Abstract
Background & Aims
Patients with baseline hepatitis C virus-RNA levels (bHCV-RNA) >6 log IU/ml or cirrhosis have a reduced probability of a sustained-virological response (SVR). We examined the relationship between bHCV-RNA, cirrhosis and SVR using a mathematical model that includes the critical-drug efficacy (εc; the efficacy required for a drug to clear HCV), the infection-rate constant (β) and the percentage of HCV-infected hepatocytes (π).
Methods
The relationship between baseline factors and SVR was evaluated in 1,000 in silico HCV-infected patients, generated by randomly assignment of realistic host and viral kinetic parameters. Model predictions were compared with clinical data from 170 non-cirrhotic and 75 cirrhotic patients.
Results
The ranges chosen for β and the viral production rate (p) resulted in bHCV-RNA levels that were in agreement with the distribution observed in US patients. Using these β and p values, higher bHCV-RNA levels led to higher εc, resulting in lower SVR rates. Alternatively, higher β values resulted in lower bHCV-RNA levels but higher π and εc, predicting lower rates of SVR. Cirrhotic patients had lower bHCV-RNA levels than non-cirrhotic patients (p=0.013) and more had bHCV-RNA levels <6 log IU/ml (p<0.001). Even cirrhotic patients with lower bHCV-RNA levels had lower SVR rates. An increase in β could explain the results observed in cirrhotic patients.
Conclusions
Our model predicts that higher bHCV-RNA levels lead to higher εc, reducing the chance of achieving SVR; cirrhotic patients have lower SVR rates because of large π values, caused by increased rates of hepatocyte infection.
Keywords: mathematical modeling, hepatitis C virus, viral replication kinetics, critical-drug efficacy, fraction of HCV-infected hepatocytes
Introduction
Approximately 3% of persons worldwide are infected with hepatitis C virus (HCV)1. Chronic HCV is one of the most common life-threatening viral infections in the United States. The disease has a lengthy natural history, progressing over decades. Current therapy consisting of pegylated interferon (PEG-IFN) and ribavirin produces sustained virologic response (SVR) rates of 50%, with no effective alternative treatment for non-responders2, 3.
Host factors as well as viral factors are associated with lower response rates to antiviral therapy (see review in4). Among genotype-1 HCV treated patients, baseline HCV-RNA (bHCV-RNA) level is a strong independent predictor of SVR. In particular, bHCV-RNA ≤ 2×106 copies/ml2, 3, 5–8 and bHCV-RNA<4×105 IU/ml9–12 have been associated with improved rates of SVR. The biological basis of this seemingly intuitive association between lower baseline virus level and SVR is unknown.
Another puzzling observation is that SVR declines with increasing duration of infection, from almost 100% within 6 months of transmission13, to approximately 50% in patients with chronic disease2, 3, and to less than 23% in patients with advanced liver disease14–17. In addition, Rodriguez-Inigo et al.18 showed that a lower percentage of HCV-infected hepatocytes (π) is also a predictor of SVR.
Viral kinetic modeling has played an important role in the analysis of HCV-RNA decay after the initiation of antiviral therapy (see review in19), and have suggested mechanisms of action for both IFN20 and ribavirin21. Recently, we introduced a model of HCV dynamics that accounts for hepatocyte proliferation22, 23. The model predicts that there is a critical-drug efficacy (εc), which therapy must reach to achieve HCV clearance and SVR. The model can also explain a wide variety of HCV-RNA kinetic profiles in treated patients (i.e., flat-partial responses, biphasic20, 24 or triphasic-viral decay25) as well as post therapy viral rebound26.
Here, using this new model22, we analyzed the relationships among εc, bHCV-RNA and π. The quantitative features of these relationships can be determined by analyzing the model behavior with different sets of parameters. Using ranges for the viral-kinetic parameters obtained from the literature, we determined these relationships for a set of 1,000 simulated HCV-infected patients generated by randomly assigning realistic parameters. We show that the major determinants of bHCV-RNA are the viral-production rate constant, p, and the infection-rate constant, β. We estimate that p=0.1–6 virions/cell/day and β=1×10−9–1×10−8 ml/day/virion, generate the distribution of bHCV-RNA values observed in chronic-HCV patients27. In addition, we show that lower bHCV-RNA values, and lower π, are associated with a lower εc and hence an enhanced probability of SVR. Finally, we show that the predictions of the model are consistent with data from cirrhotic and non-cirrhotic patients.
Methods
Mathematical model
The original viral-kinetic model of Neumann et al.20 and more recent variations25, 28 assume a source of hepatocytes but ignore explicit consideration of the proliferation of infected and uninfected cells. We recently developed a model of HCV infection22 that includes density-dependent proliferation of hepatocytes. The equations describing the dynamics of HCV infection and treatment in this model involve uninfected hepatocytes (T), productively infected hepatocytes (I) and virus (V), and are given below:
| (1) |
Uninfected and infected hepatocytes can proliferate with maximum proliferation rates rT and rI, respectively, under the control of a homeostatic process in which proliferation shuts down as the total number of hepatocytes approaches a maximum number Tmax, irrespective of whether they are infected or not29. Due to the burdens of supporting HCV replication, we assume infected cells may proliferate more slowly than uninfected cells, i.e., rI ≤ rT. The model assumes that uninfected hepatocytes are produced by differentiation of precursors at a constant rate s, die at rate dT per cell, and are infected with rate constant β. Infected hepatocytes are lost at a rate δ per cell. Viral particles (virions) are produced at rate p per infected hepatocyte and are cleared at rate c per virion. In an uninfected liver when Tmax is reached, liver size should no longer increase. To ensure this we require s≤dTTmax.
