Abstract
Disulfide bond forming (Dsb) proteins ensure correct folding and disulfide bond formation of secreted proteins. Previously, we showed that Mycobacterium tuberculosis DsbE (Mtb DsbE, Rv2878c) aids in vitro oxidative folding of proteins. Here we present structural, biochemical and gene expression analyses of another putative Mtb secreted disulfide bond isomerase protein homologous to Mtb DsbE, Mtb DsbF (Rv1677). The X-ray crystal structure of Mtb DsbF reveals a conserved thioredoxin fold although the active-site cysteines may be modeled in both oxidized and reduced forms, in contrast to the solely reduced form in Mtb DsbE. Furthermore, the shorter loop region in Mtb DsbF results in a more solvent-exposed active site. Biochemical analyses show that, similar to Mtb DsbE, Mtb DsbF can oxidatively refold reduced, unfolded hirudin and has a comparable pKa for the active-site solvent-exposed cysteine. However, contrary to Mtb DsbE, the Mtb DsbF redox potential is more oxidizing and its reduced state is more stable. From computational genomics analysis of the M. tuberculosis genome, we identified a potential Mtb DsbF interaction partner, Rv1676, a predicted peroxiredoxin. Complex formation is supported by protein co-expression studies and inferred by gene expression profiles, whereby Mtb DsbF and Rv1676 are upregulated under similar environments. Additionally, comparison of Mtb DsbF and Mtb DsbE gene expression data indicate anticorrelated gene expression patterns, suggesting that these two proteins and their functionally linked partners constitute analogous pathways that may function under different conditions.
Keywords: Mycobacterium tuberculosis, disulfide bond forming protein, structure-function, gene expression data, X-ray crystallography
Introduction
Most disulfide oxidoreductase proteins contain a conserved thioredoxin-like domain and share a common sequence motif (CxxC) at their active sites. These ubiquitous proteins have a variety of mechanistic roles including protein folding, electron transport and bioenergetics in all three kingdoms of life. The family of disulfide oxidoreductase proteins include thioredoxin, eukaryotic protein disulfide bond isomerase, glutaredoxin 1, peroxiredoxin 2 and disulfide bond forming (Dsb) proteins.
Dsb proteins are best characterized in Escherichia coli. These proteins reside in the periplasmic space of gram-negative bacteria and are necessary for correct folding of many cell envelope proteins 3. Over the last decade, Dsb proteins, and in particularly DsbA, have been shown to be involved in virulence in toxin-secreting gram-negative bacteria such as E. coli 4, Yersinia pestis 5, Shigella sp. 6 and Vibrio cholerae 7; 8. E. coli DsbA is a monomer that catalyses the oxidation of reduced, unfolded proteins 9; 10. DsbA is reoxidized by the transmembrane protein DsbB, which is in turn oxidized by components of the electron transport pathway 11; 12. Another well-characterized Dsb protein is E. coli DsbE, a monomeric thioredoxin-like protein involved in cytochrome c maturation 13. DsbE has been implicated in the reduction of thiol ether linkers to apocytochrome c prior to heme ligation by CcmF and CcmH 13; 14; 15. E. coli DsbD is a transmembrane protein spanning the cytoplasmic membrane responsible for maintaining DsbE in its reduced state 16. Finally, E. coli DsbC and DsbG are homodimers with disulfide bond isomerase activity, which are also maintained in their reduced state by the transmembrane protein DsbD 17.
In Mycobacterium tuberculosis (M. tuberculosis), the only Dsb proteins identified to-date are Mtb DsbE (Rv2878c aka MPT53) 18, its homolog annotated as Mtb DsbF (Rv1677) and its potential redox, transmembrane protein partner Mtb DsbD (Rv2874) 19. The presence of Dsb proteins in M. tuberculosis suggest these proteins are necessary for the correct folding of disulfide bond rich cell-wall associated, potential periplasmic 20 and secreted extracellular proteins. Within the M. tuberculosis proteome, it has been predicted that over 160 proteins are secreted, of which 60% may contain disulfide bonds based on their cysteine content, implying that disulfide bond forming proteins are required for correct folding of approximately 90 secreted proteins 18. M. tuberculosis secreted proteins have many different roles including involvement in virulence, pathogenicity and cell-wall maintenance; thus, interruption of their folding pathways may prevent mycobacterial infectivity and viability. As M. tuberculosis is a pathogenic bacterium responsible for tuberculosis (TB), which causes approximately 2 million deaths and 8 million new cases per year, 21; 22, the study of Dsb protein systems in M. tuberculosis may offer new insight into its virulence and provide novel anti-TB drug targets 23; 24.
Recently, we biochemically and structurally characterized a homolog of E. coli DsbE, a secreted protein, Mtb DsbE (Rv2878c) 18. We determined the crystal structure of Mtb DsbE to 1.1 Å resolution, which revealed a thioredoxin-like domain with a typical CxxC active site. The active-site cysteines in the structure of Mtb DsbE are in their reduced state. Additionally, the pKa of the active-site, solvent-exposed cysteine was determined to be approximately 2 units lower than that of gram-negative DsbE homologs. Finally, the reduced form of Mtb DsbE is more stable than the oxidized form, and Mtb DsbE is able to oxidatively refold leech hirudin. Structural and biochemical analyses imply that Mtb DsbE functions as a thiol oxidase, unlike gram-negative bacteria DsbE proteins that have been shown to be weak reductases 25. On the contrary, Mtb DsbE is functionally analogous to E. coli DsbA, folding and ensuring correct disulfide bond formation in secreted proteins, although structurally E. coli DsbA has an additional domain that caps the thioredoxin-like active site 26.
In this study, we have determined the 1.6 Å resolution structure of Mtb DsbF (Rv1677), a predicted extracellular disulfide bond forming protein homologous to Mtb DsbE. The active-site cysteines of Mtb DsbF are in both their oxidized and reduced forms. Further characterization reveals that Mtb DsbF has a redox potential of -87 mV comparable to that of E. coli DsbA (-89 to -119 mV) 27; 28, which is confirmed by its ability to refold hirudin. Additionally, we show that Mtb DsbF forms a potential transient protein complex with its genomic neighbor Rv1676, a predicted peroxiredoxin, and that these two proteins have correlated gene expression profiles suggesting that they may potentially function in the same biochemical pathway. Both Mtb DsbF and Mtb DsbE appear to be part of larger groups of coexpressed genes, suggesting the possible involvement of Mtb DsbE and Mtb DsbF in complexes or pathways. We show that the expression profiles of both Mtb DsbF and Rv1676 are inversely correlated with respect to Mtb DsbE, suggesting that they, and in turn their coexpressed partners, are induced under different conditions. Finally, we consider the environmental conditions under which Mtb DsbF and its protein partners may be expressed.
Results
The complete sequence and subsequent annotation of the M. tuberculosis genome has allowed the prediction of many genes and gene functions by homology. Rv2878c and Rv1677 were annotated as putative secreted disulfide bond forming proteins DsbE and DsbF, respectively 29. A recent study of Mtb DsbE showed that this protein was biochemically more similar to E. coli DsbA rather than to E. coli DsbE 18. This result led to the investigation of the structure, biochemistry and gene expression patterns of Mtb DsbF in an attempt to determine the function of Mtb DsbF and the pathway in which it may function.
Structural Overview of Mtb DsbF
The crystal structure of the mature form of Mtb DsbF (without its signal peptide, residues 1- 38, which was predicted to high significance by SignalP 30) consists of a thioredoxin fold, with its distinct structural motif consisting of a four-stranded β-sheet made up of β3, β4, β6 and β7 and three flanking α-helices corresponding to α3, α5 and α6 (Figure 1a). As in the structure of Mtb DsbE 18, a long α-helix (α4) and a β-strand (β5) (forming a five-stranded β-sheet) are found after the β3-α3-β4 motif of the thioredoxin fold. At the N-terminus of the structure there is an additional smaller domain, which consists of a short 310-helix (α1), two β-strands (β1 and β2) and another short 310-helix (α2). The cysteines adopt a right-handed hook conformation at the N-terminus of helix α3 as found for most active-site cysteines in the thioredoxin superfamily fold. The electron density for the two active-site cysteines, Cys81 and Cys84, is most consistent with a model in which conformations of the oxidized and reduced forms of the cysteines are observed, hence we modeled both the oxidized and reduced forms into the structure. The conformation in which the cysteines are oxidized has a distance of 2.06 ± 0.20 Å between the two Sγ atoms (Figure 1b), which is consistent with other observed disulfide bonds 31. Only the Sγ atom of Cys81 in the dithiol is exposed on the protein surface, while Sγ of Cys84 is buried. The sulfur atom of Cys81 is stabilized by weak hydrogen bonds to the amide N atom of Thr83 (3.33 Å) and the Oε1 atom of Gln146 (4.05 Å); the Sγ atom of Cys84 is hydrogen bonded to the carbonyl O atom of Ala78 (4.00 Å) and to a water molecule (3.80 Å) which is in turn hydrogen-bonded to Oε1 of Glu87 (2.59 Å) and Nε1 of Trp75 (2.50 Å). Both Cys81 and Cys84 have a hydrophobic interaction with conserved cis-Pro147 (4.42 Å and 4.80 Å respectively). Interestingly, the reduced form also appears to be present within the crystal structure with occupancy of approximately 50% (Figure 1c). The distance between the active-site cysteines modeled in the reduced form conformation for Mtb DsbF is 3.69 ± 0.10 Å. The sulfur atom of Cys81 is stabilized by hydrogen bonds to the amide N atom of Thr83 (3.78 Å) and the Oε1 atom of Gln146 (3.30 Å); the Sγ atom of Cys84 is hydrogen bonded to the carbonyl O atom of Ala78 (3.78 Å) and to a water molecule (3.45 Å) which is in turn hydrogen-bonded to Glu87 and Trp75 as described for the oxidized conformation.
