Abstract
Bardet-Biedl syndrome (BBS) is an autosomal recessively inherited ciliopathy mainly characterized by rod-cone dystrophy, postaxial polydactyly, obesity, renal tract anomalies, and hypogonadism. To date, 14 BBS genes, BBS1 to BBS14, have been identified, accounting for over 75% of mutations in BBS families. In this study, we present a consanguineous family from Pakistan with postaxial polydactyly and late-onset retinal dysfunction. Adult affected individuals did not show any renal or genital anomalies, obesity, mental retardation or learning difficulties and did thus not fulfill the proposed clinical diagnostic criteria for BBS. We mapped the disease in this family to the BBS12 locus on chromosome 4q27 and identified the novel homozygous p.S701X nonsense mutation in BBS12 in all three affected individuals of this family. We conclude that BBS12 mutations might cause a very mild phenotype, which is clinically not diagnosed by the current diagnostic criteria for BBS. Consequently, we suggest the use of less strict diagnostic criteria in familial BBS families with mild phenotypic expression.
Key Words: Bardet-Biedl syndrome, BBS12, Diagnostic criteria, Mild phenotype, Novel mutation
Introduction
Bardet-Biedl syndrome (BBS, MIM 209900) is an autosomal recessive disorder characterized by a wide spectrum of clinical features, of which the most common ones are progressive retinal degeneration (rod-cone dystrophy), obesity, postaxial polydactyly, renal tract and genital anomalies, and learning difficulties or mental retardation [Beales et al., 1999]. The phenotype of BBS is highly variable, and it has been suggested that the clinical diagnosis is established if at least four of the main manifestations are present in a patient. Clinical evaluation during early infancy remains difficult as not all of the main manifestations are congenital but may occur later during childhood. Congenital postaxial polydactyly including syndactyly and/or brachydactyly is present in approximately 70% of cases and may not affect all limbs. Developmental anomalies of the renal tract are also common and can lead to chronic renal failure [Harnett et al., 1988]. Further congenital symptoms, which can be diagnosed in early infancy, are genital anomalies such as vaginal atresia and hypoplasia of the uterus and hypogonadism in males. The appearance of a rod-cone dystrophy, which is also described as atypical retinitis pigmentosa with early macular involvement, is characteristic in BBS patients and present in over 90% of patients [Green et al., 1989]. Typically, night blindness is one of the first symptoms at the end of the first decade and the progressive retinal degeneration often causes total blindness in the course of the second decade of life. Although mental retardation is not very common in BBS patients, learning difficulties are present in over half of the patients. Truncal and rhizomelic obesity develops in the majority of BBS patients before the end of puberty. Developmental delay might be present and poor motor coordination is described in a subset of patients. Other features such as hearing loss, dental anomalies, congenital heart defects, diabetes mellitus, and hepatic fibrosis are rare [Green et al., 1989; Beales et al., 1999].
The incidence of BBS has been estimated at 1 in 150,000 to 175,000 individuals in European populations. However, populations with a high level of consanguinity or from geographically isolated regions like Newfoundland show a much higher frequency [Farag and Teebi, 1988, 1989]. BBS is a genetically heterogeneous disorder and up to date 14 genes have been identified (BBS1 to BBS14, namely BBS1, BBS2, ARL6, BBS4, BBS5, MKKS, BBS7, TTC8, BBS9, BBS10, TRIM32, BBS12, MKS1, CEP290) using traditional genome-wide mapping and positional cloning strategies in large consanguineous families or – more recently – computational comparative genomic expression methods [Katsanis et al., 2000; Mykytyn et al., 2001, 2002; Badano et al., 2003; Chiang et al., 2004; Fan et al., 2004; Li et al., 2004; Nishimura et al., 2005; Chiang et al., 2006; Stoetzel et al., 2006; Leitch et al., 2008]. While the majority of BBS patients do show a classical autosomal recessive inheritance pattern, triallelic and oligogenic inheritance was suggested in some families [Katsanis et al., 2001a]. Since then it has been suggested that genetic modifiers exist responsible for the clinical variability of phenotypic expression in BBS patients.