Chronic HCV infection is treated using pegylated IFN-α in combination with ribavirin. IFN-α acts primarily by blocking the production of new virus, although we also allow for a treatment effect in blocking de-novo infection. The efficacy of treatment in blocking virion production and reducing new infections is described by two parameters, εp and η, respectively. For example, a treatment efficacy in blocking virion production of 95% corresponds to εp=0.95. The behavior of the model under drug therapy depends on the overall drug effectiveness, ε, where 1−ε= (1−η) (−1εp). In this paper we shall simply call ε the drug effectiveness. Equations describing the model’s uninfected and infected steady states, a discussion of parameter ranges, and model simulations are included in the online supplemental material.
Patient data
Patients were included in the analysis only if they had a viral-load (genotype-1) measurement and liver biopsy in 2001 or later. All HCV-RNA measurements performed during this time period were made by a branched-chain-DNA assay and reported in IU/ml. The results of the first biopsy were used in patients who had repeat biopsies. All liver biopsies were evaluated by one clinician to reduce inter-observer variation. Fibrosis was staged on a one to four scale (termed Fib1 to Fib4), with a score of four representing cirrhosis. Duration of infection was calculated from date of first possible HCV exposure based on known risk factors such as injection drug use and transfusion prior to 1989, to date of biopsy. If major risk factors were not known or present, duration of infection was not determined. The viral load characterizing each patient was that taken at the time of liver biopsy (Table 2). If no viral load was determined at the time of biopsy, then the viral load immediately preceding the liver biopsy was used. The study was approved by the Institutional Review Board at the University of Illinois at Chicago.
Table 2.
Baseline characteristics of the patients.
| All (n = 245) | Non-cirrhotic (n=170) | Cirrhotic (n=75) | P value t-test * (non-parametric test) | |
|---|---|---|---|---|
| Male: Female, n | 149:96 | 101:69 | 48:27 | 0.49 |
| Mean age [years] | 50.9 ± 8.9 | 49.6 ± 8.6 | 53.8 ± 8.7 | 0.0006 (0.001) |
| Race, n (%) | ||||
| Caucasian | 63 (26) | 43 (25) | 20 (27) | 0.08 |
| African American | 124 (50) | 93 (55) | 31 (41) | |
| Hispanic | 58 (24) | 34 (20) | 24 (32) | |
| Mean weight [kg], | 82.9 ± 18.2 | 81.8 ± 8.6 | 85.5 ± 18.7 | 0.15 (0.14) |
| Mean body mass index [kg/m2], | 28.9 ± 6.6 | 28.4 ± 6.3 | 30.1 ± 7.1 | 0.08 (0.09) |
| Mean duration of infection [years] | 27.3 ± 8.8 | 26.4 ± 9.2@ | 29.4 ± 7.5@ | 0.04 (0.03) |
| Mean HCV RNA [log10 IU/ml], HCV RNA, | 5.9 ± 0.6 | 5.9 ± 0.7 | 5.7 ± 0.6 | 0.013 (0.017) |
| > 6 log10 IU/ml | 99 (40) | 81 (48) | 18 (24) | 0.0005 |
| < 6 log10 IU/ml | 146 (60) | 89 (52) | 57 (76) | |
For the difference between non-cirrhotic and cirrhotic patients.
We could not estimate the duration of infection in 20 cirrhotic patients and 49 non-cirrhotic patients as explained in Methods.
Statistical analysis
We used the parametric t-test and non-parametric Mann-Whitney test to compare baseline characteristics of non-cirrhotic (n=170) vs. cirrhotic (n=75) patients (Table 2). To compare categorical variables, we used the chi-square test for independence (S-plus, V.7.02, Seattle, WA).
Results
Existing viral-kinetic models have allowed for estimation of the effectiveness of anti-HCV drugs, ε, the clearance rate of free virions in serum, c, and the loss rate of virally-infected cells, δ. Yet, the viral-production rate, p, the infection-rate constant, β, and the uninfected and infected hepatocyte-proliferation-rate constants, rT and rI, respectively, are not known. We used the model in Eq. (1) and clinical data to define bounds for these unknown parameter values and to shed light on the observed relationship between bHCV-RNA and SVR, as well as the relationship between the baseline percentage of HCV-infected hepatocytes, π, and SVR rates.