Figure 1.
(a) Structure of Mtb DsbF in its oxidized form is shown as a ribbon diagram. The β-sheets, α-helices and random coil are colored blue, red and cream, respectively. The atoms for the two active-site cysteines, Cys81 and Cys84 are colored in yellow. (b) and (c) Electron density surrounding the active-site CxxC motif of Mtb DsbF where the cysteines are modeled in both the (b) oxidized and (c) reduced forms. The 2fo-fc electron density mesh (blue) and the fo-fc negative density mesh (red) are contoured at sigma 3.0. Shown are stick cartoons of the active-site where the carbon, oxygen, nitrogen and sulfur atoms are colored in white, red, blue and yellow, respectively.
Determination of the Redox Potential of Mtb DsbF and comparison with Mtb DsbE
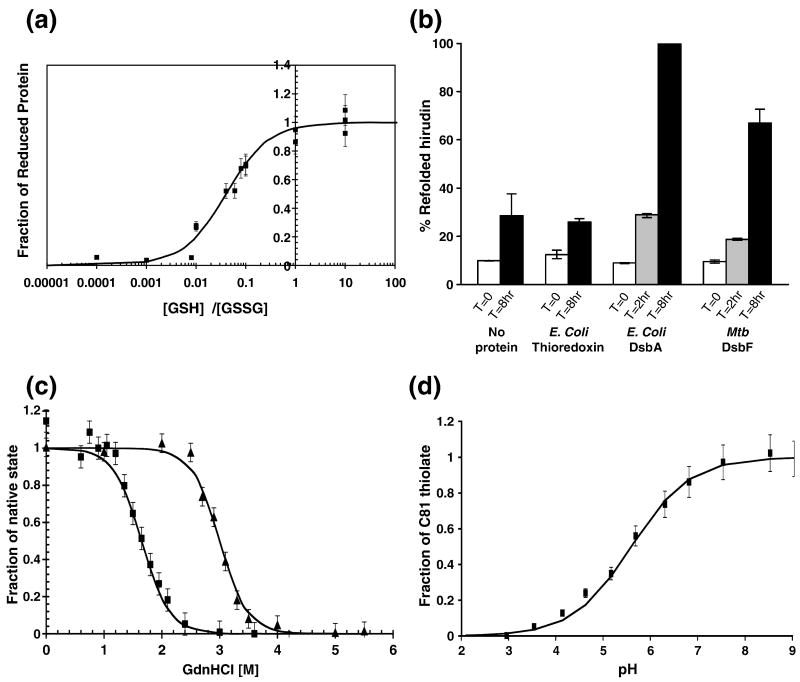
To further characterize Mtb DsbF, the redox potential relative to that of glutathione was determined. This redox potential compares the ability of reduced glutathione to transfer electrons to a protein. The Keq of Mtb DsbF is ∼40 ± 5 μM (Figure 2a). The corresponding standard redox potential (E′0) calculated for Mtb DsbF is −89 ± 9 mV. In comparison with the standard redox potential of two thiol oxidases, Mtb DsbE (−128 mV) 18 and E. coli DsbA (between −89 to −119 mV) 27; 28, and reductase E. coli thioredoxin (−269 mV) 32, the standard redox potential of Mtb DsbF suggests that it is also a thiol oxidase which has stronger oxidizing potential than its homolog Mtb DsbE.
Figure 2.
Biochemical characterization of Mtb DsbF. (a) Redox equilibrium of Mtb DsbF with glutathione. The y-axis represents the proportion of Mtb DsbF present in the reduced form at equilibrium with various mixtures of reduced (GSH) and oxidized (GSSG) glutathione (x-axis), which was measured by exploiting the difference in fluorescence (347 nm) of reduced Mtb DsbF compared to oxidized Mtb DsbF. Equilibrium concentrations of oxidized and reduced Mtb DsbF and GSH and GSSG were calculated as described 66. The equilibrium constant was calculated to be 40 ± 5 μM, which is consistent with Mtb DsbF acting as an oxidant. (b) Analysis of the refolding activity of reduced and unfolded hirudin. 5 pmol reduced, unfolded hirudin was incubated alone or with stoichiometric quantities (5 pmol) of Mtb DsbF, E. coli DsbA and E. coli thioredoxin. Samples were removed as indicated at each time point and the reactions, quenched by the addition of iodoacetamide, were analyzed by MALDI-TOF mass spectrometry. The appearance of native hirudin (m/z 6765) is represented as a percentage of the total intensities of native and carbamidomethylated hirudin. (c) GdnHCl-dependent unfolding/folding of Mtb DsbF. Guanidine hydrochloride (GdnHCl)-dependent unfolding was monitored by circular dichroism spectroscopy at 222 nm for oxidized Mtb DsbF (shown as squares) and reduced Mtb DsbF (shown as triangles). Since the reduced form of Mtb DsbF unfolds at higher concentrations of GdnHCl than the oxidized form, the reduced form of Mtb DsbF is its more stable form. (d) Determination of the pKa value of the active-site cysteine, Cys81, in Mtb DsbF. The absorption specific to reduced Mtb DsbF protein is shown as a function of pH (x-axis) compared to the fraction of Cys81 thiolate (y-axis). The pKa of Cys81 was calculated by fitting the data points as described 67. The pKa of Cys81 in Mtb DsbF is 5.6 ± 0.3.
Analysis of oxidase activity of Mtb DsbF by mass spectrometry
Reduced, unfolded hirudin from Hirudo medicinalis was used to assess the oxidative protein folding ability of Mtb DsbF. Hirudin is an inhibitor of thrombin and contains three intramolecular disulfide bridges. Mass spectrometry analysis of commercial native hirudin revealed, besides significant impurities, a major peak (m/z 6765) followed by several smaller peaks ranging up to m/z 7088, which is consistent with previous reports that hirudin is a non-homogenous protein that contains several variants 33. Reverse-phase HPLC was used to enhance the homogeneity of the m/z 6765 peak, as well as to remove contaminating proteins. The resulting mass spectrometry data for HPLC-purified hirudin indicated that the m/z 6765 peak was the predominant peak. The corresponding peaks for reduced, unfolded hirudin showed a uniform m/z increase of 6 Da. Furthermore, the addition of iodoacetamide to reduced, unfolded hirudin carbamidomethylated the free cysteines as observed by the mass increase of six 57 Da increments (+342 Da mass). The predominant peak was thus chosen to monitor the regeneration of reduced, unfolded hirudin (5 pmol) to the fully oxidized native state in the absence and presence of stoichiometric quantities of Mtb DsbF (5 pmol). At various time points, the reactions were quenched by the addition of iodoacetamide and analyzed by mass spectrometry. In the absence of Mtb DsbF, a small quantity of native hirudin was observed after 8 hrs (Figure 2b), presumably due to spontaneous, air-mediated oxidation as previously reported 18. However, when reduced, unfolded hirudin was incubated with Mtb DsbF, mass spectrometry data showed new peaks at the zero time point corresponding to the immediate quenching of the free thiols by the covalent modification with iodoacetamide molecules (57 Da increments), which decrease over the 8 hr reaction period. Conversely, the native hirudin peak increases over the same time period, suggesting that Mtb DsbF is capable of oxidizing substrate proteins, and is comparable to the oxidase activity of Mtb DsbE (data not shown), whereby at 8 hours greater than 70% of hiridin is oxidatively refolded (Figure 2b). Significantly, these data concur with the oxidase activity of E. coli DsbA, although 100% oxidation occurs after 8 hrs (Figure 2b). Additionally, when incubated with a known reductase, E. coli thioredoxin (negative control), only a small fraction of oxidized hirudin appears after 8 hrs, as seen for spontaneous air-mediated oxidation of hirudin (Figure 2b).