Mutations in BBS1–14 account for over 75% of affected families [Zaghloul and Katsanis, 2009]. In families of European descent, mutations inBBS1 and BBS10 show the highest frequency accounting for approximately 20% of BBS mutations each [Chiang et al., 2006; Stoetzel et al., 2006]. The BBS12 gene is the most recently identified gene and the encoded protein belongs to a novel branch of type 2 chaperonin superfamily, which includes also BBS6 and BBS10 [Stoetzel et al., 2007]. In general, most of the known BBS genes are highly conserved in ciliated organisms, such as Chlamydomonas, and are absent in non-ciliated ones such as Arabidopsis [Li et al., 2004]. There is molecular evidence that BBS proteins are part of primary cilia structures arising from the basal body and playing an important role in development and homeostasis of various tissues, e.g. as mechanosensors in kidney epithelium and in the organization of photoreceptor cells of the retina [Ross et al., 2005; Badano et al., 2006a]. Cilia are microtubule-based eukaryotic organelles that project from the surface of human cells. Beside performing a wide variety of functions, they play a major role in extracellular signal transduction including important biological pathways such as wnt and hedgehog signalling [Marshall and Nonaka, 2006; Saxena et al., 2007]. It is suggested that ciliary dysfunction and disturbance of the intraflagellar transport represent the main pathophysiological mechanism leading to BBS [Katsanis et al., 2001b]. For this reason, BBS is regarded as a ciliopathy.
Here we report a novel BBS12 mutation, p.S701X, found in three affected individuals of a consanguineous family from Pakistan clinically diagnosed with postaxial polydactyly and late-onset retinal dysfunction. Adult affected individuals did not show any renal and genital anomalies, obesity, learning difficulties or any other symptoms, thus indicating that mutations in BBS12 can cause a very mild phenotype which is clinically not diagnosed by the current diagnostic criteria for BBS.
Material and Methods
MR-10 family: Index patient was referred to the hospital due to bilateral polydactyly and mild vision impairment. All affected family members were clinically examined including general physical examination, ophthalmological examination, X-rays of hands and feet, and abdominal ultrasound. The study was first approved by local institutional review boards of COMSATS Institute, Islamabad, Pakistan (CIIT Biosciences Review/Ethics Committee; A.S. 24112008), and followed the Declaration of Helsinki protocols. Afterwards, blood samples were taken after written informed consent was given and DNA was isolated using standard procedures.
Linkage Analysis
Genomic DNA of the index patient of the MR-10 family (IV-3) as well as DNAs of siblings and parents (fig. 1, individuals I-1, I-2, II-1, II-2, II-3, II-4, III-2, III-3, IV-1, IV-3, IV-4, IV-5, IV-6, IV-8) were subjected to a genome-wide mapping using the Affymetrix GeneChip Human Mapping 10K Array (Affymetrix, Santa Clara, CA). This 10K array comprises a total of 10,204 single-nucleotide polymorphisms (SNPs) with a mean intermarker distance of 258 kb, equivalent to 0.36 cM. Sample processing and labelling were performed in accordance with the manufacturer's instructions (Affymetrix Mapping 10K 2.0 Assay Manual). Genotypes were provided by the GeneChip DNA Analysis Software (v.4.1, Affymetrix). Non-parametric linkage analysis using all genotypes of a chromosome simultaneously was carried out with MERLIN. Parametric linkage analysis was performed by the program ALLEGRO assuming autosomal recessive inheritance. For fine mapping analysis, available MR-10 family members were genotyped for the following markers: D4S2975, D4S2277, D4S429, D4S3039, D4S1527, D4S2920, D4S3047, D4S2921, D4S426, D11S480, D11S4122, D11S1885. Genomic localization and microsatellite sequences were obtained from the UCSC Genome Browser (http://genome.ucsc.edu/, build hg18, March 2006) and the ENSEMBL database (http://www.ensembl.org). Polymerase chain reaction (PCR) for fluorescent markers was performed on a DNA Engine Dyad Thermal Cycler (Bio-Rad, Germany) under standard PCR conditions, run on an ABI 3730 DNA Analyzer (Applied Biosystems, Germany), and evaluated with the GeneMarker 1.51 program (Soft Genetics LLC).
