Abstract
Obesity is a direct result of the accumulation of white adipose tissue (WAT). In this study, the role of autophagy in the differentiation of white adipose tissue was studied by deleting the autophagy-related 7 (atg7) gene from adipose tissue in mice. This deletion results in a striking phenotype at the cellular, tissue and whole-organism levels. Adipose tissue deposits in the mutant mice are much smaller in mass than those observed in their wild-type counterparts, and mutant adipocytes exhibit unusual morphological characteristics including multilocular lipid droplets and greatly increased numbers of mitochondria. The knockout mice are noticeably slimmer than their wild-type littermates, despite parity in food and water consumption. The mutant mice also exhibit higher basal physical activity levels and an array of metabolic changes revealed through blood tests. Most importantly, these mice show resistance to high-fat diet-induced obesity and markedly increased sensitivity to insulin. These findings establish a new function for autophagy and provide a new model system for use in the search for treatments for obesity and type II diabetes.
Keywords: atg7, adipose, knockout, obesity, diabetes
White adipocytes are highly differentiated cells that are specialized for energy storage and energy homeostasis regulation. They have a distinctive cellular structure in which nearly the entire volume of the cell is occupied by a single, large lipid droplet, while other cellular components reside in the remaining space. The process of adipocyte differentiation, or adipogenesis, has fascinated people for decades. One aspect of this process that we find particularly intriguing is how the cytoplasmic components of the fibroblast-like pre-adipocytes are degraded during adipogenesis. Because autophagy is a well-known mechanism for macromolecular degradation of intra-cellular contents, we decided to examine whether it might play a role in adipogenesis. Initial experiments conducted in our laboratory using a cellular model of adipocyte differentiation reveal that the deletion of the autophagy-related 5 (atg5) gene from primary mouse embryonic fibroblasts (MEFs) blocks adipogenesis in vitro. To investigate the implications of this finding in the whole-organism milieu, we set forth to determine what effects, if any, would result from an adipose tissue-specific autophagy deficiency.
By crossing mice bearing a flox-flanked atg7 gene with mice bearing aP2-Cre constuct (Cre under the transcriptional control of an adipose tissue-specific promoter aP2), we were able to generate off-spring in which autophagy was deficient in fat tissue. Adipocytes in adult atg7/flox/flox;ap2-Cre mice exhibit a remarkable phenotype when compared to wild-type counterparts. Under light microscopy, we immediately noticed that the tissue and cellular morphology observed in mutant WAT consisted of large numbers of smaller adipocytes filled with multilocular lipid droplets, rather than the large unilocular lipid droplet typical of wild-type white adipocytes. In the following electron microscopy studies we observed a striking difference in the number of mitochondria present in the mutant adipocytes when compared with wild-type cells. The mutant WAT cells contain far more mitochondria than control adipocytes, and these mitochondria are surrounded by significant amounts of cytoplasm. Similarly, far more mitochondria are observed in the mutant brown adipose tissue (BAT) cells compared to wild-type BAT cells. These results clearly demonstrate that autophagy is critical for normal white adipogenesis, especially for the formation of the unique unilocular lipid droplet structure and for mitochondria homeostasis control.
At the whole-organism level, the mutant mice exhibit a remarkable phenotype typified by decreased weight gain compared to their wild-type counterparts from the age of four weeks continuing until the experiment was concluded at 18 weeks. Upon dissection, there was a clear difference in fat pad mass between the mutant and wild-type mice, with mutants showing an average 80% decrease in gonadal fat pad weight. All of these changes occur independently of food intake, which is the same among wild-type and mutant mice. An even more interesting pheno-type observed in these mutant mice is that they are resistant to high-fat diet-induced obesity. When the mutant mice are fed with high-fat diet food for two months, they gain no more weight than mutant mice fed a normal balanced laboratory mouse diet. In contrast, the wild-type mice usually gain an additional 20% of their pre-experiment body weight under these feeding conditions when compared to wild-type mice fed a normal balanced diet. Perhaps the most striking and most important phenotype of these mutant mice is that, unlike many other mouse models of lipodystrophy, the nonadipose tissues of the atg7 knockout mice do not exhibit any abnormal fat deposits. Remarkably these mutant mice exhibit an increased sensitivity to insulin.
