuo. The residues were analyzed by TLC, HPLC, and liquid chromatogra-phymass spectroscopy (LC-MS).
LC2014
seed cultures of LC2014 (containing jadFGH in pUWL2014) and LC2010 (containing pUWL201) were grown at 30 °C, 220 rpm for 24 h in YEME (18) supplemented with thiostrepton (10 µg/ml). Mycelium was collected by centrifugation (3000 × g, 10 min) and transferred to d-galactose-l-isoleucine medium supplemented with UWM6 or 2,3-dehydro-UWM6. The cultures were incubated for 24 h and extracted with ethyl acetate. Cultures of LC2010 treated similarly to LC2014 served as controls.
In Vitro Bioconversion Assay with LC2014 Cell-free Extracts
LC2014 was cultured in YEME with thiostrepton (10 µg/ml) at 30 °C, 220 rpm for 3 days. After the culture was harvested and diluted appropriately with sterilized water, mycelia were collected by centrifugation (3000 × g, 10 min) and washed with a buffer containing 50 mm Tris-HCl (pH 7.4) and 10% glycerol twice. Then the mycelia were resuspended in 10 ml of lysis buffer (50 mm Tris-HCl (pH 7.4), 1 mm EDTA, 1 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, 10% glycerol) and kept on ice for 30 min. Cells were disrupted by sonication for 16 × 5 s pulses, and cell debris was removed by centrifugation (15,000 × g) at 4 °C for 20 min. The supernatant was fractionated by a 10-kDa ultra-filtration centrifugation tube and concentrated to about 1 ml. Protein concentration was monitored by Bradford dye-binding method (21). Reaction mixtures (2 ml) containing 1 mg of cell-free extracts protein, 50 mm Tris-HCl (pH 7.5, 8.0, 8.5, 9.0, and 9.5), 40 µm FAD, 500 µm NADPH, 500 µm substrates (UWM6 or 2,3-dehydro-UWM6), and 5 mm isoleucine were incubated with gentle shaking at 30 °C for 6 h. The reactions were terminated by addition of 80 µl of 1 n HCl, and then the mixture was extracted with ethyl acetate. Cell-free extract of LC2010 was prepared with the same procedure as LC2014 and used as control.
Expression and Purification of JadFGH (N-His-tagged)
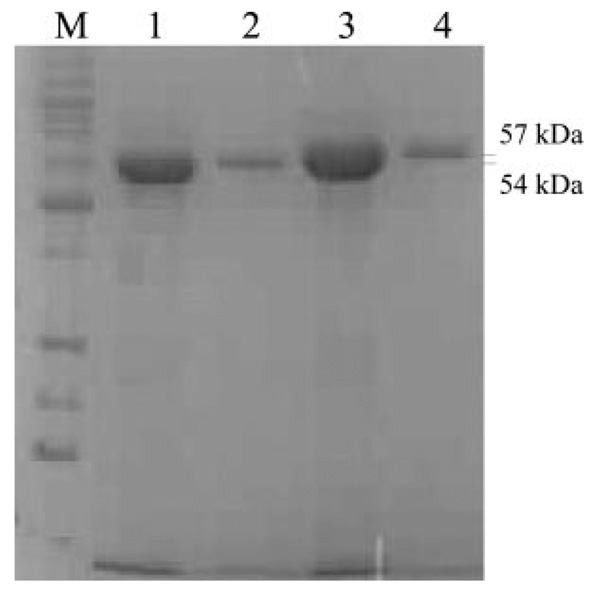
Plasmid pJV60 (containing jadF) was used as template for constructing the jadF overexpression plasmid (pRCR2). Plasmid pJV455 (containing jadFGH), constructed by inserting a 5.8-kb PstI fragment from pJV69A to pBluescript II SK(+), was used as template for constructing the jadH overexpression plasmids (pLMG1). The fragments containing jadF or jadH were generated by PCR using Pfu DNA polymerase. For pRCR2, the primers 5′-GCCGAGGGATCCCGCTCATATGACGGAGCC-3′ (jadF forward, BamHI) and 5′-TCGAAGAGGTTGAATTCGGTCAGGTAGCCG-3′ (jadF reverse, EcoRI) were used; for pLMG1, the primers 5′-GCGCTCGCTGCAGTGAGCGCCGTGACCACCACC-3′ (jadH forward, PstI), and 5′-CCGCCGGAGAATTCGTCGTGGTCGT-CACCGGGC-3′ (jadH reverse, EcoRI) were used. The genes jadF (1539 bp) and jadH (1626 bp) were inserted into the pRSETb overexpression vector, which contains the T7 promoter and an N-terminal His6-tag encoding sequence. Successful construction of the overexpression systems was confirmed by DNA sequencing and restriction analysis. Strains of E. coli BL21(DE3)plysS transformed with