Thermodynamic Properties of the Redox Forms
To compare stabilities of the different redox forms of Mtb DsbF, guanidine hydrochloride-induced unfolding and refolding (data not shown) of both oxidized and reduced forms were examined by circular dichroism. The reduced form of Mtb DsbF is more stable than that of the oxidized form, given that the reduced form of the protein denatures at a higher concentration of guanidine hydrochloride compared to the oxidized form (Figure 2c). Calculation of the free energy change (ΔΔGredox) between the reduced and oxidized form of Mtb DsbF suggests that the reduced form is 24 ± 5 kJ/mol more stable than the oxidized form. This is consistent with the trend observed for Mtb DsbE, although the reduced form of Mtb DsbE is only 12.4 ± 4 kJ/mol more stable than the oxidized form 18, suggesting greatly increased stability of the reduced form of Mtb DsbF.
Determination of the pKa (Cys81) of Mtb DsbF
As the redox potential and thermodynamic properties of the two Mtb Dsb proteins are dissimilar, for completeness, we determined the pKa of the solvent-exposed active-site cysteine of Mtb DsbF, which is associated with the redox potential of a protein. The pKa value of the Mtb DsbF Cys81 was measured by observing the change in absorption of the cysteines at 240 nm over a pH range of pH 2-9 (Figure 2d), and was determined to be 5.6 ± 0.3 which is similar to that of Mtb DsbE of 5.35 ± 0.2 18. Furthermore, the pKa of Cys81 is relatively acidic compared with the solvent-exposed active-site cysteine of a known reductase, E. coli thioredoxin (pKa of 7.5) 34, although not as acidic as E. coli DsbA, which is a known oxidase, where the pKa of the solvent-exposed active-site cysteine is 3.5 35.
Potential Protein Interaction Partner for Mtb DsbF
Comparative organization across organisms and genomic context within an organism yield information inferring functional linkages between proteins on a genome-wide scale 36; 37. M. tuberculosis genomic organization around the gene Rv1677 reveals that this gene is in the same operon as Rv1676. The final four nucleotides of Rv1676 contain both the stop-codon for Rv1676 and the start-codon for Rv1677 (Mtb DsbF). Two bioinformatics methods, gene neighbor and gene cluster, predict with high significance that these two proteins are both transcribed together and may function within a common pathway 38.
To test this hypothesis, Rv1676 and Rv1677 were cloned into a single pETDuet double expression vector in which only Mtb DsbF encodes a His6 affinity tag. The cell lysate from the double expression study was then loaded onto a Ni2+ charged HiTrap chelating column. After washing all of the non-specifically bound material from the column, the proteins of interest were eluted with an imidazole gradient from 0 – 500 mM (Figure 3a). To verify if Mtb DsbF interacts with Rv1676, the fractions from the column elution were run on an SDS-PAGE gel. A small peak from Ni2+ affinity chromatography showed both a 15 kDa protein and a second protein with a molecular mass of approximately 24 kDa (Figure 3b, lane 4). After excising and performing in-gel tryptic digestion on these bands, the peptides were eluted from the gel and identified utilizing micro-liquid chromatography with tandem mass spectrometry (μLC-MSMS) (Figure 3b). The results from the in-gel tryptic digestion mass spectrometry experiment showed that the band corresponding to the 24 kDa protein contained peptides from Rv1676. (Figure 3c). Peptides from the 15 kDa band could not be successfully identified as the mass cut-off range within the experiment did not allow for detection of the majority of the peptides from the tryptic digestion of Mtb DsbF. However, the 15 kDa protein band migrated identically to Mtb DsbF on an SDS-PAGE gel (Figure 3a, lane 3) and MALDI mass spectrometry revealed that molecular mass was consistent with that of Mtb DsbF (data not shown). Under native conditions, Rv1676 when expressed alone is insoluble whereas a small fraction of it is soluble when co-expressed with Mtb DsbF. This observation indicates that Mtb DsbF is able to maintain some soluble Rv1676, suggesting a potential transient protein complex. Interestingly, Rv1676 has been reported to localize to the mycobacterial membrane or cell-wall vicinity 39, and thus may interact with secreted Mtb DsbF. Therefore, Mtb DsbF and Rv1676 may function in a common pathway as their genes are contained within the same operon and co-expression studies infer an in vitro protein-protein interaction.
Figure 3.
Co-expression studies for Mtb DsbF with Rv1676. (a) Purified Rv1676 and Mtb DsbF complex. On the right is the chromatograph of the Ni2+ affinity column elution (0 – 500 mM imidazole) of Rv1676 and Mtb DsbF co-expression experiment and the SDS-PAGE gel shows lanes 1) protein ladder; 2) Rv1676 purified in denaturing conditions (6M urea); 3) Mtb DsbF purified under native conditions and; 4) eluted Rv1676 and Mtb DsbF complex. The eluted Rv1676 and Mtb DsbF bands correspond to a small peak before the larger elution of pure Mtb DsbF alone. (b) Mass spectrometry results from a co-elution fraction of trypsin digested Mtb DsbF and Rv1676. The peptide fragments correspond to Rv1676 with high significance. (c) Predicted trypsin peptide map of Rv1676. Highlighted in grey are the peptides identified experimentally in 3b.
Gene Expression Data
To further investigate the experimental data that Rv1676 and Mtb DsbF can interact transiently in vitro, we looked at the gene expression data from various microarray datasets for Rv1676 and Rv1677 compared with its homolog Rv2878c (encodes for Mtb DsbE), its proposed interaction partners Rv2874 18 and predicted membrane protein Rv2877c 40. Gene expression data was extracted and combined from four experimental data sets 41; 42; 43; 44 using a previously documented methodology 45. Figure 4a demonstrates that the expression of Rv1676 and Rv1677 are strongly correlated, which one would predict considering they are located within the same operon. A positive correlation was also observed for the gene expression pattern of Rv2874, Rv2877c and Rv2878c. Thus each group of proteins may represent distinct disulfide bond forming pathways. More interestingly, these two groups of proteins have anticorrelated expression patterns indicating that while one group of Dsb proteins is up-regulated, the other is down-regulated. This implies that the two Dsb groups may be distinct from one another, and are regulated, expressed and function under different environmental conditions in M. tuberculosis.
Figure 4.
Gene expression analysis. In figures 4a and 4b, red represents positive expression correlation and green represents negative correlation. The colors are shaded as to the degree of correlation, black indicates neither positive or negative correlation. Figure 4a contains the genes that encode for Rv1677 and its interaction partner Rv1676, as well as Rv2878c (Mtb DsbE) and its potential protein partners Rv2874 (DipZ, DsbD) and Rv2877c. Note Mtb DsbE (Rv2878c) and Mtb DsbF (Rv1677) and their respective protein partners have anticorrelated expression profiles. Figure 4b observes the correlated expression patterns of Rv1676, Rv1677, Rv2874, Rv2877c and Rv2878c in addition to AphC and sigma factor L systems; Rv2428 (AphC), Rv2429 (AphD) and Rv2238c (AphE), Rv0735 (SigL) and Rv0736 (anti-SigL). Note that Mtb DsbF and Aph systems have positive correlated expression patterns, which have anticorrelated expression patterns with Mtb DsbE and SigL systems (which have positively correlated expression to each other).