The distribution of baseline viral-load levels and the fraction of HCV-infected hepatocytes
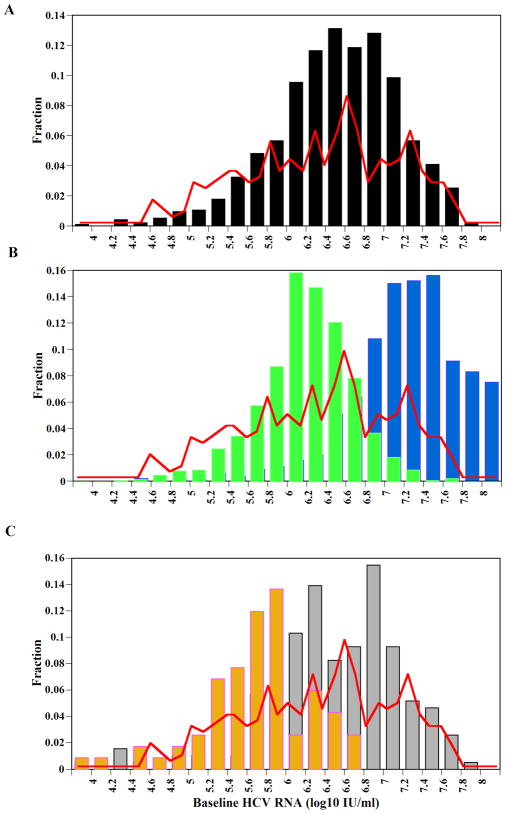
The distribution of bHCV-RNA values in the U.S. is approximately log normal (Fig. 1, red lines) with peak values between 6.5–6.8 log10 (IU/ml)27. Estimates of the percentage of hepatocytes positive for genomic HCV-RNA range from 4.8% to 87.6%, with mean values (± one standard deviation) of 40%±28% (n=10)30 and 38%±20% (n=19)31. Based on our model simulations, which generated patients with different bHCV-RNA, we found that the production of virions, p, and the infection rate, β, are the processes that most affect the bHCV-RNA distribution and the percentage of infected cells, π. We found that when p is in the range of 0.1–6 virions/day, and β is in the range of 1×10−9–1×10−8 ml/day/virions, the observed distribution of bHCV-RNA is roughly predicted (Fig. 1A, black bars), and the mean π is 58% (Table 3). Interestingly, we found that with this range of β values many patients were predicted to clear the virus when the virion clearance rate, c, or the infected cell death rate, δ, were at the high end of their ranges given in Table 1 (not shown). If we allow a larger range of values for p (e.g., 0.1–45 virions/day), with the same range of β (i.e., 1×10−9–1×10−8 ml/day/virions) then the distribution shifts to higher bHCV-RNA values (Fig. 1B, blue bars) since each infected cell produces, on average, more virions per day. In turn, higher bHCV-RNA values lead to a higher mean π=70% (Table 3). On the other hand, with a range of p=0.1–6 virions/day, larger values of the infection rate β= 1×10−7–1×10−6 ml/day/virions increase the mean π to high values, 90% (Table 3) but the distribution of bHCV-RNA, counter-intuitively, shifts to lower values than those observed (Fig. 1B, green bars). The reason for this observation is that with a high range of β values, we were able to choose the clearance rate of free virions, c, and the loss rate of infected cells, δ, over their entire feasible ranges (Table 1) without the virus being cleared. Thus, the mean values of c and δ were significantly higher (Table 3), giving rise to the lower bHCV-RNA distribution. Interestingly, the maximum proliferation rates of uninfected hepatocytes, rT, and infected hepatocytes, rI, do not play a significant role in determining the distribution of bHCV-RNA and π (not shown). Moreover, assuming that rI is not necessarily lower than rT (i.e., rT and rI chosen randomly and independently between 1 to 3 day−1) does not change our results significantly (not shown).
Figure 1. The distribution of baseline HCV-RNA levels.
(A) The predicted baseline viral load distribution from one thousand simulated patients with a range of p=0.1–6 virions/day and β=1×10−9–1×10−8 ml/day/virions (black bars) is close to the observed baseline HCV-RNA distribution in the U. S. (red line), digitized from Nainan et al.27. (B) With a larger range of p=0.1–45 virions/day and the same β=1×10−9–1×10−8 ml/day/virions the predicted HCV-RNA distribution (blue bars) is skewed right from the actual distribution (red line), and with a range of p=0.1–6 virions/day and a higher infectivity rate constant, β=1×10−7–1×10−6 ml/day/virions (green bars) it is skewed to the left due to increased rates of infected cell death, δ, and HCV clearance, c (Table 3). All other model parameters (Table 1) did not significantly change the distribution of HCV-RNA values in (A) and (B) (not shown). (C) Distribution of baseline HCV-RNA in cirrhotic (brown bars) and non-cirrhotic (gray bars) patients, from the University of Illinois at Chicago.
Table 3.
The impact of parameters p and β on total hepatocyte number and the fraction of HCV-infected hepatocytes*.
| Viral- production rate p[virions day−1] |
Infection rate constant β [ml virion−1 day−1] |
Average number of infected hepatocytes (Ī) at baseline in 1,000 in-silico patients [106 cells/ml] |
Average number of hepatocytes (Ī + T̄) at baseline in 1,000 in-silico patients [106 cells/ml] |
% infected hepatocytes at baseline Ī / Ī (+ T̄) Mean(95% CI&) |
Average infected cell death/loss rate ± SD δ (range: min – max) [1/day] |
Average virion clearance rate ± SD c (range: min – max) [1/day] |
|
|---|---|---|---|---|---|---|---|
| 0.1 – 6 | 10−9 to 10−8 | 5.25 | 8.98 | 58 (56 – 60) | 0.03 ± 0.04 (0.001 – 0.36) | 6.5 ± 5.5 (0.8 – 22.2) | Fig. 1A black bars |
| 0.1 – 6 | 10−8 to1−7 | 5.88 | 8.14 | 73 (71 – 74) | 0.09 ± 0.08 d (0.002 – 0.48) | 9.7 ± 6.0 b (0.8 – 22) | |
| 0.1 – 6 | 10−7 to 1−6 | 5.87 | 6.50 | 90 (89 – 92) | 0.13 ± 0.1 c (0.003 – 0.47) | 11.2 ± 6.1 a (0.8 – 22.2) | Fig. 1B green bars |
| 0.1 – 6 | 10−6 to 1−5 | 5.57 | 5.66 | 98 (97 – 99) | 0.14 ± 0.1 c (0.002 – 0.48) | 11.4 ± 6.2 a (0.8 – 22.2) | |
| 0.1 – 45 | 10−9 to 1−8 | 5.77 | 8.27 | 70 (68 – 72) | 0.08 ± 0.08 d (0.002 – 0.43) | 9.3 ± 6.1 b (0.8 – 22.2) | Fig. 1B blue bars |
All other model parameters were randomly chosen within the ranges given in Table 1. Since we excluded parameter sets that lead to negative baseline values of virus (V), as explained in online supplemental material, the resulting parameter values of the virus clearance rate, c, and infected cell death rate, δ, were significantly different among some groups with different p and β ranges.