pRCR2 and pLMG1 were cultured at 37 °C in Luria-Bertani (LB) medium supplemented with 50 µg/ml carbenicillin and 35 µg/ml chloramphenicol. When cultures had grown to A600 0.5–0.6, isopropyl β-d-thiogalactopy-ranoside was added to a final concentration of 1 mm; growth was continued at 18–20 °C for up to 5 h. Cells were harvested by centrifugation (4000 × g, 4 °C, 20 min). The pellets were resuspended in lysis buffer (50 mm NaH2PO4, 300 mm NaCl (pH 8.0)) and kept on ice for 0.5 h before sonication (6 × 10 s). Protamine sulfate was added to the lysate, and the insoluble debris was removed by centrifugation (10,000 × g, 4 °C, 0.5 h). The supernatant comprising His6-tagged proteins was loaded on a column of Sepharose-based TALON metal (Co2+) affinity resin previously treated with equilibration buffer (50 mm NaH2PO4, 300 mm NaCl (pH 7.0)). The column was washed with 20 volumes of equilibration buffer before purified proteins were recovered with elution buffer (50 mm NaH2PO4, 300 mm NaCl, 150 mm imidazole (pH 7.0)). Purified proteins were detected by SDS-PAGE, and their concentrations were estimated by the Bradford dye-binding method (21). The monitoring of eluates by SDS-PAGE showed that two proteins of ~54 and 57 kDa had been produced (Fig. 4a). Both of the two proteins were also identified by tryptic digestion and subsequent peptide matching of the fragments observed by electrospray ionization-MS with the data base sequence of the proteins.
FIG. 4. SDS-PAGE of purified JadF and JadH (His-tagged).
Lane M, low molecular weight protein marker; Lanes 1 and 2 are two elution fractions of His6-JadF; lanes 3 and 4 are two elution fractions of His6-JadH.
In Vitro Enzymatic Assays
Reaction mixtures (1 ml) containing freshly purified oxygenase (1 µm), 0.1 m Tris-HCl buffer (pH 8.0), FAD (4 µm), NADPH (1 µm), and substrate (400 µm) were incubated at 30 °C for 2 h. Reactions were terminated by adding 40 µl of 1 n HCl (to pH 4.0) and then extracted with ethyl acetate. As controls, mixtures of all components except the substrate were treated in the same way.
Analytical and Spectroscopic Procedures
TLC was used as described earlier (7). Jadomycins A and B, UWM6, 2,3-dehydro-UWM6, and rabelomycin were monitored by HPLC on a Waters 717 system with a Waters 2487 dual λ UV detector and an inertsil ODS-3 column (GL sciences, 5 µm, 4.6 × 250 mm). The elution solvents were: water with 0.1% trifluoroacetic acid (solvent A) and acetonitrile with 0.1% trifluoroacetic acid (solvent B). The percentage of solvent B (flow rate: 1.0 ml/min) changed linearly from 50 to 100% between 0 and 20 min, stayed at 100% for 3 min, and then decreased to 50% between 23 and 23.5 min. For LC-MS analysis, an Agilent 1100 system was used, maintaining the parameters described above for HPLC and for the electrospray ionization-MS component using Finnigan LCQ DecaXP ion trap mass spectrometry (Thermo Finnigan, San Jose, CA) with a spray voltage of 4.5 kv and a heated transfer capillary temperature set to 275 °C. The mass spectrometer scanned from m/z = 100–600 in positive/negative ion mode.
