Abstract
Cardiac sodium channel are protein complexes that are expressed in the sarcolemma of cardiomyocytes to carry a large inward depolarizing current (INa) during phase 0 of the cardiac action potential. The importance of INa for normal cardiac electrical activity is reflected by the high incidence of arrhythmias in cardiac sodium channelopathies, i.e., arrhythmogenic diseases in patients with mutations in SCN5A, the gene responsible for the pore-forming ion-conducting α-subunit, or in genes that encode the ancillary β-subunits or regulatory proteins of the cardiac sodium channel. While clinical and genetic studies have laid the foundation for our understanding of cardiac sodium channelopathies by establishing links between arrhythmogenic diseases and mutations in genes that encode various subunits of the cardiac sodium channel, biophysical studies (particularly in heterologous expression systems and transgenic mouse models) have provided insights into the mechanisms by which INa dysfunction causes disease in such channelopathies. It is now recognized that mutations that increase INa delay cardiac repolarization, prolong action potential duration, and cause long QT syndrome, while mutations that reduce INa decrease cardiac excitability, reduce electrical conduction velocity, and induce Brugada syndrome, progressive cardiac conduction disease, sick sinus syndrome, or combinations thereof. Recently, mutation-induced INa dysfunction was also linked to dilated cardiomyopathy, atrial fibrillation, and sudden infant death syndrome. This review describes the structure and function of the cardiac sodium channel and its various subunits, summarizes major cardiac sodium channelopathies and the current knowledge concerning their genetic background and underlying molecular mechanisms, and discusses recent advances in the discovery of mutation-specific therapies in the management of these channelopathies.
Keywords: Arrhythmia, Action potential, Cardiac electrophysiology, Cardiomyocyte, Ion channels
Introduction
Cardiac sodium channels are transmembrane proteins that are located in the sarcolemma of atrial and ventricular myocytes and the Purkinje fibers. They enable a large and rapid influx of sodium ions (sodium current; INa) during the rapid upstroke (phase 0) of the cardiac action potential. INa underlies the initiation and propagation of action potentials, and, by doing so, it determines cardiac excitability and conduction velocity of electrical stimuli through the heart (Fig. 1a, b) [9]. To a lesser extent, cardiac sodium channels are also present in the sinoatrial node and the atrioventricular node, where they contribute to pacemaker activity [45]. The importance of sodium channels for normal electrical activity of the heart is emphasized in arrhythmogenic diseases caused by inherited or de novo (mostly heterozygous) mutations in SCN5A, the gene that encodes the cardiac sodium channel (“cardiac sodium channelopathies”). These diseases include long QT syndrome type 3 (LQT-3), Brugada syndrome (BrS), progressive cardiac conduction disease (PCCD), dilated cardiomyopathy (DCM), sick sinus syndrome (SSS), atrial fibrillation (AFib), sudden infant death syndrome (SIDS), and overlap syndromes [5]. In overlap syndromes, patients display overlapping clinical symptoms or arrhythmia forms of more than one disease [42]. Although less prevalent, mutations in genes encoding proteins that regulate the expression or function of the sodium channel may also cause these diseases [5].
Fig. 1.