Discussion
Structural Comparison of Mtb DsbF and Mtb DsbE
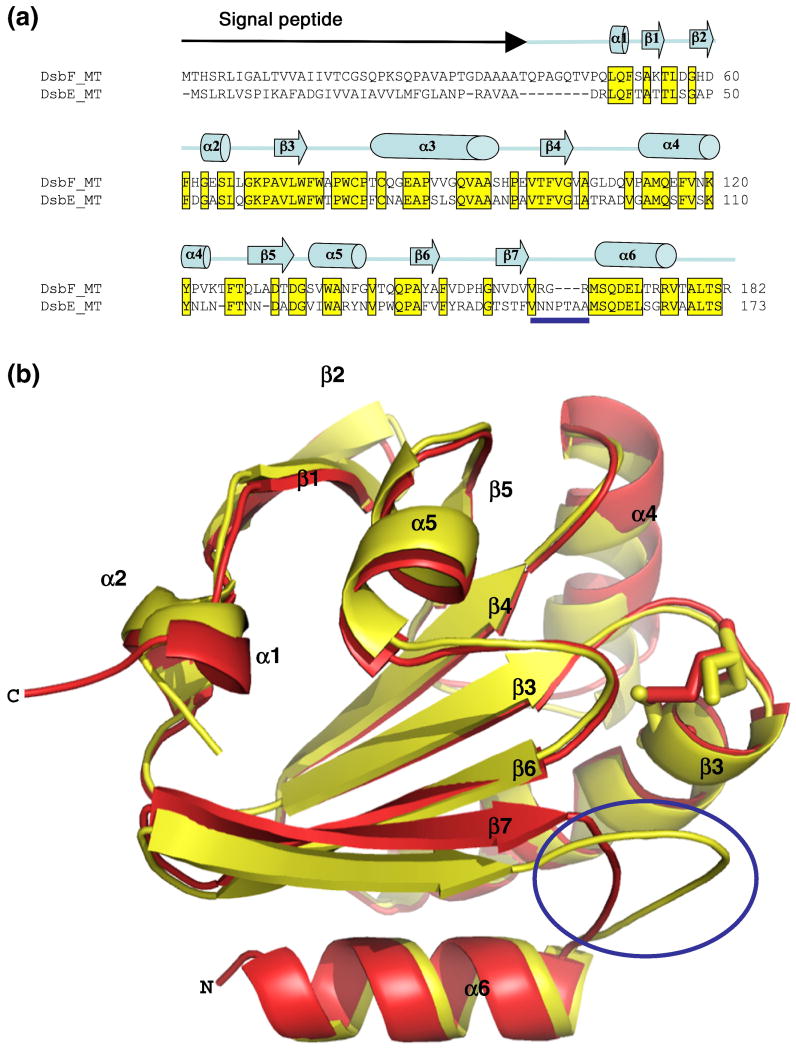
Mtb DsbF and Mtb DsbE are extremely similar in both sequence and structure but there are two striking differences between their structures. The sequence identity between Mtb DsbF and Mtb DsbE is 55.4% (Figure 5a), and the crystal structures of Mtb DsbF and Mtb DsbE can be superimposed with a root mean square derivation of 1.0 Å for 127 Cα atoms. The two structural differences are 1) the active-site cysteines are in different redox states in the crystal form; and 2) one of the loop regions in Mtb DsbE is extended compared to Mtb DsbF (Figure 5b).
Figure 5.
Overall sequence and structural comparison of Mtb DsbF and Mtb DsbE. (a) Sequence alignment between Mtb DsbE and DsbF. The signal peptides for Mtb DsbE (Rv2878c) and Mtb DsbF (Rv1677) are depicted on the top line as a black arrow. The sequences are 54% identical and these residues are boxed in yellow. Residues underlined with the blue bar correspond to the loop region between β7 and α6. Secondary structure elements are depicted on the top line. (b) Superimposition of the structures of Mtb DsbF (red) and Mtb DsbE (yellow) which are shown as ribbon diagrams. The active-site cysteines are shown in sticks with the same respective coloring schemes: Mtb DsbF active-site oxidized cysteines, Cys81 and Cys84, are shown as red sticks and Mtb DsbE active-site reduced cysteines, Cys36 and Cys39, are shown in yellow sticks. The most prominent difference between the two structures is the loop region between β7 and α6, (circled). In the Mtb DsbE structure (yellow) the loop is longer and appears to protect the active-site CxxC motif from solvent accessibility, whereas in the Mtb DsbF structure (red) this loop is shorter which allows greater accessibility to its active-site.
Within the active-site motif CxxC of the Mtb DsbF structure, Cys81 and Cys84 are in both oxidized and reduced states whereas in the Mtb DsbE structure the active cysteines Cys36 and Cys39 are in their reduced state, 3.69 Å distance between the two Sγ atoms 18 (Figure 5b). The loop between β-strand 7 (β7) and α-helix 6 (α6) is extended by three extra amino acids (inserted in between Arg162 and Gly163 in Mtb DsbF) in Mtb DsbE compared to Mtb DsbF (Figures 5a and 5b). In the Mtb DsbE structure, this extended loop (P118TAA121) is stabilized due to a hydrophobic interaction with Phe38 within the active-site motif, CPFC (Figure 6a), thus decreasing solvent accessibility of the active site compared to that of Mtb DsbF (Figures 6c and 6d, respectively). It should be noted that this loop in the Mtb DsbE structure also plays a role in forming a crystallographic homodimeric interface 18. In contrast, the crystal packing within the Mtb DsbF structure reveals no crystallographic dimeric interface. Additionally, within the shorter loop region, NH1 of Arg164 forms a hydrogen bond with the OH group of Thr83 (3.35 Å) within the active-site CPTC motif (Figure 6b), revealing a more solvent-exposed active site compared to that of Mtb DsbE (Figures 6d and 6c, respectively).
Figure 6.
Loop region, molecular surface representation and active-site comparisons of Mtb DsbF and Mtb DsbE. In (a), (b), (e) and (f) Mtb DsbE and Mtb DsbF are shown as ribbon diagrams in blue and purple, respectively. Stick models represent the pertinent amino acid residues where nitrogen, oxygen and sulfurs atoms are shown in blue, red and yellow, respectively. Hydrogen bonding is depicted with broken black lines. The images were generated in PYMOL. (a) The extended loop region in Mtb DsbE forms a cleft through a hydrophobic interaction with Phe38, which results in a more concave architecture surrounding the active-site. (b) The shorter loop region in Mtb DsbF stabilizes the active-site loop by a hydrogen bond between Arg164 and Thr83. Electrostatic molecular surface representation of (c) Mtb DsbE and (d) Mtb DsbF with the solvent-exposed active-site cysteines indicated. The overall position is equivalent to the active-sites shown in (a) and (b). Positive and negative electrostatic potentials are shown in blue and red, respectively generated in PYMOL. The figure shows that Mtb DsbE has active-site is more solvent protected than Mtb DsbF. Also the surface surrounding the active-site of Mtb DsbF is more positively charged than that of Mtb DsbE. (e) and (f) show the conserved amino acid (Glu-Trp) pair in Mtb DsbE and Mtb DsbF, respectively. The hydrogen bond between Trp30 and Glu42 in the Mtb DsbE structure maintains a conformation in which the active-site residues are in their reduced form. Whereas, there is an extensive hydrogen bonding network between the amino acid pair (Trp75 and Glu87) and Tyr142, and which acts upon the active-site Cys81 facilitated by a water molecular in Mtb DsbF.
Structural analyses of the two Mtb Dsb proteins along with gram-negative B. japonicum DsbE (reductase) and E. coli DsbA (oxidase), implicate an amino acid pair that contributes to the stability of the reduced and oxidized forms of the Dsb proteins. In the reduced form of Mtb DsbE, the amino acid pair Trp30 from β-strand 3 (β3) and Glu42 from α-helix 3 (α3) forms a hydrogen bond between Nε1 of Trp30 and Oε1 of Glu42 (2.8 Å), which is flanked by the active-site residues as well as a hydrophobic residue Phe103 from β-strand 4 (β4), Figure 6e. It was proposed previously that this interaction contributes to the stability of the active-site loop to form a conformation where the reduced thiol form is favored 18. This amino acid (Trp-Glu) pair is conserved throughout mycobacterial and gram-positive DsbE homologs, although these residues in the mixed redox state Mtb DsbF structure form a very weak hydrogen bond between Nε1 of Trp75 and Oε1 of Glu87 (4.44 Å) due to the interactions with Tyr142 (Figure 6f) that is superimposable on Mtb DsbE Phe103. The OH group of Tyr142 hydrogen bonds to both Nε1 of Trp75 and Oε1 of Glu87 (3.6 and 2.6 Å respectively), preventing a stronger hydrogen bond between Glu87 and Trp75. Additionally, a water molecule hydrogen bonds to all three residues: Nε1 of Trp75 (2.5 Å), OH of Tyr142 (3.8 Å) and Oε1 of Glu87 (2.6 Å), and also to the reduced thiol form of Cys84 (3.5 Å), Figure 6f. In the structures of Mtb DsbE, E. coli DsbA and B. japonicum DsbE, this extensive hydrogen-bonding network is not observed although the hydrogen-bonded amino acid pair across the β-strand and the α-helix is conserved. Additionally, the amino pair in the structure of E. coli DsbA, Glu37 and Lys58, is ∼0.75 Å closer to each other in the more stable, reduced form compared to the oxidized form 46. In comparison with B. japonicum DsbE 47, the disulfide form is possibly stabilized by the hydrogen bond between Asn86 and Glu98 (3.1 Å). One should note that within the structures of E. coli DsbC and DsbG 48 there are no corresponding amino acid pairs that form hydrogen bonds across the β-strand and α-helix containing the active-site cysteines. Both these structures have been observed with a mixture of reduced and oxidized forms and, similar to Mtb DsbF, these proteins are more stable in their reduced forms (Figure 2c). In summary, the weak hydrogen bond between the amino acid pair Trp75 and Glu87 may, in part, contribute to the mixed redox state within the Mtb DsbF structure.