RESULTS
Analyses of Protein Sequences
The amino acid sequences of JadF and JadH are similar (45.8% identity) and resemble those of other FAD-dependent aromatic hydroxylases, e.g. UrdE (44.7% identity with JadF and 54.9% identity with JadH) (7, 12) and several oxygenases implicated in aromatic polyketide ring cleavage reactions. For example, MtmOIV, postulated to be a Baeyer-Villiger monooxygenase (22, 23) responsible for the fourth ring cleavage in the aureolic acid polyketide mithramycin, shares 37.3% identity with JadF and 38.5% identity with JadH. The oxygenase Cm-mOIV (39.0% identity with JadF and 37.5% with JadH) is suggested to function like MtmOIV in the gene cluster for biosynthesis of the aureolic acid polyketide chromomycin (24), and the oxygenase GrhO6 (35.3% identical to JadF and 36.4% to JadH) is postulated to catalyze ring cleavage in the rubro-mycin-like polyketide, griseorhodin (25). Three motifs conserved in FAD- and NAD(P)H-dependent monooxygenases are found in JadF and JadH: a dinucleotide binding motif characterized by a Rossmann fold (GxGxxG/A) interacting with the adenosine of FAD (26, 27), a GD motif responsible for binding the FAD flavin moiety (28) and a DG motif involved in both FAD and NAD(P)H binding (Fig. 1) (29).
The overall sequence of JadG is consistent with a protein constructed from two halves (amino acids 1–112 and 113–225) sharing a high degree (22.8%) of sequence identity and implying an evolution event via gene duplication. The two halves are each similar to anthrone oxygenases such as ActVA-Orf6 (17.5 and 32.8% amino acid identity) (30, 31) and TcmH (21.9 and 32.2% amino acid identity) (32), suggesting that JadG catalyzes similar monooxygenation in jadomycin biosynthesis.
Evaluation of the jadH Disruption Mutant (VS668)
Replacement of the expected hybridization signal at 5.3 kb in wild-type S. venezuelae genomic DNA with the signal at 6.4 kb in DNA from the insertionally inactivated apramycin-resistant and thiostrepton-sensitive transformant were consistent with disruption of jadH in VS668 (Fig. 2b). Growth of the transformant under conditions promoting jadomycin production by the wild-type resulted in excretion of a yellow compound with RF 0.69 (TLC) and retention time 6.733 min (HPLC). Extraction, purification, and structure analysis of the metabolite suggested that it was 2,3-dehydro-UWM6 (6) (20).
Construction ofajadA In-frame Deletion Mutant
To facilitate the choice of appropriate substrates for bioconversion experiments, a jadA in-frame deletion mutant (CH56) was constructed by deleting a 1.2-kb internal fragment from the jadA sequence in VS662a, a strain in which the regulation of jadomycin biosynthesis has been disturbed by inserting an apramycin resistance cassette into the jadR2 repressor gene (13). Phenotypically, the jadR2 mutant (VS662a) differs from the wild type in producing jadomycin B without external stimulation and in producing twice the yield of the wild type when stressed with ethanol. Since S. venezuelae strains derived from VS662a should potentially be more efficient than the wild-type in bioconversion assays, VS662a protoplasts transformed with pHK400A were screened for a thiostrepton-resistant pheno-type. The strains selected were propagated for three generations on MYM agar. Of 24 thiostrepton-sensitive strains examined under growth conditions supporting jadomycin B production, 13 still produced this product, while 11 had lost the ability. PCR examination of the 11 clones proved jadA deletion occurred (Fig. 3b). The deletion of jadA was also verified by Southern blotting (Fig. 3c). One of the 11 mutants was chosen and designated CH56. HPLC analysis showed no jadomycin B production by CH56 in the first 24 h after ethanol treatment, but a small amount (about 3% of that in VS662a) was detected when the fermentation was prolonged to 48 h. The “leakage” phenomenon may be explained by a partial complementation of the jadA function by another ketosynthase α gene in S. venezuelae ISP5230.