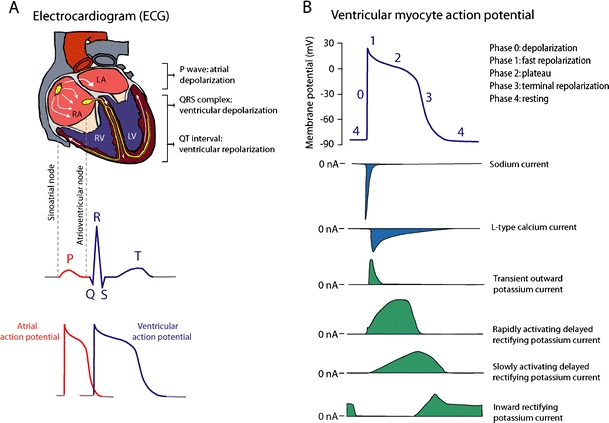
The cardiac electrical activity and cardiac ion currents. a The electrical activity of the heart is represented on the surface electrocardiogram (ECG), and results from coordinated action potential generation in individual cardiomyocytes. The electrical activity starts by the spontaneous generation of action potentials in pacemaker cells in the sinoatrial node. Propagation of these action potentials creates an excitation wave through the atria, leading to atrial depolarization. After traveling through the atrioventricular node, the excitation wave reaches the ventricles, and leads to ventricular depolarization. b The cardiac action potential is generated by transmembrane inwardly and outwardly directed ion currents. The inward (depolarizing) sodium and calcium currents are pointed downwards and colored blue. The outward (repolarizing) potassium currents are pointed upwards and colored green
The effects of SCN5A mutations on the normal structure, expression, and function of cardiac sodium channels are routinely investigated by cloning and expression of channel proteins in heterologous systems (e.g., human embryonic kidney cells, Chinese hamster ovary cells, Xenopus oocytes). The effects of mutations in genes encoding the regulatory proteins are studied by co-expression of the mutant protein with the normal sodium channel protein. Additionally, genetically engineered mice carrying mutations in a specific gene of interest are increasingly used to study the effect of mutations in the native environment of the myocyte [17]. These expression models, in conjunction with various experimental techniques (e.g., electron crystallography to study channel structure, polymerase chain reaction to study mRNA expression, Western blot and immunohistochemistry to study protein expression, co-immunoprecipitation to study protein–protein associations, and patch-clamp technique to measure currents) have greatly enhanced our understanding of the role of sodium channel dysfunction in the pathophysiology of cardiac sodium channelopathies. Furthermore, they have provided basic rationale for gene-specific and even mutation-specific approaches in the clinical management of subject with such channelopathies. This review briefly describes the structure and function of the cardiac sodium channel and its modulation by regulatory proteins. Moreover, it discusses the well-recognized cardiac sodium channelopathies and the nature and role of sodium channel dysfunction in the underlying mechanisms of these diseases.
The cardiac sodium channel
Voltage-gated sodium channels are dynamic transmembrane proteins in excitable cells (e.g., cardiomyocytes, skeletal muscle cells, neurons) that open and close to conduct ions. They are large molecular complexes containing a pore-forming ion-conducting α-subunit and ancillary β-subunits, and several regulatory proteins [1, 9, 29, 33, 39, 48, 77, 89]. The α-subunit is called Nav1.5, and is encoded by SCN5A. It is organized into a cytoplasmic N terminus, four homologous domains (DI–DIV) that are connected to each other by cytoplasmic linkers, and a cytoplasmic C terminus. Each domain consists of six transmembrane α-helical segments (S1–S6), connected to each other by alternating extracellular and cytoplasmic loops. The four domains fold around an ion-conducting pore, which is lined by the extracellular loops (P-loops) between S5 and S6 segments (Fig. 2a, b). Some amino acids in the P-loops confer the selectivity for sodium ions. Gating represents time-dependent transitions between distinct conformational states of the channel protein due to molecular movements in response to membrane potential changes (“voltage-dependent gating”) [9]. When cardiomyocytes are excited by electrical stimuli from adjacent cells or by artificially applied stimuli during patch-clamp experiments, their resting membrane potential (approximately −85 mV) depolarizes. This triggers an outward movement of the positively charged S4 segments (voltage sensors), which leads to channel activation (i.e., opening of the pore). Inactivation (closing of the pore) starts simultaneously with activation, but since inactivation is slower than activation, channels remain transiently open to conduct INa during phase 0 of the action potential (Fig. 3a, b). Inactivation comprises different conformational states, including fast, intermediate, and slow inactivation. Fast inactivation is coupled to activation and initiated by the outward movement of the S4 segment of DIV. This triggers the amino acids isoleucine, phenylalanine, and methionine (IFM motif; “the lid”) and the neighboring glycine and proline (“the hinges”) in the DIII–DIV linker to occlude the pore by binding to multiple amino acids in the cytoplasmic loops between the S4 and S5 segments of DIII and DIV (“the dock”) [9]. The molecular movements leading to slow inactivation are less well understood. However, mutations in the P-loops, the S6 segments and the C terminus have been reported to affect this state. In any case, more than 99% of sodium channels are inactivated at the end of action potential phase 1, and can be reactivated only after recovery from inactivation during action potential phase 4. Slow inactivation requires much longer recovery times than fast inactivation. Finally, a very small fraction of sodium channels may reactivate during action potential phase 3. The current through these channels (<1% of the peak INa) is called the window current, since it arises when the sarcolemma reaches a potential that is depolarized sufficiently to reactivate some channels, but not enough to cause complete inactivation (Fig. 3c, d). The voltage range for the window current is very restricted and narrow in healthy hearts, granting it a small role during the cardiac action potential [41].