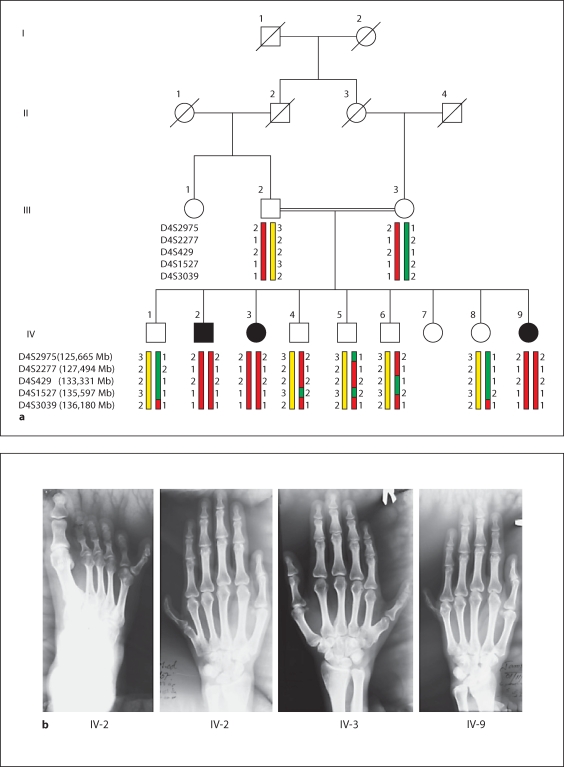
Fig. 1.
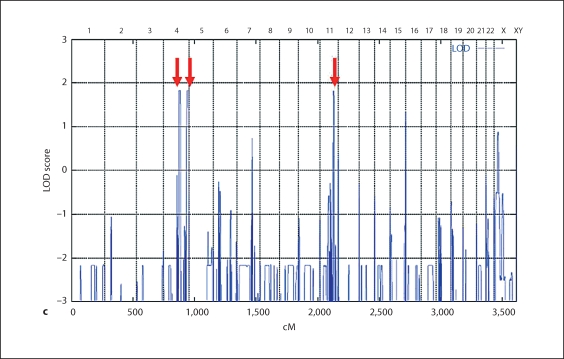
MR-10 family maps to the BBS12 locus on chromosome 4q27. a Pedigree of the MR-10 family and haplotypes of the 4q27 region. The disease-associated haplotype is shown in red. b X-rays of affected individuals showing the postaxial polydactyly of hands and feet. Continued MR-10 family maps to the BBS12 locus on chromosome 4q27. c Schematic view of genome-wide LOD score calculations. The arrows indicate regions of maximum LOD scores of 1.83.
Mutation Analysis
The following genes of the critical region were tested for mutations in the index patient of family MR-10: FGF2, BBS7, BBS12, NUDT6, SPATA5, and SPRY. Primer sequences and PCR protocols are available upon request. The 2,144 bp of the coding sequence of exon 2 of the BBS12 gene (123,873,307–123,885,548 Mb, UCSC Human Genome Browser, hg 18 assembly) was sequenced in the index patients (IV-2, IV-3, IV-9). 20 ng of genomic DNA from patients was used to amplify the 4 amplicons of exon 2 by a touchdown PCR protocol on a DNA Engine Dyad Thermal Cycler (Bio-Rad, Germany). PCR fragments were purified and directly sequenced from both sides using the ABI BigDye Terminator v1.1 Cycle Sequencing Kit and the ABI 3730 DNA Analyzer (Applied Biosystems). Co-segregation of the mutation in the family was investigated by direct sequencing of all available family members and by a PCR/enzyme digestion method using ApoI (data not shown). In addition, the mutation was tested in 147 Pakistani control individuals by PCR/enzyme digestion method.