In Vivo Bioconversion Assays
CH56
UWM6, 2,3-dehydro-UWM6, and rabelomycin were tested as substrates for bioconversion by incubation with the culture of CH56 (jadA in-frame deletion mutant) and examination of the broths for recognizable products. HPLC analysis of the incubation mixture showed that 2,3-dehydro-UWM6 disappeared rapidly, and two new peaks, corresponding to jadomycin B and jadomycin A, appeared in a ratio similar to that in wild-type S. venezuelae ISP5230 (the titer of jadomycin A was 5–15% that of jadomycin B). The identities of the peaks (confirmed by LC-MS) indicated that 2,3-dehydro-UWM6 was efficiently converted to jadomycins. During incubation of UWM6 with strain CH56, the substrate peak disappeared completely, giving rabelomycin and small amounts of jadomycins A and B. The ratio between jadomycins A and B was also similar to that in wild-type S. venezuelae ISP5230. The low efficiency of this bioconversion might be due to the instability of the substrate, which is converted to rabelomycin by spontaneous dehydration and oxygenation (8). However, no jadomycins were detected when rabelomycin was the substrate, although the amount of rabelomycin decreased (Table I).
Table I.
Bioconversion assays in vivo and in vitro
| Strain/Enzyme | Substrate | Main products | In vivo/In vitro |
|---|---|---|---|
| CH56 | UWM6 | Jadomycin A/B | In vivo |
| CH56 | 2,3-Dehydro-UWM6 | Jadomycin A/B | In vivo |
| CH56 | Rabelomycin | Unidentified product | In vivo |
| LC2014 | UWM6 | Jadomycin A and rabelomycin | In vivo |
| LC2014 | 2,3-Dehydro-UWM6 | Jadomycin A | In vivo |
| LC2014 | UWM6 | Jadomycin A and rabelomycin | In vitro |
| LC2014 | 2,3-Dehydro-UWM6 | Jadomycin A and dehydrorabelomycin | In vitro |
| JadH | UWM6 | Rabelomycin | In vitro |
| JadH | 2,3-Dehydro-UWM6 | Dehydrorabelomycin | In vitro |
| JadH | rabelomycin | Rabelomycin | In vitro |
LC2014
To elucidate the combined functions of JadFGH, UWM6 and 2,3-dehydro-UWM6 were used as substrates for conversion by LC2014 (jadFGH expressed). When UWM6 was incubated with a culture of LC2014, its HPLC peak disappeared within 24 h; a small peak corresponding to jadomycin A appeared and was shown by LC-MS to have the expected molecular weight (419 Da); a peak corresponding to rabelomycin was also observed. In cultures of LC2010, UWM6 also disappeared, but instead of jadomycin A, it was mainly converted to rabelomycin. In contrast, 2,3-dehydro-UWM6 was unchanged in cultures of LC2010, while in LC2014 it quickly gave jadomycin A, as expected.
In Vitro Bioconversion Assays with LC2014 Cell-free Extracts
At pH 7.0 and 7.5, no obvious conversion of the substrates (UWM6 and 2,3-dehydro-UWM6) was observed. New peaks emerged when the pH of the reaction mixtures were increased to 8.0, 8.5, and 9.0; the optimum pH was 8.5. When assayed at this pH, UWM6 was mainly converted to rabelomycin; a small peak corresponding to jadomycin A was also observed. In the case of 2,3-dehydro-UWM6, rapid consumption and conversion to jadomycin A and dehydrorabelomycin were observed after 6-h incubation at pH 8.5, 30 °C. Both UWM6 and 2,3-dehydro-UWM6 were depleted in 6 h; and the amount of jadomycin A converted from UWM6 was only about 2% of that from 2,3-dehydro-UWM6. In the control experiment with LC2010 cell-free extract, the decrease of UWM6 and 2,3-dehydro-UWM6 was not obvious, and jadomycin A was not observed. The bio-conversions by LC2014 (in vivo and in vitro) indicate that post-PKS modification of UWM6 to jadomycin A is brought about by the co-presence of JadF, JadG and JadH (Table I) but do not eliminate the possibility of a functional contribution from the host.
In Vitro Enzyme Assays
To elucidate the catalytic activity of JadF and JadH, which were suspected to be bifunctional oxygenase/dehydrases (see “Discussion”), the two enzymes (N-terminal His-tagged) were expressed in E. coli and purified for enzyme assays (see Table I) under defined conditions (1 µm enzyme concentration, 0.1 m Tris-HCl buffer (pH 8.0), 4 µm FAD, 1 µm NADPH, 400 µm substrate, 30 °C, 2-h incubation). Rabelomycin, UWM6, and 2,3-dehydro-UWM6 were used as potential substrates. Extraction and analysis of the reaction mixtures by LC-MS gave the following results (Table I): 1) JadF did not show any activity toward rabelomycin, UWM6 or 2,3-dehydro-UWM6. 2) for JadH, both UWM6 and 2,3-dehydro-UWM6 underwent 4a,12b-dehydration and C-12 monooxygenation and were converted to rabelomycin and dehydrorabelomycin, respectively. Rabelomycin remained unchanged. 3) In controls without supplemented enzymes, the substrates remained unchanged.