Fig. 2.
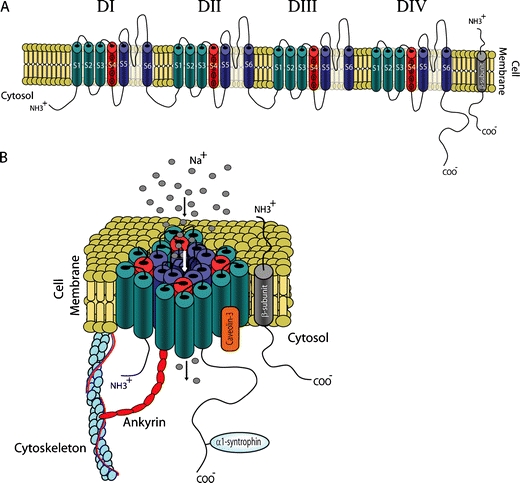
Molecular structure of the cardiac sodium channel. a Cartoon of the α-subunit (Nav1.5) and the β-subunit of the cardiac sodium channel. Nav1.5 consists of four domains (DI–DIV), each containing six transmembrane segments (S1–S6); S4 segments are positively charged and act as voltage sensors. The β-subunit consists of one single transmembrane segment. b The four domains of Nav1.5 fold around an ion-conducting pore, which is lined by the loops between the S5 and S6 segments. The expression and function of Nav1.5 is regulated by β-subunits and several directly or indirectly interacting regulatory proteins
Fig. 3.
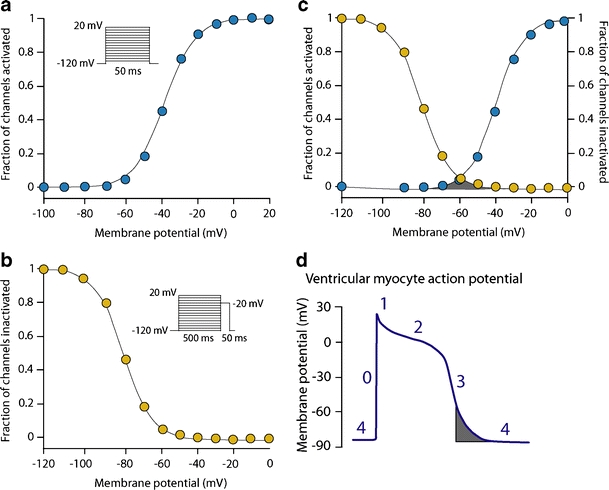
Voltage-dependent activation and inactivation of the cardiac sodium channel. a Using the patch-clamp technique, the membrane potential dependence of activation is studied by applying 50 ms depolarizing voltage steps from a holding potential of −120 mV (inset). The activation curve is obtained by dividing the amplitude of the resulting sodium current at each voltage step by the maximum peak sodium current amplitude, and plotting versus the corresponding voltage. b The membrane potential dependence of inactivation is studied by applying 500 ms depolarizing voltage steps from a holding potential of −120 mV to inactivate the channels (prepulse). Next, the fraction of channels that is not inactivated by the prepulse is measured by applying a voltage step to 20 mV (test pulse). The inactivation curve is obtained by dividing the amplitude of sodium current at each test pulse by the maximum peak sodium current amplitude, and plotting versus the corresponding prepulse voltage. c The window current (gray area) arises when the sarcolemma reaches a potential that is depolarized sufficiently to reactivate some channels, but not enough to cause complete inactivation. d The voltage range for the window current is normally narrow and achieved during phase 3 of the ventricular action potential
Although Nav1.5 is sufficient to generate sodium current in heterologous expression systems, the obtained current is quite different from INa present in isolated cardiomyocytes. This may be due to the absence of ancillary β-subunits and regulatory proteins in heterologous systems (Fig. 2b). So far, four β-subunits are known in the heart (β1 to β4), which are encoded by four genes (SCN1B to SCN4B) [1, 29, 33, 48, 89]. The β-subunits are proteins with an extracellular N terminus, one transmembrane segment, and a cytoplasmic C terminus. They increase the expression of Nav1.5 in the sarcolemma, augment the amplitude of INa, modulate its gating properties, and play