Results
Clinical Evaluation of the Family
The three affected individuals of family MR-10 from Pakistan (fig. 1a) were born to consanguineous parents (first-degree cousins). Congenital postaxial polydactyly of hands and feet were observed in various expressions (fig. 1b). In addition, progressive night blindness started between 13–15 years of age in all the patients. Individuals IV-2, IV-3, and IV-9 are now 30, 27, and 19 years old, respectively, and all of them have developed severe night blindness, while vision is only mildly impaired in daytime. Ophthalmological examination was performed and an atypical retinitis pigmentosa (RP, rod-cone dystrophy), myopia and astigmatism were diagnosed. Patients’ history neither showed developmental delay in childhood nor mental retardation or learning difficulties. Obesity was not present in all three affected individuals either. No organ anomaly was found in ultrasound; especially no renal tract abnormalities were seen. Furthermore, physical examination showed hypodontia but did not show any genital anomalies or additional symptoms. General neurological testing was normal. No signs for hypogonadism was present and individual IV-2 was fertile and had 4 children. A summary of clinical findings is listed in table 1. The initial diagnosis of an autosomal recessively inherited disorder characterized by postaxial polydactyly and atypical RP was given. It is of interest to note that a second, independent autosomal recessive disorder, namely non-syndromic mental retardation (MR), was present in the family, but none of the BBS patients was affected by MR (individuals IV-1, IV-5, IV-6, IV-8 had MR). The molecular basis of the independent MR phenotype will be analyzed in an upcoming study.
Table 1.
Clinical findings in three affected individuals of family MR-10 listed according to the clinical criteria for the diagnosis of BBS [Beales et al., 1999]
| IV-2 | IV-3 | IV-9 | |
|---|---|---|---|
| Major criteria | |||
| Rod-cone dystrophy | + (14 years) | + (15 years) | + (13 years) |
| Postaxial polydactyly | + | + | + |
| Truncal obesity | − | − | − |
| Hypogonadism | − | − | − |
| Renal anomalies | − | − | − |
| Minor criteria | |||
| Speech disorder/delay | − | − | − |
| Development delay | − | − | − |
| Behaviour | normal | normal | normal |
| Ataxia/imbalance | − | − | − |
| Diabetes mellitus | − | − | − |
| Congenital heart defects | − | − | − |
| Liver disease | − | − | − |
| Hearing loss | − | − | − |
| Facial features | − | − | − |
| Situs inversus | − | − | − |
| Hirschsprung's disease | − | − | − |
| Polyuria/polydipsia | − | − | − |
| Hypodontia | + | + | + |
| Anosmia | − | − | − |
Linkage to the BBS12 Locus on 4q27 and Identification of a Novel BBS12 Mutation
In order to map the recessive disorder characterized by postaxial polydactyly and rod-cone dystrophy in the MR-10 family, we performed homozygosity mapping using the 10K array. Genome-wide LOD score calculations resulted in maximum LOD scores of 1.83 at three different chromosomal regions located on chromosome 4q27–4q31.22, 4q35.1–4q35.2, and 11q23.1–11q23.3, respectively (fig. 1c). Genotyping of additional microsatellite markers of these regions clearly excluded the two loci on 4q35.1–4q35.2 and 11q23.1–11q23.3 (data not shown). For locus 4q27–4q31.22, marker analysis confirmed homozygosity and the critical region was defined by SNPs rs1390560 (located at 122.07 Mb) and rs1343812 (143.70 Mb) (fig. 2a).
Fig. 2.