The results were not materially altered by varying the reaction times, reaction temperature (37 °C), or pH (6.5–9.5). The concentration of dithiothreitol or variation of cofactors (NADH, NAD+, NADP+) also had no affect.
DISCUSSION
With CH56 (a jadA ketosynthase in frame deletion mutant of S. venezuelae), UWM6 and 2,3-dehydro-UWM6 were identified as convertible intermediates in jadomycin biosynthesis. In the biosynthesis of jadomycin A, the conversion from UWM6 requires two dehydrations (at positions 2,3- and 4a,12b-), an oxygenation at C-12 and cleavage and recyclization of ring B with insertion of an amino acid to generate the fused phenoxazinone ring system. Conversion of UWM6 to jadomycin A by LC2014 (in vivo/in vitro) indicates that all the reactions in this pathway are catalyzed by functions provided on JadF, JadG, and JadH. The 3-hydroxyl group remains in rabelomycin when jadF is inactivated, while the 4a-hydroxyl remains in 2,3-de-hydro-UWM6 when jadH is inactivated, indicating that both proposed oxygenases, JadF and JadH, also possess dehydrase activity: JadF catalyzes 2,3-dehydration and JadH catalyzes 4a,12b-dehydration. The JadH dehydration function was verified by in vitro enzymatic conversion assays: UWM6 and 2,3-dehydro-UWM6 were dehydrated at 4a,12b- to rabelomycin and dehydrorabelomycin, respectively, by purified His-tagged JadH. Bifunctional hydroxylase/dehydrase has been reported previously, e.g. CYP71E1 is a cytochrome P450-dependent hydroxylase/dehydrase involved in the biosynthesis of the cya-nogenic glucoside dhurrin in Sorghum bicolor (33). To our knowledge, JadH is the first example of a bifunctional FAD-dependent oxygenase/dehydrase in polyketide biosynthesis. Considering that UWM6 is converted to jadomycin A by LC2014, and the similarity between JadF and JadH, JadF is also suspected to be a bifunctional oxygenase/dehydrase. However, His-tagged JadF cannot convert UWM6 in enzyme assays. To eliminate potential interference of JadF activity by His-tag, we prepared native JadF by removing the N-terminal His-tag with enterokinase and investigated the activity of purified JadF with the same potential substrates as used for His-tagged JadF. Similarly, no conversion was detected after incubated at 30 °C for 2 h,2 suggesting that the purified JadF cannot catalyze 2,3-dehydration. This may be due to the dehydration activity of JadF demands an appropriate context (together with JadGH?). Evidence in support of this inference is that, besides dehydrorabelomycin (7) and l-digitoxosyl-dehy-dorabelomycin (8), rabelomycin was also accumulated in the jadG in-frame deletion mutant (20), indicating the dehydration activity of JadF partially depends on its interaction with JadG. Post-PKS aromatization is a common phenomenon in angucy-cline and other aromatic polyketide biosynthetic pathways, e.g. in kinamycin (9) and landomycin biosyntheses (34–37). The high similarities between the oxygenase enzymes involved in these biosyntheses (e.g. KinO2 shows 69.2% identity and LanM shows 43.9% identity to JadH) possibly indicate that bifunctionality is a general property of these aromatic polyketide-modifying oxygenases.
Sequence analyses and functional characterization of JadG homologues in other PKS gene clusters implicate JadG as an anthrone oxygenase, here catalyzing C-12 monooxygenation. However, the jadG inactivation mutant accumulated three C-12 monooxygenated compounds (20), and VS668 (jadH disrupted) mainly yielded 2,3-dehydro-UWM6, an angucyclic compound without quinone moiety, implying JadH, instead of JadG, catalyzed the C-12 monooxygenation (Fig. 5). Enzymatic studies also revealed that C-12 monooxygenation can be completed when UWM6 or 2,3-dehydro-UWM6 was converted by JadH. However, we cannot rule out the possibility that JadG can catalyze C-12 monooxygenation because the monooxygen-ation can take place spontaneously after 4a,12b-dehydration (8), here catalyzed by JadH. Similar spontaneous monooxygen-ations were also observed in shunt products (aloesaponarin II and 3,8-dihydroxy-1-ethylanthraquinone-2-carboxylic acid) of actinorhodin, whose quinone moiety was actually formed by the JadG homologue, ActVA-Orf6 (38, 39).