crucial roles in the interaction of Nav1.5 proteins to extracellular matrix molecules, cytoplasmic cytoskeleton apparatus, and components of cardiac intercellular junctions (e.g., cadherins, connexins). The β-subunits may also contribute to the preferential localization of Nav1.5 proteins in the intercalated disks. Nav1.5 proteins may also directly interact with several regulatory proteins, including enzymes involved in their glycosylation and phosphorylation (e.g., Ca2+/calmodulin-dependent protein kinase II) [1, 77], adaptor proteins that connect them to the cytoskeleton (e.g., ankyrins) [77], and proteins that mediate their trafficking from the endoplasmic reticulum (ER) to the sarcolemma (e.g., glycerol-3-phosphate dehydrogenase 1-like protein) [39]. Of note, mutations in genes encoding β-subunits and regulatory proteins are occasionally found in patients with clinical phenotypes similar to arrhythmogenic diseases caused by SCN5A mutations. This reflects the importance of these proteins for the normal functioning of the cardiac sodium channel [5].
Long QT syndrome type 3
Long QT syndrome (LQTS) is a disease characterized by prolonged QT intervals on the surface electrocardiogram (ECG), and increased risk for sudden death due to ventricular tachyarrhythmias, in particular torsades de pointes. Prolonged QT intervals reflect increased action potential durations in ventricular myocytes, and correspond with delayed ventricular repolarization (Fig. 4a, b). Heritable LQTS is classified into different types, and each type is linked to mutations in a gene encoding a protein that is directly (ion channel) or indirectly (β-subunit or regulatory protein) involved in repolarization [50]. LQT-3 is linked to mutations in SCN5A, and covers approximately 13% of all genotyped individuals with LQTS [31, 87]. So far, more than 80 SCN5A mutations have been identified in patients with LQT-3, and nearly 50% of them have been studied heterologously [93]. Most of these mutations are missense mutations, and are found to cause sodium channel gain-of-function, by disrupting fast inactivation and thereby causing an abnormal sustained (or persistent) non-inactivating sodium current (Isus or Ipst; Fig. 4c, d) [13]. Mutations causing Isus are mainly clustered in Nav1.5 regions that are involved in fast inactivation (i.e., S4 segment of DIV, the DIII–DIV linker, and the cytoplasmic loops between the S4 and S5 segments of DIII and DIV), or in regions that stabilize fast inactivation (e.g., the C terminus) [31, 52, 76, 93]. The C terminus probably interacts with the DIII–DIV linker to stabilize the occlusion of the pore during inactivation [60].
Fig. 4.