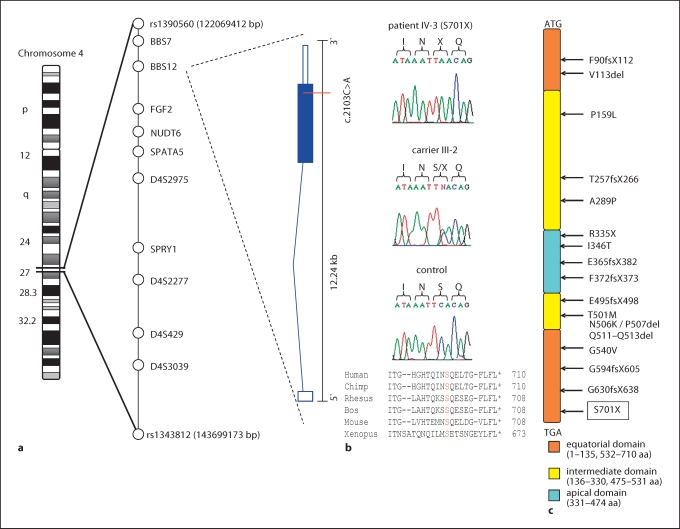
Identification of the novel p.S701X mutation in BBS12. a Ideogram of chromosome 4 and localisation of the linked region on 4q27. SNPs defining the critical region, analysed markers, and tested candidate genes are listed within the critical region. Illustration of the genomic organisation and transcript composition of BBS12. The position of the c.2103C>A mutation is indicated. b Sequence chromatograms of c.2103C>A mutation in the index patient (IV-3), heterozygous carrier (III-2), and a healthy control. Amino-acid (aa) alignment of the truncated C-terminal end of BBS12 proteins from different species. c Overview of BBS12 protein structure and location of previously identified mutations.
The 21.63-Mb critical region is gene rich and contains approximately 80 known and predicted genes. In total, we tested 6 highly relevant candidate genes of this region: FGF2, BBS7, BBS12, NUDT6, SPATA5, and SPRY1. Sequencing of the coding exon 2 of BBS12 in affected individuals identified a novel homozygous c.2103C>A mutation (fig. 2b), which is predicted to insert a stop codon at position 701 of the BBS12 protein (p.S701X). This nonsense mutation is located at the very C-terminal end of the protein and the truncation leads to a loss of 10 amino acids (fig. 2c). Co-segregation of the p.S701X mutation with the disease in the family could be confirmed. All affected individuals were homozygous for the p.S701X mutation and parents were heterozygous carriers. In addition, the mutation was not found in 147 healthy controls from Pakistan.
We compared the localisation of this novel BBS12 mutation with mutations described in the original gene identification study [Stoetzel et al., 2007]. The p.S701X mutation is located close to the C-terminus in the equatorial domain of the BBS12 protein (fig. 2b, c). Of the 17 pathogenic mutations described so far, which include missense, nonsense, and frameshift mutations, the p.S701X is the most C-terminal one.
Discussion
It is well known that a large clinical variability of phenotypic expression exists in Bardet-Biedl syndrome, both between and within families. In addition, clinical diagnosis is hampered by the fact that not all symptoms are congenital but may develop later during childhood or within the second decade of life. Therefore, especially in early infancy clinical diagnosis is often difficult. Diagnostic criteria for the clinical diagnosis of BBS have been suggested [Beales et al., 1999] that were modified by Tobin and Beales in 2007 using a sub-classification of symptoms in primary and secondary features [Tobin and Beales, 2007]. Primary features are rod-cone dystrophy, postaxial polydactyly, obesity, hypogonadism, and renal anomalies, whereas secondary features include speech disorder/delay, development delay, diabetes mellitus, congenital heart disease, liver disease, situs inversus and facial dysmorphism. It was suggested that the clinical diagnosis of BBS can be made in a patient if four primary features or three primary plus two secondary features are present. Using this classification, diagnosis would have been missed in all three adult affected individuals in our family, because only 2 primary (polydactyly and rod-cone dystrophy) and 1 secondary criterium (hypodontia) are present. It is interesting to point out that this very mild form of BBS was present in all three affected individuals, suggesting a generally mild phenotypic expression of the disease in this particular family. Therefore, strict diagnostic criteria in BBS might lead to false-negative classification. For this reason, we propose that molecular genetic testing for BBS might be considered in familial forms even if the diagnostic criteria are not fulfilled. However, the phenotype observed in the three affected individuals of our family clearly represents symptoms known to be caused by dysfunction of cilia. Many human genetic disorders have been described which are collectively called the ciliopathies, and symptoms associated with ciliary dysfunction include alterations of left-right body axis, polydactyly, kidney, liver and pancreas anomalies, retinal degeneration and anosmia, neural tube defects as well as cognitive defects.