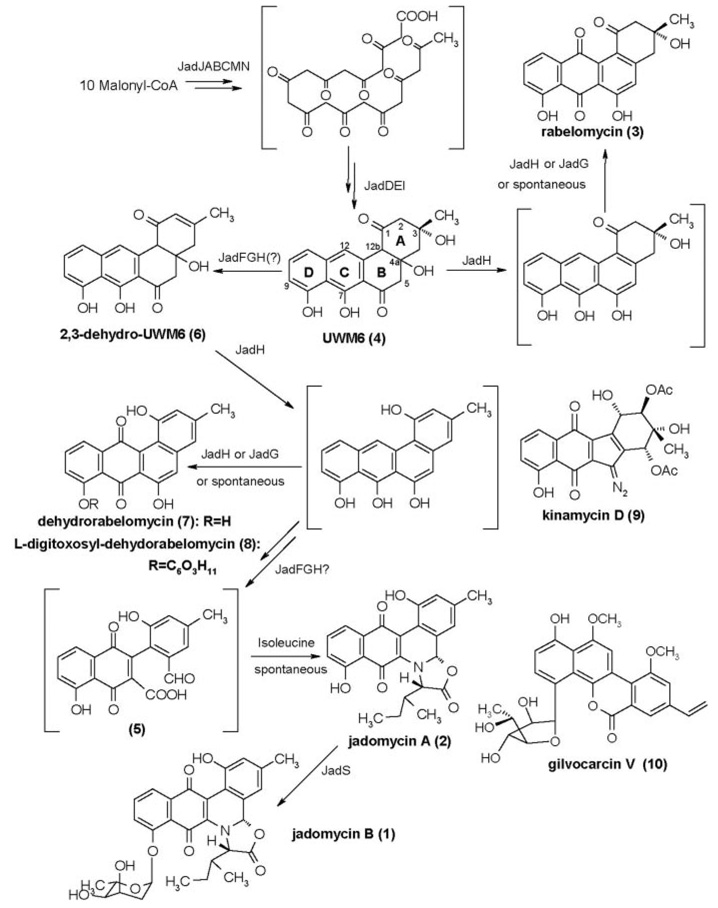
FIG. 5.
Proposed biosynthetic pathway of jadomycin and selected structures of angucycline polyketides, whose biosyntheses require similar ring B cleavages of angucyclinone precursors.
In vivo and in vitro bioconversion experiments with LC2014 imply that ring B cleavage can occur when JadF, JadG, and JadH are all present. Both JadF and JadH show high similarity to MtmOIV, a proposed Baeyer-Villiger mono-oxygenase responsible for ring cleavage reaction in mithra-mycin biosynthesis, indicating JadF and/or JadH are the likely enzyme(s) to catalyze ring B cleavage in jadomycin. The requirement of JadG for ring B cleavage is unexpected but strongly supported by jadG inactivation experiment (three ring B intact compounds accumulated). To summarize, although genetic and biochemical experiments proved Jad-FGH are sufficient to ring B cleavage, and JadG is undoubtedly needed, available evidence still does not allow us to pin point the enzyme(s) directly responsible for this oxidative ring cleavage reaction (Fig. 5).