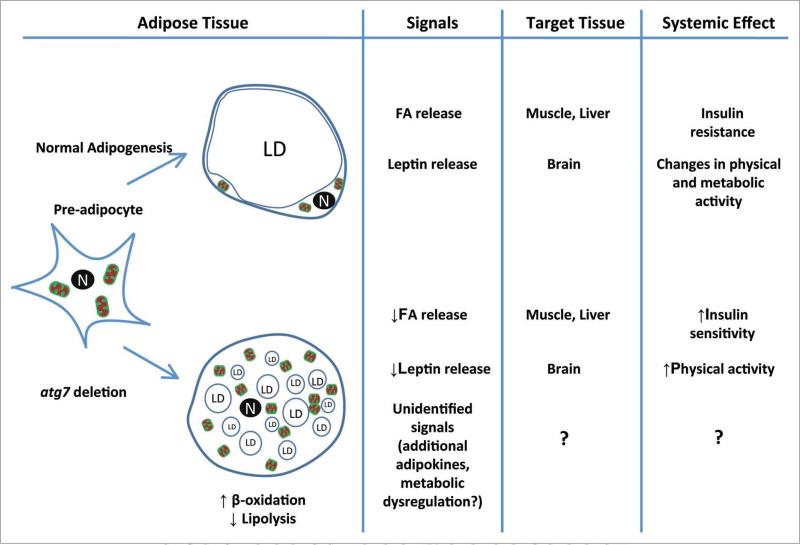
The unique combination of “fitness” phenotypes observed in these mice begs for an explanation. It is clear that by inactivating autophagy activity in adipocytes we have created a “healthier” adipose tissue. The observed phenotype likely results from altered metabolic features of the mutant adipose tissue and/or alterations in communication between mutant adipose tissue and the other parts of the body. Mutant WAT exhibits some morphological features of BAT; however, it does not express UCP1 protein or other brown fat specific proteins. Nevertheless, the change in fatty acid metabolism in these cells is apparent. Consistent with the increased number of mitochondria, we observed a marked increased in the rates of fatty acid (FA) β-oxidation in mutant adipocytes, along with a marked increase in the rate of free fatty acid (FFA) clearance from plasma in response to insulin administration. Interestingly, these mutant adipocytes also showed much slower rates of hormone-induced lipolysis. The change of fatty acid metabolism in adipose tissue may lead to a systemic alteration of fatty acid homeostasis, as evidenced by lower levels of plasma FFA under normal dietary conditions observed in the mutant mice. Fatty acids are powerful modulators of insulin sensitivity in peripheral tissues like myocytes, and the decrease in circulating FFA could lead to increased insulin sensitivity. In addition to its function as a depot for energy storage, WAT also acts as an endocrine organ, secreting adipokines that regulate systemic energy homeostasis. Autophagy-deficient adipose tissue apparently has an altered spectrum of adipokine secretion. Such alterations (drastically reduced leptin secretion, for example) could potentially lead to behavior changes (i.e., increased physical activity) in the mutant animal. It should be kept in mind that our current information on the mutant mouse model cannot explain all of the favorable metabolic phenotypes observed. Teasing out these unknown variables will be of great interest for the study of metabolic controls. Figure 1 is a general working model that illustrates how defective autophagy may have an impact upon adipose tissue development, and how the resulting adipose tissue may in turn exert an affect on lipid metabolism and insulin response at the systemic level.
Figure 1.
A model for autophagy-deficient adipogenesis and its effects. n, nucleus; Ld, lipid droplet; FA, fatty acid.
Recent work in the field of adipogenesis indicates that about 10% of adipocytes may be turned over annually in human adults. It is our hope that the findings in this study may open new avenues of research for the potential treatment of obesity and type II diabetes. By further dissecting the cellular and systemic circuits by which autophagy inhibition affects adipogenesis and ultimately leads to the beneficial metabolic phenotype we observed, we will continue to enhance our knowledge of obesity and insulin resistance and may therefore make progress towards the more effective management of these conditions.
Footnotes
Punctum to: Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. PNAS 2009; 106:19860–5; PMID: 19910529; DOI: 10.1073/pnas.0906048106.