SD, one standard deviation.
CI, confidence interval.
, In post-hoc analyses, groups marked with the same superscript letter were not significantly different from each other; but were significantly different (p<0.01 allowing for Bonferroni correction) from all the other groups in the same column.
Table 1.
Model parameter values.
| Parameter | Parameter definition | [Units] | Parameter ranges from literature a |
|---|---|---|---|
| Tmax | total (normalized) hepatocyte number | [107 cells ml−1 ] | 0.4 – 1.3 |
| s | maximum de novo hepatocyte influx rate | [cells ml−1 day−1 ] | 1 < s ≤ dTmax |
| d | uninfected hepatocyte death rate | [day−1] | 0.001 – 0.014 ( t1/2 =50 – 700 days)b |
| δ | HCV-infected hepatocyte loss rate | [day−1] | d ≤ δ ≤ 0.5 ( t1/2 >1.4 days) |
| c | HCV RNA clearance rate | [day−1] | 0.8 – 22 ( t1/2 =0.7 – 20.8 hr)b |
| p | HCV production rate per cell | [virions day−1] | 0.1 – 45 |
| β | infection rate constant | [virions−1ml day−1] | 1×10−9 – 1×10−5 |
| rT | maximum uninfected hepatocyte proliferation rate | [day−1] | 1.0 – 3.0 (t2=0.2 – 0.7 days)b |
| rI | maximum infected hepatocyte proliferation rate | [day−1] | 10% to 100% of rT |
As explained in the online supplemental material.
t1/2, half life; t2, doubling time.
The relationship between critical drug efficacy, baseline HCV RNA level and infection-rate constant, β
When it was thought that HIV might be eradicated by antiretroviral therapy, the notion of a critical-drug efficacy, εc, was introduced32, 33. The critical-drug efficacy was defined such that if the efficacy of a drug, ε, was larger than this value, ε > εc, viral levels would continually decline on therapy ultimately leading to eradication, whereas if ε < εc viral loads would initially decline but ultimately they would stabilize at a non-zero steady state despite continued therapy. We recently, showed that the concept of εc applies to HCV kinetic models in which target cells levels are allowed to vary22. For the model given in Eq. (1) and its steady state solutions described in the online supplemental material (also see Table 1 for parameter definitions) one can show22
| (2) |
where the total number of uninfected hepatocytes, before infection, is T̄0 (see Eq. S2 in online supplemental material). Thus, one can define successful drug therapy, i.e., sustained decrease in viral load that ultimately can eradicate the virus and lead to a sustained virological response, as those therapies for which ε >εc throughout the course of treatment.
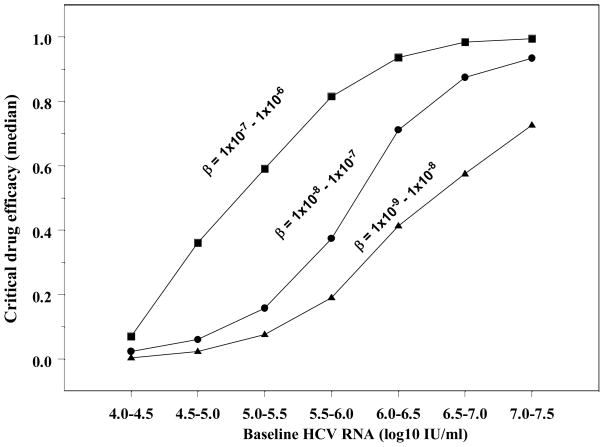
As above, we calculated εc, (Eq. 2) and the bHCV-RNA (Eq. S3 in online supplemental material) for 1,000 in-silico patients by randomly choosing sets of parameter values within the ranges given in Table 1 for each such “patient”. We found that εc required for successful antiviral therapy, tends to increase with bHCV-RNA (Fig. 2). Moreover, for larger values of β, which lead to a higher fraction of infected cells, π (Table 3), the relationship between εc and bHCV-RNA is shifted to the left (Fig. 2). Thus for the same bHCV-RNA, larger β leads to higher εc and therefore to a lower chance to achieve SVR (Fig. 2).
Figure 2. Critical-drug efficacy, baseline HCV-RNA level and infection-rate constant, β.