The biosyntheses of kinamycin (34, 40) and gilvocarcin V (10) (41) demand ring B cleavage processes similar to that proposed for jadomycin. The associated catalytic enzymes for kinamycin may involve KinOR (63.2% identity with JadF), KinG (43.7% identity with JadG), and KinO2 (69.2% identity with JadH); the candidate enzymes for gilvocarcin V are GilOIV (31.3% identity with JadF), GilOII (51.1% identity with JadG), and GilOI (39.8% identity with JadH). Interestingly, the kinamycin biosynthetic gene cluster possesses an additional oxygenase gene (kinO1) encoding a protein highly similar to both JadF (41.8% identity) and JadH (50.3% identity). This may account for the special ring B rearrangement style after C–C bond cleavage in kinamycin. In the kinamycin pathway, dehydrora-belomycin was proven to be a convertible intermediate to a ring B-cleaved product (42) (Fig. 5). In gilvocarcin V biosynthesis, homorabelomycin was shown to be shunt product, whereas the 2,3-dehydro-UWM6 homologue, as well as a novel pregilvocar-cin-o-quinone, were proved to be convertible intermediates. These compounds were obtained after inactivation of gilOIV (2,3-dehydro-UWM6 homologue) and gilOI (pregilvocarcin-o-quinone), respectively. The two enzymes were suggested to cooperate in a sequential manner to achieve the C–C bond cleavage (43). Further investigation of the biosynthesis of these polyketides may help in understanding the complicated ring cleavage reactions and assist in generating new biologically active compounds.
In brief, by investigating post-PKS reactions leading to the jadomycin aglycone, we demonstrate JadF, JadG, and JadH together can complete the post-PKS modifications from UWM6 to jadomycin A and identified the post-PKS aromatization reaction catalyzed by bifunctional oxygenase/dehydrase.
Footnotes
This work was supported by grants from the Chinese Academy of Sciences (to K.-Q. Y.) and partially by a grant from the Kentucky Lung Cancer Research Foundation and by National Institutes of Health Grant CA 91901 (to J. R.).
The abbreviations used are: PKS, polyketide synthase; LC-MS, liquid chromatography-mass spectroscopy.
Y.-H. Chen and K.-Q. Yang, unpublished data.
REFERENCES
- 1.Hopwood DA. Chem. Rev. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 2.Shen B. Top. Curr. Chem. 2000;209:1–51. [Google Scholar]
- 3.Rawlings BJ. Nat. Prod. Rep. 1999;16:425–484. doi: 10.1039/a900566h. [DOI] [PubMed] [Google Scholar]
- 4.Hutchinson CR, Fujii I. Annu. Rev. Microbiol. 1995;49:201–238. doi: 10.1146/annurev.mi.49.100195.001221. [DOI] [PubMed] [Google Scholar]
- 5.Rix U, Fischer C, Remsing LL, Rohr J. Nat. Prod. Rep. 2002;19:542–580. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- 6.Han L, Yang KQ, Ramalingam E, Mosher RH, Vining LC. Microbiology. 1994;140:3379–3389. doi: 10.1099/13500872-140-12-3379. [DOI] [PubMed] [Google Scholar]
- 7.Yang KQ, Han L, Ayer SW, Vining LC. Microbiology. 1996;142:123–132. doi: 10.1099/13500872-142-1-123. [DOI] [PubMed] [Google Scholar]
- 8.Kulowski K, Pienkowski EW, Han L, Yang KQ, Vining LC, Hutchinson CR. J. Am. Chem. Soc. 1999;121:1786–1794. [Google Scholar]
- 9.Ayer SW, McInnes AG, Thibault P, Walter JA, Doull JL, Parnell T, Vining LC. Tetrahedron Lett. 1991;32:6301–6304. [Google Scholar]
- 10.Doull JL, Singh AK, Hoare M, Ayer SW. J. Ind. Microbiol. 1994;13:120–125. doi: 10.1007/BF01584109. [DOI] [PubMed] [Google Scholar]
- 11.Rix U, Zheng JT, Remsing LL, Greenwell L, Yang KQ, Rohr J. J. Am. Chem. Soc. 2004;126:4496–4497. doi: 10.1021/ja031724o. [DOI] [PubMed] [Google Scholar]
- 12.McVey J. M.Sc. dissertation. Canada: Dalhousie University; 1998. Characterization of the Downstream Genes for Jadomycin B Biosynthesis in Streptomyces venezuelae ISP5230. [Google Scholar]