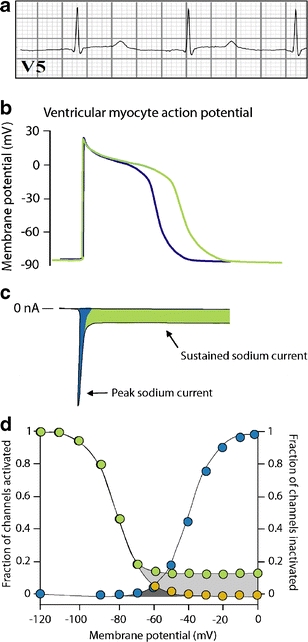
Long QT syndrome type 3. a Prolonged QT intervals on the surface ECG of an individual with LQT-3. b QT interval prolongation results from delayed repolarization of ventricular action potentials. c Delayed repolarization in LQT-3 is often due to the presence of abnormal sustained non-inactivating sodium current (green area). d Sustained current results from incomplete inactivation of the sodium channels (green circles)
Other less common mechanisms of SCN5A mutations to cause LQT-3 include increased window current [86, 90], slower inactivation [66, 90], faster recovery from inactivation [2, 19], and larger peak INa density [64]. The window current increases when inactivation of mutant sodium channels occurs at more positive (depolarized) potentials (i.e., delayed inactivation), while activation is not changed. This widens the voltage range during which the sodium channel may reactivate without inactivation (Fig. 5a) [86, 90]. Slower inactivation allows longer channel openings, and causes a slowly inactivating sodium current (Fig. 5b) [66, 90]. To distinguish it from Isus (which does not inactivate), this current is called the late sodium current (INaL) in this review. However, in the literature, the terms Isus, Ipst, and INaL are often used interchangeably. Comparable to Isus, both the window current and INaL exert their effects during phases 2 and 3 of the action potential, where normally no or very small sodium current is present. However, the other two mechanisms (i.e., faster recovery from inactivation and larger peak INa density) involve increased influx of sodium ions during phase 0 of the action potential (Fig. 5c). Faster recovery from inactivation leads to larger peak INa by increasing the fraction of channels available for activation during subsequent depolarizations. Finally, larger INa density may result from increased expression of mutant Nav1.5 through enhanced mRNA translation or protein trafficking to the sarcolemma, decreased protein degradation, or altered modulation by β-subunits and regulatory proteins. Importantly, one single SCN5A mutation may cause several changes in the expression and/or gating properties of the resulting sodium channels. Regardless of the mechanism, increased sodium current (Isus, window current, INaL, or peak INa) upsets the balance between depolarizing and repolarizing currents in favor of depolarization. The resulting delay in the repolarization process triggers early afterdepolarizations (i.e., re-activation of L-type calcium channel during phases 2 or 3 of the action potential), especially in Purkinje fiber myocytes where action potential durations are intrinsically longer [22, 27]. Early afterdepolarizations are believed to induce torsades de pointes [92].
Fig. 5.
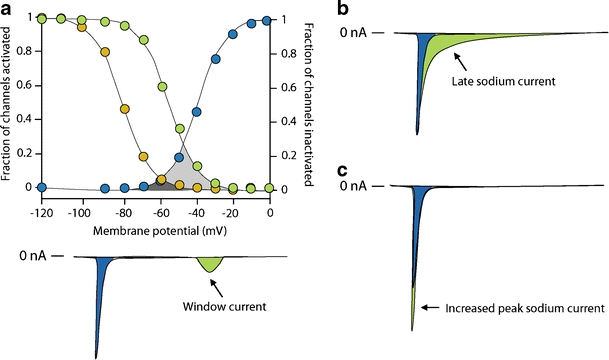
Alternative mechanisms of sodium channel gain-of-function in long QT syndrome type 3. a Increased window current due to delayed inactivation of cardiac sodium channels (green circles). Increased windows current is carried at potentials corresponding to phases 2 and 3 of the ventricular action potential (green area), remote from the peak sodium current during phase 0 (blue area). b Slower inactivation creates a late sodium current (green area). c Increased peak sodium current