We randomly chose one thousand parameter sets within the ranges given in Table 1, except p=0.1–6 virions/day (see Figure 1). We then calculated the critical-drug efficacy (εc) and virus steady-state level (V̄) using Eqs. (2) and (Eq. S3 in online supplemental material), respectively, for different ranges of the infection rate constant, β. These different ranges affect the percentage of hepatocytes that are HCV-infected, as shown in Table 3, - high fraction of infected cells (filled squares), moderately-high fraction of infected cells (filled circles), and moderate fraction of infected cells (filled triangles)). Here we plot the median εc in groups of in-silico patients that have baseline viral loads that differ by 0.5log10 HCV-RNA IU/ml, and found that this median increases as a sigmoid function of baseline viral-load. Thus, higher baseline HCV-RNA levels correlate with higher median εc.
Analysis of patient data
Pal et al.34 showed that patients with advanced liver disease have a higher fraction of productively infected hepatocytes, π, than patients with no or mild liver disease. Others showed that advanced liver disease patients have a lower chance of SVR14–17, in agreement with our model predictions assuming that advanced liver disease correlates with high π as shown by Pal et al.34. Further, although not part of our formal model, one can envision that in patients with a large fraction of infected hepatocytes inflammatory responses will be greater than in patients with a low π, and hence such patients would tend to have more damaged tissue and fibrotic repair, thus providing a causal link between high π and advanced liver disease.
When we compared baseline viral loads between non-cirrhotic and cirrhotic patients we found that those in non-cirrhotic patients were significantly higher (5.9±0.7 log10) than those in cirrhotic patients (5.7±0.6 log10) (p=0.013). Furthermore, a higher proportion (p=0.0005) of cirrhotic patients had a bHCV-RNA<6 log IU/ml compared to non-cirrhotic patients (Table 2 and Fig. 1C). From our model simulations, we found that an increased infection rate, β, may be the best explanation of the above features: high π, slightly lower baseline viral loads, and low SVR for patients with advanced liver disease.
Discussion
A higher baseline HCV-RNA (bHCV-RNA) is associated with a higher rate of treatment failure with (PEG) IFN-α alone or in combination with ribavirin. Large clinical trials indicated that patients with bHCV-RNA>4×105 to 6×105 IU/ml have a lower chance of SVR compared with patients with bHCV-RNA below this value2, 3, 5–12. Recently, we showed the pivotal role of the critical drug efficacy, εc, in explaining HCV-RNA kinetic profiles under treatment22. When the drug effectiveness, ε, is higher than the critical value, εc, HCV can be eliminated; however, when ε<εc our model predicts that SVR will not be attained. Here we have shown that εc is positively correlated with bHCV-RNA. Thus, patients with high bHCV-RNA are intrinsically more difficult to treat and require higher drug efficacies to achieve SVR.
We have shown that εc increases if the magnitude of the infection-rate constant, β, is increased, which in turn increases the fraction of HCV-infected hepatocytes, π. This may explain why patients with higher π are also difficult to treat18. In our model, we assume that all hepatocytes, Tmax, are susceptible to HCV infection. We found that π=0.02%–99% in our 1,000 in-silico patients. This range is close to the percentage of hepatocytes positive for genomic HCV-RNA (range 0.04%–88%) found in liver biopsies from patients and chimpanzees with chronic HCV18, 31, 35, 36. However, the detection of genomic HCV-RNA in hepatocytes does not necessarily indicate that these cells are productively infected. Since HCV replication creates negative strand-HCV-RNA, the detection of both positive and negative strand RNA provides a more accurate assessment of productive HCV-infection in the liver. While Agnello et al.31 detected that a high percentage (3%–83%) of hepatocytes harbor negative-strand HCV-RNA in HCV-infected patients and chimpanzees, others36 found negative strand RNA in only a low percentage (4%–25%) of hepatocytes in human subjects. These conflicting results highlight the current limitation of not having precise data on π. Nevertheless, one way to reconcile our prediction of an extremely wide range of levels of HCV-infected hepatocytes with observations suggesting lower levels of infected cells is to assume that not all hepatocytes are susceptible to infection (i.e., reducing Tmax in our model). For HBV infection, London and Blumberg previously suggested that the state of hepatocyte differentiation could affect their susceptibility to infection37. If this also is the case for HCV infection, then our estimate of the HCV-RNA production rate per cell, p (i.e., 0.1–6 virions/day), should be considered a minimal estimate because there could be fewer infected cells responsible for the observed level of viremia. The mean daily production of virions in our 1,000 in-silico patients is 3×1011 virion/day (ranging from 6×108 to 1×1012), which is somewhat smaller than prior estimates20, consistent with production rates of 0.1–6 virions/day being a minimal estimate.
Almost 100% of treated patients obtain SVR when treatment is initiated early (within 6 months) after HCV infection13. However, during the chronic phase of infection only about 50% of patients achieve SVR2, 3. Moreover, lower SVR rates (<29%) were observed in patients with cirrhosis14, 15, 17. The reduction in SVR rates throughout the course of infection could be explained, in part, with our theory if the infection rate, β, increased with duration of disease. Lower values of β, early in infection, reduce the critical drug efficacy and hence would yield higher SVR rates. Also, as discussed above, lower β values lead to higher chance of clearance of infection in in-silico patients with large infected cell loss rates and/or large viral clearance rates. This might explain why some infected patients spontaneously clear the virus during the acute phase. Then, due to within patient viral/host evolution, β might increase from the acute phase to the chronic stage and further increase as HCV-related liver disease develops. Of note, it is not clear yet why only some HCV-infected patients develop advanced liver disease and cirrhosis. Further investigations will be necessary to test this hypothesis in-vivo.