Biochemical analysis reveals that the physicochemical properties of Mtb DsbF is similar to Mtb DsbE 18. Additionally, the in vitro activity assay indicates that Mtb DsbF, as with Mtb DsbE and E. coli DsbA, is capable of refolding reduced, unfolded hirudin, suggesting that it functions as a thiol oxidase. The reduced form of Mtb DsbF is more energetically stable compared to that of Mtb DsbE and the redox potential of Mtb DsbF active-site cysteines is −89 mV compared to −123 mV observed for Mtb DsbE 18, which is consistent with Mtb DsbF being a stronger oxidant than Mtb DsbE. The extensive hydrogen bonding network surrounding the solvent-protected Cys84 observed in the Mtb DsbF structure compared to the Mtb DsbE structure (Figures 6f and 6e respectively) could possibly stabilize the thiol form of Cys84 and thus favors Mtb DsbF's reduced state. Additionally, electrostatic surface potential analysis of the two Mtb Dsb proteins suggests that the surface potential surrounding the active site of Mtb DsbF is more positively charged facilitating the greater oxidizing redox potential and stabilizing the thiol form of the active site, whereas in the Mtb DsbE structure the surface potential suggests a mixed acidic and basic nature surrounding the active site which may lower the oxidizing redox potential compared to Mtb DsbF (Figures 6c and 6d).
Implications of transient in vitro interaction of Mtb DsbF and Rv1676
Co-expression studies, as well as their localization within the mycobacterial membrane or cell-wall, suggest that Mtb DsbF may aid in correct folding of Rv1676, a predicted peroxiredoxin. Peroxiredoxins are a ubiquitous family of antioxidant enzymes that have peroxidase activity and can be regulated by changes in phosphorylation, redox and possibly oligomerization states 49. Peroxiredoxins have been shown to interact with thioredoxin-like folds 50 and more importantly, several lines of evidence have documented the ability of disulfide bond forming proteins to assist in folding peroxidases 51; 52. Thus, an interaction between Mtb DsbF and Rv1676 is not unprecedented within the capacity of Mtb DsbF's potential role in ensuring correct folding of Rv1676 in the membrane or cell-wall vicinity. The well-characterized M. tuberculosis Ahp system involved in antioxidant defense contains thioredoxin-like proteins and peroxiredoxins 53. The gene expression patterns of AhpC (Rv2428) 54, a peroxiredoxin alkyl hydroperoxide reductase, its adaptor protein AhpD (Rv2429) 53, also annotated as an alkyl hydroperoxide reductase D, and a probable peroxiredoxin AhpE (Rv2238c) 55 were observed in addition to the Mtb Dsb systems (Figure 4b). Interestingly, the positively correlated genes within the Ahp system also have positive expression patterns with Mtb DsbF and Rv1676. These data suggest that, under oxidative stress, Mtb DsbF may potentially play a crucial role in detoxification whereby it may facilitate the correct folding of Rv1676. In contrast, they have anticorrelated expression patterns to the homolog of Mtb DsbF, Mtb DsbE and its potential redox partners.
M. tuberculosis extracytoplasmic sigma factors play a major role in altering patterns of gene expression to allow adaptation to stress responses during infection of its host, and upon entry into stationary phase 56; 57. One of these sigma factors, SigL (Rv0735), upregulates polyketide synthases and secreted/membrane proteins, including the pair of proteins, Mtb DsbE (Rv2878c) and Rv2877c. Additionally, it has been demonstrated that a sigL mutant of M. tuberculosis is severely attenuated in a mouse model, suggesting that SigL (Rv0735) plays a role in virulence 40; 57. Also, SigL has an anti-sigma factor, Rv0736 57. By association through correlated expression patterns with SigL and anti-SigL, Mtb DsbE may be involved in virulence and invasion whereas Mtb DsbF may be up-regulated under oxidative stress (Figure 4b). Our observation that two homologous proteins which have similar functions under distinct cellular conditions is not unprecedented. In plants, it has been shown that different thioredoxin isoforms function in different biological pathways, have different differential gene expression patterns, and different protein accumulation patterns in pea tissues, even though their sequence similarity is 60% identical at the amino acid level 58. Thus, although Mtb DsbE and Mtb DsbF have both been shown to possess oxidase activity in vitro, the gene expression data infer that they may possibly function in separate biological pathways.
We have presented a study of Mtb DsbF which, despite its strong sequence homology and biochemical and structural similarity to Mtb DsbE, appears to function in distinct cellular contexts from Mtb DsbE, as shown by analysis of interacting partners and correlated gene expression. Mtb DsbE and Mtb DsbF likely assist in correct folding of disparate sets of disulfide bond containing secreted or cell-wall associated proteins in response to varying cellular conditions. This highlights the increasing need for understanding components of biological systems in terms of their context as well as simple homology relationships.
Methods and Materials
Cloning of Rv1676 and Rv1677
M. tuberculosis Rv1676 and Rv1677 genes were amplified from M. tuberculosis H37Rv genomic DNA using KOD HotStart Polymerase Kit (Novagen). For Rv1676, the 5′ primer (5′-GCCATATGGCTTGCCCTGAATGGGAAATTAGTCGATCG-3′) starts with the NdeI restriction site, which includes nucleotides 2-33 of Rv1676. The 3′ primer (5′-GCCTCGAGTCACTGAGTGCCCTTACCTC-3′) ends with the XhoI restriction site and contains the remaining 24 nucleotides of Rv1676 including the stop-codon. For Rv1677, the 5′ primer (5′-GCGGATCCGCCACCCAGGTGCCGGCGGGCCAAACC-3′) starts with the BamHI restriction site which includes nucleotides 115-141 of Rv1677. The 3′ primer (5′-GCAAGCTTTCAACGGCTGGTTAACGCCGAGACGCGCCGCGTCAG-3′) ends with the HindIII restriction site and contains the remaining 36 nucleotides of Rv1677 including the stop-codon. The resulting products were excised from a 2% agarose gel and purified using a gel extraction kit. The PCR products were ligated into a linearized blunt vector, pCR-BluntII-TOPO (Invitrogen), and then transformed into OneShot TOP10 E. coli cells (Invitrogen). Presence of the correct genes were confirmed by DNA sequencing (Davis sequencing, Davis, CA).
Construction of the Expression Vector For Rv1676
Rv1676 was double-digested from the blunt vector with NdeI and XhoI, and the plasmid pET28a(+) (Novagen) was digested with the same restriction enzymes. The cut Rv1676 gene was then ligated into cut pET28a(+) vector and transformed into E. coli BL21(DE3) cells (Novagen). Presence of the correct gene was confirmed by sequencing (Davis sequencing, Davis, CA).
Construction of the Expression Vector For Rv1677
Rv1677 was double-digested from the blunt vector with BamHI and HindIII, and the plasmid pETDuet-1 (Novagen) was digested with the same restriction enzymes. The cut Rv1677 gene was then ligated into cut pETDuet vector and transformed into E. coli BL21(DE3) cells. Presence of the correct gene was confirmed by sequencing (Davis sequencing, Davis, CA) using primers specific to the pETDuet vector.
Construction of the Co-Expression Vector For Rv1676 and Rv1677
Both full-length Rv1676 from the blunt vector and pETDuet-Rv1677 were double-digested with NdeI and XhoI. Cut Rv1676 was ligated into cut pETDuet-Rv1677 vector and transformed into E. coli BL21(DE3) cells. Presence of the correct gene was confirmed by sequencing (Davis sequencing, Davis, CA) using primers specific to the pETDuet vector.
Overexpression, Purification and Crystallization of Mtb DsbF (Rv1677)
Mtb DsbF was purified from expression of pETDuet plasmid containing Rv1677 gene using BL21(DE3) cells. Cells were grown aerobically at 37°C in LB medium containing 100 μg/mL ampicillin. Protein expression was induced by addition of 1 mM isopropyl-beta-D-thiogalactoside at an AOD600 of ∼1.0 and cells harvested 4 h after induction. Cell harvesting, disruption and protein purification utilized the same protocols as described for Rv3607c 59. The purified protein at 15 mg/ml was dialyzed into 50mM Tris/HCl pH 7.4 and 350mM NaCl for crystallization trials. The protein crystallized in 0.1 M Na-Malonate pH 7.0, 2.04 M Na-Citrate and 5% LDAO. Crystals were swiped through 1:1 crystallization condition and 50% glycerol, and diffraction data was collected at 70 K. Complete data sets were collected from single crystals.
Data collection, structure determination and refinement
Mtb DsbF native crystal diffracted to 1.6 Å with a unit cell dimensions of 100.1 × 100.1 × 30.1 Å with one monomer per asymmetric unit in space group P42212. After autoindexing, images were indexed/integrated/reduced using DENZO and SCALEPACK 60. Data collection statistics are presented in Table 1. The initial phases for the Mtb DsbF structure were determined by stochastic evolutionary programmed molecular replacement method (EPMR) 61 using a search model of Mtb DsbE (PDB code 1LU4) 18. The EPMR solution was used for refinement carried out in CNS and model building which was carried out in O. The final rounds of refinement and addition of water molecules were carried out in SHELXL (http://shelx.uni-ac.gwdg.de/SHELX/). The CxxC active-site motif was in both its oxidized and reduced form, and was modeled as such. The final structure was complete from Val47 to Thr177 (which was modeled as Ala), where residues 39-46 and 178-182 are disordered and thus are missing from the model. The final data and refinement statistics are shown in Table 1, Rwork and Rfree were 14.4 and 20.0, respectively. The stereochemistry and geometry of each Mtb DsbF monomer was validated with PROCHECK 62 and ERRAT 63, and was found to be acceptable (i.e. no residues in the disallowed region of φ,ψ, space for Mtb DsbF model).