Using homozygosity mapping, we linked the disease in our family to the BBS12 locus on chromosome 4q27 and identified the novel nonsense mutation p.S701X in BBS12. There is convincing evidence for a causative nature of this mutation: (i) genome-wide mapping identified only one homozygous locus in this consanguineous family, which could be confirmed by subsequent marker analysis; (ii) the mutation co-segregated with the disease in the family and was absent in 147 healthy and ethnically matched control chromosomes; (iii) the nonsense mutation is predicted to have a clear effect on the BBS12 protein, truncating – even though −10 amino acids from the conserved C-terminal end of the equatorial domain of the protein. It is estimated that BBS12 mutations account for up to 5% of families with BBS [Stoetzel et al., 2007]. One frameshift mutation has yet been described to alter the equatorial domain of BBS12, namely p.G594fsX605 (fig. 2c).
Although clear genotype-phenotype correlations could not yet be established in BBS, mutations in BBS3 seem to cause more often polydactyly of all four limbs, while patients with BBS4 mutations more frequently show polydactyly only of the upper limbs. Moreover, BBS2-associated cases do not frequently suffer from obesity, whereas patients carrying BBS3 mutations often develop early-onset obesity [Carmi et al., 1995; Beales et al., 1997; Bruford et al., 1997]. Polygenic inheritance or genetic modifiers might be an explanation for phenotypic variability in BBS. Moreover, epistatic interactions might play an important role in phenotypic variability [Badano and Katsanis, 2002]. In this context, it was demonstrated that certain synergistic effects can modify the penetrance or expressivity of phenotype. Badano et al. reported some kind of evidence for a novel gene called MGC1203, which assists epistatically to BBS1 mutations and also enhances the phenotype in a zebrafish model [Badano et al., 2006b].
No obvious phenotypic difference of BBS12 patients compared to other BBS patients was reported in the original study, although no detailed information about the clinical presentation of the families was given [Stoetzel et al., 2007]. Nevertheless, our study showed that mutations in BBS12 can cause a very mild phenotype that is clinically not diagnosed by the proposed diagnostic criteria. Only mild phenotypic effects of BBS12 suppression were also reported in a zebrafish model [Stoetzel et al., 2007]. In the future, it will be interesting and necessary to further delineate the phenotypic expression in other BBS12 families.
In conclusion, we provide evidence for a very mild, familial BBS12 phenotype. Clinical diagnosis would have been missed when applying the suggested diagnostic criteria for BBS. For this reason, we propose the use of less strict diagnostic criteria in familial BBS cases that might also influence the molecular testing strategies of BBS.
Acknowledgements
We are grateful to the family for their participation in the study. This work was supported by the German Federal Ministry of Education and Research (BMBF) by grant number 01GM0880 to B.W.
Footnotes
B.P. and A.M. contributed equally to this work.