In the current study, patients with advanced liver disease had slightly lower baseline viral loads than those without cirrhosis (p=0.013). Our model suggests that the best way to explain the lower SVR rates in cirrhotic patients who have lower baseline viral loads is that these patients have larger values for the infection rate, β. Another hypothesis would be that in advanced disease there are fewer hepatocytes available for infection (i.e., Tmax is lower) leading to lower viral loads. However, inspection of the critical drug efficacy, εc, expression (Eq. 2) shows that a lower Tmax by itself also results in a lower critical drug efficacy, and thus it should be easier to achieve SVR, which is not observed in clinical practice. Therefore, we prefer the former explanation.
One clinical implication of our results is that factors that increase β might contribute to progression to cirrhosis. These could include viral factors, such as the ability to tolerate mutations that lead to high infectivity, and host factors that increase hepatocyte susceptibility to infection. With regard to antiviral therapy, the model indicates that the use of drugs with higher critical drug effectiveness will be important in improving SVR in patients with cirrhosis. Preliminary investigations of new small molecules for the treatment of HCV appear promising in this regard38. The model also suggests that the development of agents capable of reducing β by blocking viral entry into hepatocytes could be particularly beneficial in slowing down the development of advanced liver disease.
Two other potential implications of our study are worth mentioning. First, large studies have shown lower rates of SVR in African Americans (~28%) as compared to Caucasian Americans (~52%)39–41. It was suggested, via modeling of HCV kinetics42 and by analyzing the CD4+ T-cell response43, that, compared to Caucasian Americans, African Americans are immune tolerant, implying that they have a lower loss rate of infected cells, δ. Our results suggest that a lower δ could contribute to a high εc in African Americans (see Eq. 2), partially explaining their lower SVR rates. Second, low baseline levels of interferon-γ inducible protein 10 kDa (IP-10), are predictive of SVR44. If HCV-infected hepatocytes are the main cellular source of IP-10, as suggested by Harvey et al.45, then low levels of IP-10 may indicate a low π. Our theory suggests that this leads to a lower εc and hence a higher chance of SVR. Thus, low IP-10 may not be directly related to treatment success but instead may be a marker for patients with lower levels of infected hepatocytes and thus of εc. Further investigations will be necessary to test these hypotheses in-vivo.
According to our model, lower SVR rates in patients with advanced liver disease could be linked to a high critical drug efficacy, εc, and a high percentage of HCV-infected hepatocytes, π. In patients with IFN effectiveness, ε, lower than εc, an initial viral decline followed by a flat-final phase is predicted, i.e., HCV RNA remains positive throughout therapy46. In patients with ε>εc, a final phase decline is predicted that will ultimately lead to viral clearance if treatment is long enough. However, the final phase decline is predicted to be slower in patients with high π than in patients with low π, when ε is not close to 1 (manuscript submitted), and therefore longer treatment may be needed to achieve SVR.
Cirrhosis is a chronic liver disease that causes damage to liver tissue in which the normal architecture of the liver is distorted and its function is impaired. In some cases the liver is greatly reduced in volume, being no more than one-third the normal size47. Thus, it is reasonable to assume that the number of hepatocytes is reduced in an end-stage cirrhotic liver compared to a non-cirrhotic liver. Interestingly, our model predicts that larger infection rates, β, cause an approximately 30% to 40% reduction in hepatocytes (see T + I in Table 3).
In summary, we predict the distribution of HCV-RNA values observed in the US and suggest that patients with low baseline HCV-RNA levels have lower critical-drug efficacies and hence an enhanced probability of SVR, in agreement with large clinical studies. Moreover, our model suggests that a larger infection rate, β, may lead to: (i) lower baseline viral loads as seen in our cirrhotic patients, (ii) a higher fraction of HCV-infected cells, as seen in advanced disease34, and (iii) a higher critical drug effectiveness, resulting in a lower chance of SVR. Finally, our model can be used to show, in a highly quantitative way, how changing viral/host parameters through pharmacological or immunological interventions can change viral load and lead to SVR.
Supplementary Material
Acknowledgments
This research was performed under the auspices of the U.S. Department of Energy under contract DE-AC52-06NA25396 and supported by National Institutes of Health (NIH) grants RR06555, AI28433, AI065256 (to A.S.P.), and P20-RR18754 (to R.M.R and H.D). H.D is supported by the University of Illinois Gastrointestinal and Liver Disease (GILD) Association.
Footnotes
No conflicts of interest exist.