Arrhythmic events in LQT-3 usually occur at rest or during sleep when the heart rate is slow. Accordingly, Isus is larger at slower stimulus frequencies, suggesting that the degree of Isus may be a strong determinant for arrhythmias to occur [70]. This is confirmed by a report of homozygous carriers of an SCN5A mutation who displayed longer QT intervals and more frequent arrhythmias than heterozygous carriers of the same mutation. Consistently, homozygous expression of the mutation in heterologous system caused larger Isus densities than heterozygous expression of the mutation [40]. The definite role of Isus in LQT-3 is further reflected by the effectiveness of drugs that inhibit Isus in the treatment of patients with LQT-3. Such drugs (e.g., ranolazine, mexiletine, flecainide) shorten QT intervals in patients with LQT-3 by preferentially blocking Isus over peak INa [11, 51, 69]. However, this effect may be mutation specific. Mexiletine has been shown to be especially effective in patients with SCN5A mutations that shift inactivation toward more negative potentials (i.e., earlier inactivation) [66]. Moreover, in high concentrations these drugs may also block peak INa, and exert pro-arrhythmic effects by decreasing cardiac excitability and slowing electrical conduction velocity. Finally, beta-blockers (the cornerstone therapy in most long QT syndrome patients, particularly those with LQTS type 1 and, somewhat less so, in LQTS type 2) seem to be less effective in LQT-3 [70]. This may be due to their pro-arrhythmic effect by slowing the heart rate and consequently increasing Isus, which opposes their anti-arrhythmic effect (i.e., preferential block of Isus by direct binding to Nav1.5). The anti-arrhythmic effect of beta-blockers in LQT-3 may also be mutation-specific (e.g., when inactivation occurs at more negative potentials), and limited to some agents (propranolol, carvedilol, but not metoprolol) [10].
LQTS and mutations in sodium channel regulatory proteins
Three less common types of LQTS are caused by mutations in genes encoding proteins that regulate the expression or function of Nav1.5 proteins. A mutation in SCN4B, encoding the β4-subunit, has been linked to LQTS type 10. When co-expressed heterologously with SCN5A, the mutation shifted the inactivation toward more positive potentials, but did not change the activation. This resulted in increased window currents at membrane potentials corresponding to the phase 3 of the action potential [48]. Mutations in CAV3, encoding caveolin-3, are linked to LQTS type 9 (LQT-9). Caveolin-3 co-localizes and interacts with Nav1.5 proteins at the sarcolemma of cardiomyocytes (Fig. 2b). When co-expressed with SCN5A, the mutant caveolin-3 proteins induce Isus through a yet unknown mechanism [81]. Finally, a mutation in SNTA1 has been linked to LQTS type 12. SNTA1 encodes α1-syntrophin, a cytoplasmic adaptor protein that enables the interaction between Nav1.5, nitric oxide synthase (NOS), and sarcolemmal calcium ATPase (PMCA). PMCA inhibits nitric oxide synthesis by NOS. By inducing nitrosylation of Nav1.5 proteins, nitric oxide decreases channel inactivation and increases Isus densities. The mutation in SNTA1 disrupted the interaction between Nav1.5 and PMCA, and consequently caused increased Nav1.5 nitrosylation and Isus densities [77].