References
- Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet. 2002;3:779–789. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- Badano JL, Ansley SJ, Leitch CC, Lewis RA, Lupski JR, Katsanis N. Identification of a novel Bardet-Biedl syndrome protein, BBS, that shares structural features with BBS1 and BBS2. Am J Hum Genet. 2003;72:650–658. doi: 10.1086/368204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006a;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Badano JL, Leitch CC, Ansley SJ, May-Simera H, Lawson S, et al. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006b;439:326–330. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- Beales PL, Warner AM, Hitman GA, Thakker R, Flinter FA. Bardet-Biedl syndrome: a molecular and phenotypic study of 18 families. J Med Genet. 1997;34:92–98. doi: 10.1136/jmg.34.2.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet. 1999;36:437–446. [PMC free article] [PubMed] [Google Scholar]
- Bruford EA, Riise R, Teague PW, Porter K, Thomson KL, et al. Linkage mapping in 29 Bardet-Biedl syndrome families confirms loci in chromosomal regions 11q13, 15q22.3–q23, and 16q21. Genomics. 1997;41:93–99. doi: 10.1006/geno.1997.4613. [DOI] [PubMed] [Google Scholar]
- Carmi R, Elbedour K, Stone EM, Sheffield VC. Phenotypic differences among patients with Bardet-Biedl syndrome linked to three different chromosome loci. Am J Med Genet. 1995;59:199–203. doi: 10.1002/ajmg.1320590216. [DOI] [PubMed] [Google Scholar]
- Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, et al. Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3) Am J Hum Genet. 2004;75:475–484. doi: 10.1086/423903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AP, Beck JS, Yen HJ, Tayeh MK, Scheetz TE, et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc Natl Acad Sci USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, et al. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- Farag TI, Teebi AS. Bardet-Biedl and Laurence-Moon syndromes in a mixed Arab population. Clin Genet. 1988;33:78–82. doi: 10.1111/j.1399-0004.1988.tb03414.x. [DOI] [PubMed] [Google Scholar]
- Farag TI, Teebi AS. High incidence of Bardet Biedl syndrome among the Bedouin. Clin Genet. 1989;36:463–464. doi: 10.1111/j.1399-0004.1989.tb03378.x. [DOI] [PubMed] [Google Scholar]
- Green JS, Parfrey PS, Harnett JD, Farid NR, Cramer BC, et al. The cardinal manifestations of Bardet-Biedl syndrome, a form of Laurence-Moon-Biedl syndrome. N Engl J Med. 1989;321:1002–1009. doi: 10.1056/NEJM198910123211503. [DOI] [PubMed] [Google Scholar]
- Harnett JD, Green JS, Cramer BC, Johnson G, Chafe L, et al. The spectrum of renal disease in Laurence-Moon-Biedl syndrome. N Engl J Med. 1988;319:615–618. doi: 10.1056/NEJM198809083191005. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, et al. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet. 2000;26:67–70. doi: 10.1038/79201. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, et al. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001a;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- Katsanis N, Lupski JR, Beales PL. Exploring the molecular basis of Bardet-Biedl syndrome. Hum Mol Genet. 2001b;10:2293–2299. doi: 10.1093/hmg/10.20.2293. [DOI] [PubMed] [Google Scholar]
- Leitch CC, Zaghloul NA, Davis EE, Stoetzel C, Diaz-Font A, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Nonaka S. Cilia: tuning in to the cell's antenna. Curr Biol. 2006;16:R604–R614. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, et al. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet. 2001;28:188–191. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, et al. Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet. 2002;31:435–438. doi: 10.1038/ng935. [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Searby CC, Berg EM, Ferguson AL, et al. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am J Hum Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AJ, May-Simera H, Eichers ER, Kai M, Hill J, et al. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet. 2005;37:1135–1140. doi: 10.1038/ng1644. [DOI] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Laurier V, Davis EE, Muller J, Rix S, et al. BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet. 2006;38:521–524. doi: 10.1038/ng1771. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Muller J, Laurier V, Davis EE, Zaghloul NA, et al. Identification of a novel BBS gene (BBS12) highlights the major role of a vertebrate-specific branch of chaperonin-related proteins in Bardet-Biedl syndrome. Am J Hum Genet. 2007;80:1–11. doi: 10.1086/510256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin JL, Beales PL. Bardet-Biedl syndrome: beyond the cilium. Pediatr Nephrol. 2007;22:926–936. doi: 10.1007/s00467-007-0435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. J Clin Invest. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]