References
- 1.NIH. National Institutes of Health Consensus Development Conference: management of hepatitis C: 2002. Hepatology. 2002;36:S3–20. doi: 10.1053/jhep.2002.37117. [DOI] [PubMed] [Google Scholar]
- 2.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 3.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 4.Mihm U, Herrmann E, Sarrazin C, et al. Review article: predicting response in hepatitis C virus therapy. Aliment Pharmacol Ther. 2006;23:1043–54. doi: 10.1111/j.1365-2036.2006.02863.x. [DOI] [PubMed] [Google Scholar]
- 5.Poynard T, Marcellin P, Lee SS, et al. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT) Lancet. 1998;352:1426–1432. doi: 10.1016/s0140-6736(98)07124-4. [see comments] [DOI] [PubMed] [Google Scholar]
- 6.Lindsay KL, Trepo C, Heintges T, et al. A randomized, double-blind trial comparing pegylated interferon alfa-2b to interferon alfa-2b as initial treatment for chronic hepatitis C. Hepatology. 2001;34:395–403. doi: 10.1053/jhep.2001.26371. [DOI] [PubMed] [Google Scholar]
- 7.McHutchison JG, Gordon SC, Schiff ER, et al. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;19(339):1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 8.Zeuzem S, Feinman SV, Rasenack J, et al. Peginterferon alfa-2a in patients with chronic hepatitis C. N Engl J Med. 2000;343:1666–72. doi: 10.1056/NEJM200012073432301. [DOI] [PubMed] [Google Scholar]
- 9.Bronowicki JP, Ouzan D, Asselah T, et al. Effect of ribavirin in genotype 1 patients with hepatitis C responding to pegylated interferon alfa-2a plus ribavirin. Gastroenterology. 2006;131:1040–8. doi: 10.1053/j.gastro.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 10.Berg T, Wagner M, Hinrichsen H, et al. Definition of a pre-treatment viral load cut-off for an optimized prediction of treatment outcome in patients with genotype 1 infection receiving either 48 or 72 weeks of peginterferon alfa-2a plus ribavirin. Hepatology. 2006;44:321A. [Google Scholar]
- 11.Zeuzem S, Fried MW, Reddy KR, et al. Improving the clinical relevance of pre-treatment viral load as a predictor of sustained virological response (SVR) in patients infected with hepatitis C genotype 1 treated with peginterferon alfa-2a (40KD) (PEGASYSR) plus ribavirin (COPEGUSR) Hepatology. 2006;44:267A–268A. [Google Scholar]
- 12.Zehnter E, Mauss S, John C, et al. Better prediction of SVR in patients with HCV genotype 1 (G1) with peginterferon alfa-2a (PEGASYS) plus ribavirin: Improving differentiation between low (LVL) and high baseline viral load (HVL) Hepatology. 2006;44:328A. [Google Scholar]
- 13.Jaeckel E, Cornberg M, Wedemeyer H, et al. Treatment of acute hepatitis C with interferon alfa-2b. N Engl J Med. 2001;345:1452–1457. doi: 10.1056/NEJMoa011232. [DOI] [PubMed] [Google Scholar]
- 14.Everson GT, Hoefs JC, Seeff LB, et al. Impact of disease severity on outcome of antiviral therapy for chronic hepatitis C: Lessons from the HALT-C trial. Hepatology. 2006;44:1675–84. doi: 10.1002/hep.21440. [DOI] [PubMed] [Google Scholar]
- 15.Iacobellis A, Siciliano M, Perri F, et al. Peginterferon alfa-2b and ribavirin in patients with hepatitis C virus and decompensated cirrhosis: a controlled study. J Hepatol. 2007;46:206–12. doi: 10.1016/j.jhep.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 16.Westin J, Lagging M, Dhillon AP, et al. Impact of hepatic steatosis on viral kinetics and treatment outcome during antiviral treatment of chronic HCV infection. J Viral Hepat. 2007;14:29–35. doi: 10.1111/j.1365-2893.2006.00777.x. [DOI] [PubMed] [Google Scholar]
- 17.Medeiros-Filho JE, de Carvalho Mello IM, Pinho JR, et al. Differences in viral kinetics between genotypes 1 and 3 of hepatitis C virus and between cirrhotic and non-cirrhotic patients during antiviral therapy. World J Gastroenterol. 2006;12:7271–7. doi: 10.3748/wjg.v12.i45.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez-Inigo E, Lopez-Alcorocho JM, Bartolome J, et al. Percentage of hepatitis C virus-infected hepatocytes is a better predictor of response than serum viremia levels. J Mol Diagn. 2005;7:535–43. doi: 10.1016/S1525-1578(10)60585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perelson AS, Herrmann E, Micol F, et al. New kinetic models for the hepatitis C virus. Hepatology. 2005;42:749–54. doi: 10.1002/hep.20882. [DOI] [PubMed] [Google Scholar]
- 20.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–7. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 21.Dixit NM, Layden-Almer JE, Layden TJ, et al. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004;432:922–4. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
- 22.Dahari H, Ribeiro RM, Perelson AS. Triphasic decline of hepatitis C virus RNA during antiviral therapy. Hepatology. 2007;46:16–21. doi: 10.1002/hep.21657. [DOI] [PubMed] [Google Scholar]
- 23.Dahari H, Lo A, Ribeiro RM, et al. Modeling hepatitis C virus dynamics: Liver regeneration and critical drug efficacy. J Theor Biol. 2007;247:371–81. doi: 10.1016/j.jtbi.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawlotsky JM, Dahari H, Neumann AU, et al. Antiviral action of ribavirin in chronic hepatitis C. Gastroenterology. 2004;126:703–14. doi: 10.1053/j.gastro.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann E, Lee JH, Marinos G, et al. Effect of ribavirin on hepatitis C viral kinetics in patients treated with pegylated interferon. Hepatology. 2003;37:1351–8. doi: 10.1053/jhep.2003.50218. [DOI] [PubMed] [Google Scholar]