Table 1.
X-ray diffraction data collection and atomic refinement for Rv1677 in its double conformation of its oxidized and reduced forms from M. tuberculosis
| Space group | P42212 |
| Unit cell dimensions (Å) | 100.12 × 100.12 × 30.09 |
| pH of crystallization condition | 7.0 |
| Model includes residues | 47–177 |
| Data set | |
| Wavelength (Å) | 1.00 |
| Resolution range (Å) | 90–1.6 |
| Unique reflections (total) | 23,073 (229,537) |
| Completeness (%) | 100 (100) |
| Rmergea | 6.8 (59.7) |
| I/σ | 40.68 (3.85) |
| Model refinement | |
| Resolution range (Å) | 10–1.6 |
| No. of reflections (working/free) | 16,851/1799 |
| No. of protein atoms | 1046 |
| No. of water molecules | 109 |
| Rwork/Rfreeb (%) | 14.4/20.0 (25.1/33.5) |
| RMSD | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.3 |
| Ramachandran plot (%) | |
| Most favorable region | 93.7 |
| Additional allowed region | 6.3 |
| Disallowed region | 0 |
| PDB ID | 3IOS |
Statistics for the highest-resolution shell are given in parentheses.
Rmerge = Σ|I−〈I〉|/ΣI.
Rwork = Σ|Fobs−Fcalc|/ΣFobs. Rfree was computed identically except where all reflections belong to a test set of 5% randomly selected data.
Oxidation and Reduction of Mtb DsbF
To oxidize Mtb DsbF, 50 mM of oxidized glutathioine (GSSG) was added to Mtb DsbF in 0.5 M NaCl and 0.1 M Tris.HCl, pH 7.4, and incubated for 1 hour at room temperature. The oxidized protein was then isolated by gel filtration chromatography in its original buffer. To reduce Mtb DsbF, 100 mM DTT was added to Mtb DsbF in 0.5 M NaCl and 0.1 M Tris.HCl, pH 7.4, and incubated overnight at 4°C. The reduced protein was then isolated by gel filtration chromatography in its original buffer.
Redox Properties of Mtb DsbF - Comparison to Glutathione
The in vitro redox state of Mtb DsbF was assayed as described previously 64; 65; 66. In this assay, the change in fluorescence intensity (excitation wavelength 280 nm) was measured at the wavelength of maximum emission (356 nm for Mtb DsbF). Experiments were carried out in 100 mM sodium phosphate, pH 7.0, and 1.0 mM EDTA. Oxidized and reduced 5 μM Mtb DsbF were incubated at 25°C in the presence of 0.1 mM GSSG and varying concentration of GSH (0 - 10 mM) for 12 hr before recording the fluorescence emission on a Spex Fluorolog (Jubin Yvon-Spex). The equilibrium concentrations of GSH and GSSG were calculated as described 18. The equilibrium constant Keq was estimated from nonlinear regression analysis of the data according to the Nernst equation, and from the equilibrium constant and by using the glutathione standard potential (E′0GSH/GSSG = -240 mV) 32 the standard redox potential (E′0) was calculated as described 18.
Determination of pKa of Cys81
The pH-dependent ionization of the Cys81 thiol (solvent-exposed) was followed by the specific absorbance of the thiolate anion at 240 nm as described earlier 25. As a control, the pH-dependent absorbance for the oxidized form of Mtb DsbF was recorded. To avoid precipitation artifacts and to minimize buffer absorbance, a buffer system consisting of 10 mM K2PO4, 10 mM boric acid, 10 mM sodium succinate, 1 mM EDTA and 200 mM KCl (containing 100 μM DTT for the reduced protein) was used. The pH (initial value, 8.5) was lowered to 2.2 by stepwise addition of aliquots of 0.1 M HCl, and the absorbance at 240 and 280 nm was recorded and corrected for the volume increase. Samples had an average initial protein concentration of approximately 30 μM. The pH dependence of the thiolate-specific absorbance signal was fitted according to the Henderson-Hasselbalch equation as described previously 18.
Determination of unfolding/folding equilibrium
The reversible guanidine hydrochloride (GdnHCl)-induced unfolding/folding of Mtb DsbF was performed by measuring the CD ellipticities at 222 nm 67. The spectra of the reduced form were recorded in the presence of 0.5 mM DTT. For unfolding equilibrium, Mtb DsbF (final concentration of 7 μM) was dissolved in different concentrations of GdnHCl and incubated for 3 h at 25°C. Data were analyzed according to the two-state assumption 27; 68. The standard changes of folding free energy and the difference in stability between the oxidized and reduced forms of DsbF protein were calculated as described previously 18.
Oxidase activity of Mtb DsbF - refolding of hirudin
Commercial Hirudo medicinalis hirudin (Sigma) was purified to remove contaminants by reverse-phase HPLC on a polymeric column (PLRP/S; 300A, 5 μm bead; 2 mm × 15 cm) at a flow rate of 100 μL/min. Solvents A and B used for reverse-phase HPLC were 0.1% trifluoroacetic acid in water and acetonitrile, respectively. HPLC purified hirudin was reduced and unfolded as described 69, and then reduced, unfolded hirudin (5 pmol) was refolded with stoichiometric amounts of oxidized Mtb DsbF (5 pmol) in 100 mM ammonium bicarbonate, pH 8.0 at 25 °C. Each 10 μL reaction was quenched at different time points by incubating initially with 50 μL 6M GdnHCl for 10 minutes at 50 °C followed by 0.5 μL 100 mM iodoacetamide for 15 mins at 25 °C. The samples were then desalted with ZipTipC4 (Millipore) before analysis with MALDI-TOF (Voyager). Experiments were repeated no protein added and with similar quantities (5 pmol) of Mtb DsbE, E. coli DsbA and E. coli thioredoxin as positive and negative controls, respectively.
Expression and purification of Rv1676
Rv1676 was purified from expression of pET28a(+) plasmid containing Rv1676 gene using BL21(DE3) cells. Cells were grown aerobically at both 18°C and 37°C in LB medium containing 100 μg/mL kanamycin. Protein expression was induced by addition of 1 mM isopropyl-beta-D-thiogalactoside at an AOD600 of ∼1.0 and cells harvested between 2h, 4h and overnight after induction. Cell harvesting, disruption, solubility tests and protein purification in 6M GdnHCl were carried out as described previously 70. Attempts were made to refold the Rv1676 by step-wise dialysis from 6M GdnHCl to native buffer (50 mM Tris pH 7.4, 150 mM NaCl).
Co-expression of Rv1676 and Rv1677 (DsbF)
The pETDuet co-expression plasmid containing Rv1676 and Rv1677 was transformed into BL21(DE3) cells. Cells were grown aerobically at 37°C in LB medium to an AOD600 of ∼0.8 then protein expression was induced by addition of 1 mM IPTG and the cells were harvested 4 h after induction. Cells were harvested, lyzed and the supernatant extracted as for Rv1677. The supernatant was loaded onto a Ni2+ charged HiTrap chelating column. The column was washed with 20 mM Hepes, pH 7.8 and 150 mM NaCl, and eluted with a linear gradient of imidazole from 0 to 500 mM in 20 mM Hepes pH 7.8 and 150 mM NaCl. The fractions were analyzed by SDS-PAGE gel. The fraction that corresponds to both Rv1676 and Rv1677 proteins were analyzed by mass spectrometry.
Analysis of tryptic peptide sequence tags by micro-liquid chromatography tandem mass spectrometry (μLC-MSMS)
Samples were analyzed by μLC-MSMS with data-dependent acquisition (LCQ-DECA, ThermoFinnigan, San Jose, California) after dissolution in 5 μL 70 % acetic acid (v/v). A reverse-phase column (200 μm × 10 cm; PLRP/S 5 μm, 300 Å; Michrom Biosciences, San Jose) was equilibrated for 10 minutes at 1.5 μL/min with 95% A, 5% B (A, 0.1% formic acid in water; B, 0.1% formic acid in acetonitrile) prior to sample injection. A linear gradient was initiated 10 min after sample injection ramping to 60% A, 40% B after 50 minutes and 20% A, 80% B after 65 minutes. Column eluent was directed to a coated glass electrospray emitter (TaperTip, TT150-50-50-CE-5, New Objective) at 3.3 kV for ionization without nebulizer gas. The mass spectrometer was operated in ‘triple-play’ mode with a survey scan (400-1500 m/z), data-dependent zoom scan, and MSMS with exclusion of singly-charged ions. Individual sequencing experiments were matched to a custom M. tuberculosis sequence database downloaded from The Sanger Center using Sequest software (ThermoFinnigan, San Jose). The search was run under the ‘no enzyme’ mode to identify non-tryptic peptides. The results of Sequest searches were carefully scrutinized. MSMS spectra of doubly charged ions with cross correlation scores (Xcorr) greater than 2.8 and triply charged ions with scores over 3.2 were examined manually. Some noisy spectra were discarded despite high Xcorr scores. Non-tryptic peptide returns were retained only if the data had particularly high signal to noise.