- 13.Yang KQ, Han L, Vining LC. J. Bacteriol. 1995;177:6111–6117. doi: 10.1128/jb.177.21.6111-6117.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacNeil DJ, Gewain KM, Rudy CL, Dezeny G, Gibbons PH, MacNeil T. Gene (Amst.) 1992;111:61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 15.Hopwood DA, Kieser T, Wright HM, Bibb MJ. J. Gen. Microbiol. 1982;129:2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- 16.Doumith M, Weingarten P, Wehmeier UF, Salah-Bey K, Benhamou B, Capdevila C, Michel JM, Piepersberg W, Raynal MC. Mol. Gen. Genet. 2000;264:477–485. doi: 10.1007/s004380000329. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Laboratory, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 18.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. [Google Scholar]
- 19.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rix U, Wang C, Chen Y, Lipata FM, Remsing Rix LL, Greenwell LM, Vining LC, Yang K, Rohr J. ChemBioChem. 2005;6:838–845. doi: 10.1002/cbic.200400395. [DOI] [PubMed] [Google Scholar]
- 21.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 22.Prado L, Fernandez E, Weißbach U, Blanco G, Quirós LM, Braña AF, Méndez C, Rohr J, Salas JA. Chem. Biol. 1999;6:19–30. doi: 10.1016/s1074-5521(99)80017-9. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez D, Quirós LM, Braña AF, Salas JA. J. Bacteriol. 2003;185:3962–3965. doi: 10.1128/JB.185.13.3962-3965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menéndez N, Nur-e-Alam M, Bra[caron]na AF, Rohr J, Salas JA, Méndez C. Chem. Biol. 2004;11:21–32. doi: 10.1016/j.chembiol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Li A, Piel J. Chem. Biol. 2002;9:1017–1026. doi: 10.1016/s1074-5521(02)00223-5. [DOI] [PubMed] [Google Scholar]
- 26.Wieregna RK, Terpstra P, Hol WGJ. J. Mol. Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 27.Vallon O. Proteins. 2000;38:95–114. doi: 10.1002/(sici)1097-0134(20000101)38:1<95::aid-prot10>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 28.Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. J. Mol. Biol. 1990;212:135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 29.Eppink MHM, Schreuder HA, Van Berkel WJH. Protein Sci. 1997;6:2454–2458. doi: 10.1002/pro.5560061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kendrew SG, Hopwood DA, Marsh ENG. J. Bacteriol. 1997;179:4305–4310. doi: 10.1128/jb.179.13.4305-4310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciara G, Kendrew SG, Miele AE, Marsh NG, Federici L, Malatesta F, Schimperna G, Savino C, Vallone B. EMBOJ. 2003;22:205–215. doi: 10.1093/emboj/cdg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen B, Hutchinson CR. Biochemistry. 1993;32:6656–6663. doi: 10.1021/bi00077a019. [DOI] [PubMed] [Google Scholar]
- 33.Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA. Plant Mol. Biol. 1998;36:393–405. doi: 10.1023/a:1005915507497. [DOI] [PubMed] [Google Scholar]
- 34.Gould SJ. Chem. Rev. 1997;97:2499–2509. doi: 10.1021/cr9600215. [DOI] [PubMed] [Google Scholar]
- 35.Weber S, Zolke C, Rohr J, Beale JM. J. Org. Chem. 1994;59:4211–4214. [Google Scholar]
- 36.Westrich L, Domann S, Faust B, Bedford D, Hopwood DA, Bechthold A. FEMS Microbiol. Lett. 1999;170:381–387. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]
- 37.Ostash B, Rix U, Remsing Rix LL, Liu T, Lombó F, Luzhetskyy A, Gromyko O, Wang CC, Braña AF, Méndez C, Salas JA, Fedorenko V, Rohr J. Chem. Biol. 2004;11:547–555. doi: 10.1016/j.chembiol.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 38.McDaniel R, Khosla SE, Hopwood DA, Khosla C. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 39.McDaniel R, Khosla SE, Hopwood DA, Khosla C. Nature. 1995;375:549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 40.Gould SJ, Hong ST, Carney JR. J. Antibiot. 1998;51:50–57. doi: 10.7164/antibiotics.51.50. [DOI] [PubMed] [Google Scholar]
- 41.Fischer C, Lipata F, Rohr J. J. Am. Chem. Soc. 2003;125:7818–7819. doi: 10.1021/ja034781q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seaton PJ, Gould SJ. J. Am. Chem. Soc. 1987;109:5282–5284. [Google Scholar]
- 43.Liu T, Fischer C, Beninga C, Rohr J. J. Am. Chem. Soc. 2004;126:12262–12263. doi: 10.1021/ja0467521. [DOI] [PubMed] [Google Scholar]