- 26.Sentjens RE, Weegink CJ, Beld MG, et al. Viral kinetics of hepatitis C virus RNA in patients with chronic hepatitis C treated with 18 MU of interferon alpha daily. Eur J Gastroenterol Hepatol. 2002;14:833–40. doi: 10.1097/00042737-200208000-00004. [DOI] [PubMed] [Google Scholar]
- 27.Nainan OV, Alter MJ, Kruszon-Moran D, et al. Hepatitis C virus genotypes and viral concentrations in participants of a general population survey in the United States. Gastroenterology. 2006;131:478–84. doi: 10.1053/j.gastro.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 28.Colombatto P, Civitano L, Oliveri F, et al. Sustained response to interferon-ribavirin combination therapy predicted by a model of hepatitis C virus dynamics using both HCV RNA and alanine aminotransferase. Antivir Ther. 2003;8:519–30. [PubMed] [Google Scholar]
- 29.Dahari H, Major M, Zhang X, et al. Mathematical modeling of primary hepatitis C infection: Noncytolytic clearance and early blockage of virion production. Gastroenterology. 2005;128:1056–66. doi: 10.1053/j.gastro.2005.01.049. [DOI] [PubMed] [Google Scholar]
- 30.Gosalvez J, Rodriguez-Inigo E, Ramiro-Diaz JL, et al. Relative quantification and mapping of hepatitis C virus by in situ hybridization and digital image analysis. Hepatology. 1998;27:1428–34. doi: 10.1002/hep.510270534. [DOI] [PubMed] [Google Scholar]
- 31.Agnello V, Abel G, Knight GB, et al. Detection of widespread hepatocyte infection in chronic hepatitis C. Hepatology. 1998;28:573–84. doi: 10.1002/hep.510280240. [DOI] [PubMed] [Google Scholar]
- 32.Wein LM, D’Amato RM, Perelson AS. Mathematical analysis of antiretroviral therapy aimed at HIV-1 eradication or maintenance of low viral loads. J Theor Biol. 1998;192:81–98. doi: 10.1006/jtbi.1997.0622. [DOI] [PubMed] [Google Scholar]
- 33.Callaway DS, Perelson AS. HIV-1 infection and low steady state viral loads. Bull Math Biol. 2002;64:29–64. doi: 10.1006/bulm.2001.0266. [DOI] [PubMed] [Google Scholar]
- 34.Pal S, Shuhart MC, Thomassen L, et al. Intrahepatic hepatitis C virus replication correlates with chronic hepatitis C disease severity in vivo. J Virol. 2006;80:2280–90. doi: 10.1128/JVI.80.5.2280-2290.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez-Inigo E, Bartolome J, de Lucas S, et al. Histological damage in chronic hepatitis C is not related to the extent of infection in the liver. Am J Pathol. 1999;154:1877–81. doi: 10.1016/S0002-9440(10)65445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang M, Marquardt AP, Wood BL, et al. In situ distribution of hepatitis C virus replicative-intermediate RNA in hepatic tissue and its correlation with liver disease. J Virol. 2000;74:944–55. doi: 10.1128/jvi.74.2.944-955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.London WT, Blumberg BS. A cellular-model of the role of hepatitis-B virus in the pathogenesis of primary hepatocellular-carcinoma. Hepatology. 1982;2:10S–14S. [Google Scholar]
- 38.Zeuzem S, Nelson DR, Marcellin P. Dynamic evolution of therapy for chronic hepatitis C: how will novel agents be incorporated into the standard of care? Antivir Ther. 2008;13:747–60. [PubMed] [Google Scholar]
- 39.Conjeevaram HS, Fried MW, Jeffers LJ, et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470–7. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 40.Jeffers LJ, Cassidy W, Howell CD, et al. Peginterferon alfa-2a (40 kd) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology. 2004;39:1702–8. doi: 10.1002/hep.20212. [DOI] [PubMed] [Google Scholar]
- 41.Muir AJ, Bornstein JD, Killenberg PG. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med. 2004;350:2265–71. doi: 10.1056/NEJMoa032502. [DOI] [PubMed] [Google Scholar]
- 42.Layden-Almer JE, Ribeiro RM, Wiley T, et al. Viral dynamics and response differences in HCV-infected African American and white patients treated with IFN and ribavirin. Hepatology. 2003;37:1343–50. doi: 10.1053/jhep.2003.50217. [DOI] [PubMed] [Google Scholar]
- 43.Rosen HR, Weston SJ, Im K, et al. Selective decrease in hepatitis C virus-specific immunity among African Americans and outcome of antiviral therapy. Hepatology. 2007;46:350–8. doi: 10.1002/hep.21714. [DOI] [PubMed] [Google Scholar]
- 44.Lagging M, Romero AI, Westin J, et al. IP-10 predicts viral response and therapeutic outcome in difficult-to-treat patients with HCV genotype 1 infection. Hepatology. 2006;44:1617–25. doi: 10.1002/hep.21407. [DOI] [PubMed] [Google Scholar]
- 45.Harvey CE, Post JJ, Palladinetti P, et al. Expression of the chemokine IP-10 (CXCL10) by hepatocytes in chronic hepatitis C virus infection correlates with histological severity and lobular inflammation. J Leukoc Biol. 2003;74:360–9. doi: 10.1189/jlb.0303093. [DOI] [PubMed] [Google Scholar]
- 46.Dahari H, Perelson AS. Hepatitis C virus RNA kinetics: Drug efficacy and the rate of HCV-infected cells loss. World J Gastroenterol. 2007;13:3020–1. doi: 10.3748/wjg.v13.i21.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolf D. Evaluation of the Size, Shape, and Consistency of the Liver. Butterworth. 1990 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.