Genome-wide analysis of Mtb gene coexpression
This methodology has been previously described45, so in brief, four published gene expression datasets from M. tuberculosis strain H37Rv were collected 41; 42; 43; 44 from the Gene Expression Omnibus (GEO) (PMID: 17099226). Gene expression data from the four studies were represented as a matrix where the rows were genes and the columns were experiments. To construct an expression vector for a gene, the data from each of the four studies were concatenated. Correlation coefficients of gene expression vectors were calculated for all possible pairs of genes. To obtain a correlation coefficient for genes x and y over N experiments, the Pearson correlation coefficient, rxy, was calculated as:
where xi and yi are the log expression values reported in the GEO data file, μx and μy are the means of the values in the combined vector, sx and sy are the standard deviations. For each pair of genes analyzed, combined expression vectors were truncated to include only those experiments for which measurements were obtained for both genes. Thus N was adjusted for each pair of genes assessed. In all, 553 experiments were included for the analysis of 3,925 Mtb genes to infer some 7,700,000 pairwise coexpression relationships between pairs of genes.
Acknowledgments
This work has been supported by a grant from the National Institutes of Health, DOE-BER and HHMI (for D.E.) and National Institutes of Health (P01AIO68135, subcontract for C.W.G.) The authors thank Dr John T. Belisle, Colorado State University, NIH, NIAID Contract NO1 A1-75320 for generous supply of M. tuberculosis H37Rv genomic DNA. We also thank Drs. Michael Sawaya and Duilio Cascio for invaluable help with data collection and general crystallography. Finally, special thanks to Morgan Beeby for helpful comments.
Abbreviations used
- Mtb
Mycobacterium tuberculosis
- E. coli
Escherichia coli
- Rv number
Sanger center notation for each gene in Mtb
- TB
tuberculosis
- Dsb
disulfide bond forming proteins
- rmsd
root mean square deviation function
- TFA
trifluoroacetic acid
- IPTG
isopropyl-beta-D-thiogalactoside
Footnotes
Protein Data Bank accession number
The atomic coordinates and structure factors for the crystal structure of Mtb DsbF modeled with its active-site cysteines in both their reduced and oxidized states have been deposited with the Protein Data Bank (RCSB, http://www.rcsb.org/pdb) as entry 3IOS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martin JL. Thioredoxin--a fold for all reasons. Structure. 1995;3:245–50. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 2.Schroder E, Ponting CP. Evidence that peroxiredoxins are novel members of the thioredoxin fold superfamily. Protein Sci. 1998;7:2465–8. doi: 10.1002/pro.5560071125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–71. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang HZ, Donnenberg MS. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:787–97. doi: 10.1046/j.1365-2958.1996.431403.x. [DOI] [PubMed] [Google Scholar]
- 5.Zav'yalov VP, Chernovskaya TV, Chapman DA, Karlyshev AV, MacIntyre S, Zavialov AV, Vasiliev AM, Denesyuk AI, Zav'yalova GA, Dudich IV, Korpela T, Abramov VM. Influence of the conserved disulphide bond, exposed to the putative binding pocket, on the structure and function of the immunoglobulin-like molecular chaperone Caf1M of Yersinia pestis. Biochem J. 1997;324(Pt 2):571–8. doi: 10.1042/bj3240571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J, Kroll JS. DsbA: a protein-folding catalyst contributing to bacterial virulence. Microbes Infect. 1999;1:1221–8. doi: 10.1016/s1286-4579(99)00239-7. [DOI] [PubMed] [Google Scholar]
- 7.Hu SH, Peek JA, Rattigan E, Taylor RK, Martin JL. Structure of TcpG, the DsbA protein folding catalyst from Vibrio cholerae. J Mol Biol. 1997;268:137–46. doi: 10.1006/jmbi.1997.0940. [DOI] [PubMed] [Google Scholar]
- 8.Manning PA. The tcp gene cluster of Vibrio cholerae. Gene. 1997;192:63–70. doi: 10.1016/s0378-1119(97)00036-x. [DOI] [PubMed] [Google Scholar]
- 9.Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–9. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 10.Joly JC, Swartz JR. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry. 1997;36:10067–72. doi: 10.1021/bi9707739. [DOI] [PubMed] [Google Scholar]
- 11.Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–27. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 12.Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci U S A. 1993;90:7084–8. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grovc J, Busby S, Cole J. The role of the genes nrf EFG and ccmFH in cytochrome c biosynthesis in Escherichia coli. Mol Gen Genet. 1996;252:332–41. doi: 10.1007/BF02173779. [DOI] [PubMed] [Google Scholar]
- 14.Reid E, Cole J, Eaves DJ. The Escherichia coli CcmG protein fulfils a specific role in cytochrome c assembly. Biochem J. 2001;355:51–8. doi: 10.1042/0264-6021:3550051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabianek RA, Hofer T, Thony-Meyer L. Characterization of the Escherichia coli CcmH protein reveals new insights into the redox pathway required for cytochrome c maturation. Arch Microbiol. 1999;171:92–100. doi: 10.1007/s002030050683. [DOI] [PubMed] [Google Scholar]
- 16.Stewart EJ, Katzen F, Beckwith J. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. Embo J. 1999;18:5963–71. doi: 10.1093/emboj/18.21.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci. 2006;31:455–64. doi: 10.1016/j.tibs.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Goulding CW, Apostol MI, Gleiter S, Parseghian A, Bardwell J, Gennaro M, Eisenberg D. Gram-positive DsbE proteins function differently from Gram-negative DsbE homologs. A structure to function analysis of DsbE from Mycobacterium tuberculosis. J Biol Chem. 2004;279:3516–24. doi: 10.1074/jbc.M311833200. [DOI] [PubMed] [Google Scholar]
- 19.Goldstone D, Baker EN, Metcalf P. Crystallization and preliminary diffraction studies of the C-terminal domain of the DipZ homologue from Mycobacterium tuberculosis. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2005;61:243–5. doi: 10.1107/S1744309105001909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci U S A. 2008;105:3963–7. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raviglione MC, Snider DE, Jr, Kochi A. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. Jama. 1995;273:220–6. [PubMed] [Google Scholar]
- 22.Pablos-Mendez A, Raviglione MC, Laszlo A, Binkin N, Rieder HL, Bustreo F, Cohn DL, Lambregts-van Weezenbeek CS, Kim SJ, Chaulet P, Nunn P. Global surveillance for antituberculosis-drug resistance, 1994-1997. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N Engl J Med. 1998;338:1641–9. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 23.Goulding CW, Perry LJ, Anderson D, Sawaya MR, Cascio D, Apostol MI, Chan S, Parseghian A, Wang SS, Wu Y, Cassano V, Gill HS, Eisenberg D. Structural genomics of Mycobacterium tuberculosis: a preliminary report of progress at UCLA. Biophys Chem. 2003;105:361–70. doi: 10.1016/s0301-4622(03)00101-7. [DOI] [PubMed] [Google Scholar]
- 24.Goulding CW, Apostol M, Anderson DH, Gill HS, Smith CV, Kuo MR, Yang JK, Waldo GS, Suh SW, Chauhan R, Kale A, Bachhawat N, Mande SC, Johnston JM, Lott JS, Baker EN, Arcus VL, Leys D, McLean KJ, Munro AW, Berendzen J, Sharma V, Park MS, Eisenberg D, Sacchettini J, Alber T, Rupp B, Jacobs W, Jr, Terwilliger TC. The TB structural genomics consortium: providing a structural foundation for drug discovery. Curr Drug Targets Infect Disord. 2002;2:121–41. doi: 10.2174/1568005023342551. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, Hu H, Xu G. Biochemical characterization of the thioredoxin domain of Escherichia coli DsbE protein reveals a weak reductant. Biochem Biophys Res Commun. 2001;283:849–53. doi: 10.1006/bbrc.2001.4876. [DOI] [PubMed] [Google Scholar]
- 26.Martin JL, Bardwell JC, Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993;365:464–8. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- 27.Wunderlich M, Glockshuber R. Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci. 1993;2:717–26. doi: 10.1002/pro.5560020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zapun A, Bardwell JC, Creighton TE. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry. 1993;32:5083–92. doi: 10.1021/bi00070a016. [DOI] [PubMed] [Google Scholar]
- 29.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 30.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–71. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 31.Musil D, Zucic D, Turk D, Engh RA, Mayr I, Huber R, Popovic T, Turk V, Towatari T, Katunuma N, et al. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. Embo J. 1991;10:2321–30. doi: 10.1002/j.1460-2075.1991.tb07771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aslund F, Berndt KD, Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–6. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 33.Van Dorsselaer A, Lepage P, Bitsch F, Whitechurch O, Riehl-Bellon N, Fraisse D, Green B, Roitsch C. Mass spectrometry analyses of recombinant hirudins (7 kDa) Biochemistry. 1989;28:2949–56. doi: 10.1021/bi00433a031. [DOI] [PubMed] [Google Scholar]
- 34.Chivers PT, Prehoda KE, Volkman BF, Kim BM, Markley JL, Raines RT. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry. 1997;36:14985–91. doi: 10.1021/bi970071j. [DOI] [PubMed] [Google Scholar]
- 35.Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JC. Why is DsbA such an oxidizing disulfide catalyst? Cell. 1995;83:947–55. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 36.Strong M, Mallick P, Pellegrini M, Thompson MJ, Eisenberg D. Inference of protein function and protein linkages in Mycobacterium tuberculosis based on prokaryotic genome organization: a combined computational approach. Genome Biol. 2003;4:R59. doi: 10.1186/gb-2003-4-9-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–6. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 38.Bowers PM, Pellegrini M, Thompson MJ, Fierro J, Yeates TO, Eisenberg D. Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol. 2004;5:R35. doi: 10.1186/gb-2004-5-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu S, Chen J, Dobos KM, Bradbury EM, Belisle JT, Chen X. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol Cell Proteomics. 2003;2:1284–96. doi: 10.1074/mcp.M300060-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Hahn MY, Raman S, Anaya M, Husson RN. The Mycobacterium tuberculosis extracytoplasmic-function sigma factor SigL regulates polyketide synthases and secreted or membrane proteins and is required for virulence. J Bacteriol. 2005;187:7062–71. doi: 10.1128/JB.187.20.7062-7071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., 3rd The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–84. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 42.Boshoff HI, Reed MB, Barry CE, 3rd, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–93. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 43.Gao Q, Kripke K, Arinc Z, Voskuil M, Small P. Comparative expression studies of a complex phenotype: cord formation in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2004;84:188–96. doi: 10.1016/j.tube.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 44.Gao Q, Kripke KE, Saldanha AJ, Yan W, Holmes S, Small PM. Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology. 2005;151:5–14. doi: 10.1099/mic.0.27539-0. [DOI] [PubMed] [Google Scholar]
- 45.Riley R, Pellegrini M, Eisenberg D. Identifying cognate binding pairs among a large set of paralogs: the case of PE/PPE proteins of Mycobacterium tuberculosis. PLoS Comput Biol. 2008;4:e1000174. doi: 10.1371/journal.pcbi.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guddat LW, Bardwell JC, Martin JL. Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization. Structure. 1998;6:757–67. doi: 10.1016/s0969-2126(98)00077-x. [DOI] [PubMed] [Google Scholar]
- 47.Edeling MA, Guddat LW, Fabianek RA, Thony-Meyer L, Martin JL. Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment. Structure. 2002;10:973–9. doi: 10.1016/s0969-2126(02)00794-3. [DOI] [PubMed] [Google Scholar]
- 48.Heras B, Edeling MA, Schirra HJ, Raina S, Martin JL. Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide. Proc Natl Acad Sci U S A. 2004;101:8876–81. doi: 10.1073/pnas.0402769101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 50.Barranco-Medina S, Krell T, Bernier-Villamor L, Sevilla F, Lazaro JJ, Dietz KJ. Hexameric oligomerization of mitochondrial peroxiredoxin PrxIIF and formation of an ultrahigh affinity complex with its electron donor thioredoxin Trx-o. J Exp Bot. 2008;59:3259–69. doi: 10.1093/jxb/ern177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurokawa Y, Yanagi H, Yura T. Overexpression of protein disulfide isomerase DsbC stabilizes multiple-disulfide-bonded recombinant protein produced and transported to the periplasm in Escherichia coli. Appl Environ Microbiol. 2000;66:3960–5. doi: 10.1128/aem.66.9.3960-3965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyazaki-Imamura C, Oohira K, Kitagawa R, Nakano H, Yamane T, Takahashi H. Improvement of H2O2 stability of manganese peroxidase by combinatorial mutagenesis and high-throughput screening using in vitro expression with protein disulfide isomerase. Protein Eng. 2003;16:423–8. doi: 10.1093/protein/gzg054. [DOI] [PubMed] [Google Scholar]
- 53.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–7. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 54.Guimaraes BG, Souchon H, Honore N, Saint-Joanis B, Brosch R, Shepard W, Cole ST, Alzari PM. Structure and mechanism of the alkyl hydroperoxidase AhpC, a key element of the Mycobacterium tuberculosis defense system against oxidative stress. J Biol Chem. 2005;280:25735–42. doi: 10.1074/jbc.M503076200. [DOI] [PubMed] [Google Scholar]
- 55.Li S, Peterson NA, Kim MY, Kim CY, Hung LW, Yu M, Lekin T, Segelke BW, Lott JS, Baker EN. Crystal Structure of AhpE from Mycobacterium tuberculosis, a 1-Cys peroxiredoxin. J Mol Biol. 2005;346:1035–46. doi: 10.1016/j.jmb.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 56.Rodrigue S, Provvedi R, Jacques PE, Gaudreau L, Manganelli R. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol Rev. 2006;30:926–41. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 57.Dainese E, Rodrigue S, Delogu G, Provvedi R, Laflamme L, Brzezinski R, Fadda G, Smith I, Gaudreau L, Palu G, Manganelli R. Posttranslational regulation of Mycobacterium tuberculosis extracytoplasmic-function sigma factor sigma L and roles in virulence and in global regulation of gene expression. Infect Immun. 2006;74:2457–61. doi: 10.1128/IAI.74.4.2457-2461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Traverso JA, Vignols F, Cazalis R, Pulido A, Sahrawy M, Cejudo FJ, Meyer Y, Chueca A. PsTRXh1 and PsTRXh2 are both pea h-type thioredoxins with antagonistic behavior in redox imbalances. Plant Physiol. 2007;143:300–11. doi: 10.1104/pp.106.089524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goulding CW, Apostol MI, Sawaya MR, Phillips M, Parseghian A, Eisenberg D. Regulation by oligomerization in a mycobacterial folate biosynthetic enzyme. J Mol Biol. 2005;349:61–72. doi: 10.1016/j.jmb.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 60.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 61.Kissinger CR, Gehlhaar DK, Smith BA, Bouzida D. Molecular replacement by evolutionary search. Acta Crystallogr D Biol Crystallogr. 2001;57:1474–9. doi: 10.1107/s0907444901012458. [DOI] [PubMed] [Google Scholar]
- 62.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–67. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 63.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–9. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loferer H, Wunderlich M, Hennecke H, Glockshuber R. A bacterial thioredoxin-like protein that is exposed to the periplasm has redox properties comparable with those of cytoplasmic thioredoxins. J Biol Chem. 1995;270:26178–83. doi: 10.1074/jbc.270.44.26178. [DOI] [PubMed] [Google Scholar]
- 65.Rossmann R, Stern D, Loferer H, Jacobi A, Glockshuber R, Hennecke H. Replacement of Pro109 by His in TlpA, a thioredoxin-like protein from Bradyrhizobium japonicum, alters its redox properties but not its in vivo functions. FEBS Lett. 1997;406:249–54. doi: 10.1016/s0014-5793(97)00270-6. [DOI] [PubMed] [Google Scholar]
- 66.Bessette PH, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci U S A. 1999;96:13703–8. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hennecke J, Spleiss C, Glockshuber R. Influence of acidic residues and the kink in the active-site helix on the properties of the disulfide oxidoreductase DsbA. J Biol Chem. 1997;272:189–95. doi: 10.1074/jbc.272.1.189. [DOI] [PubMed] [Google Scholar]
- 68.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 69.Hennecke J, Sebbel P, Glockshuber R. Random circular permutation of DsbA reveals segments that are essential for protein folding and stability. J Mol Biol. 1999;286:1197–215. doi: 10.1006/jmbi.1998.2531. [DOI] [PubMed] [Google Scholar]
- 70.Goulding CW, Perry LJ. Protein production in Escherichia coli for structural studies by X-ray crystallography. J Struct Biol. 2003;142:133–43. doi: 10.1016/s1047-8477(03)00044-3. [DOI] [PubMed] [Google Scholar]