Brugada syndrome
BrS is diagnosed in subject with high risk for sudden death due to ventricular tachyarrhythmias (polymorphic ventricular tachycardia and ventricular fibrillation), accompanied by typical coved-type ST segment elevation in the right-precordial ECG leads V1 to V3 [16]. The ECG changes are often concealed, but can be unmasked by a drug challenge test using Class 1A or 1C anti-arrhythmic drugs (e.g., ajmaline, flecainide, pilsicainide), which block the cardiac sodium channel (Fig. 6a) [91]. This indicates the crucial role of INa inhibition in the pathophysiology of BrS. Accordingly, 10–30% of subject with BrS carry a mutation in SCN5A, including missense mutations, nonsense mutations, and nucleotide deletions or insertions [91, 93]. The latter may alter mRNA splicing or create a premature stop codon by shifting the open reading frame, and result in truncated Nav1.5 proteins. More than 100 SCN5A mutations are linked to BrS. Virtually all mutations that are heterologously expressed (<50%) lead to sodium channel loss-of-function (i.e., reduced peak INa; Fig. 6b). The underlying mechanisms include decreased expression of Nav1.5 proteins [78], expression of non-functional channels [35], and altered gating properties [6, 15, 21, 73]. Decreased sarcolemmal expression of Nav1.5 proteins results from premature degradation of the mutant proteins by the quality control system in the ER (Fig. 7a) [78]. Expression may also be decreased because mutant Nav1.5 proteins fail to interact with β-subunits or regulatory proteins, which mediate their normal localization on the sarcolemma. Some mutant Nav1.5 proteins pass the ER quality control system and traffic normally to the sarcolemma, but form channels that conduct no or very small INa (non-functional) [35]. The third mechanism (altered gating properties) comprises delayed activation (i.e., activation at more positive potentials) [73], earlier inactivation (i.e., inactivation at more negative potentials) [15], faster inactivation [21], and enhanced slow inactivation [6]. Delayed activation, earlier inactivation, and faster inactivation reduce INa by decreasing the probability of the channels to reside in the activated state (Fig. 7b, c). Enhanced slow inactivation means that mutant channels preferentially enter into the slow inactivation state. As mentioned earlier, the slow inactivation state requires relatively long recovery times during the action potential phase 4. At fast heart rates, phase 4 becomes too short for such channels to recover completely from slow inactivation. This leads to an accumulation of the channels in the slow inactivation state, and INa reduction. Although BrS-linked SCN5A mutations are in general randomly located in various regions of Nav1.5, mutations that cause enhanced slow inactivation are mainly located in the P-loops, the S6 segments, and the C terminus [93].
Fig. 6.
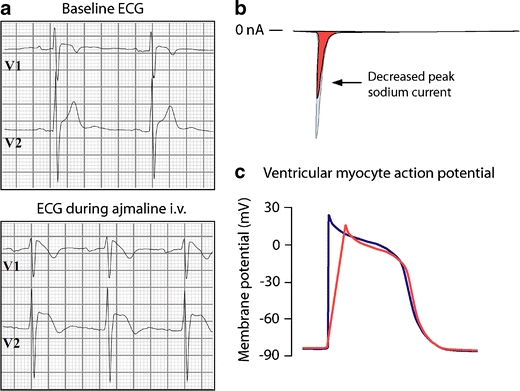
Brugada syndrome. a Coved-type ST segment elevation in the right-precordial ECG leads V1 and V2 after intravenous (i.v.) administration of sodium channel blocking drug ajmaline in an individual with BrS. b BrS-linked SCN5A mutations often lead to peak sodium current reduction. c Reduced peak sodium current decreases the upstroke velocity of action potential phase 0, which slows cardiac electrical conduction
Fig. 7.
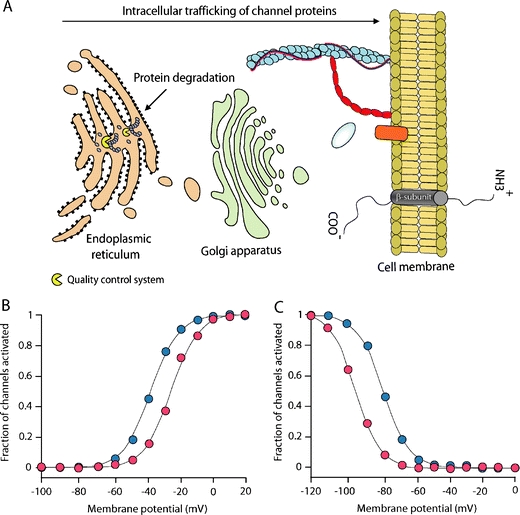
Mechanisms of sodium channel loss-of-function in Brugada syndrome, progressive cardiac conduction disease, and sick sinus syndrome. a Decreased sarcolemmal expression of Nav1.5 due to premature degradation of mutant channel proteins by the quality control system in the endoplasmic reticulum. b Delayed activation of cardiac sodium channels (red circles). c Earlier inactivation of cardiac sodium channels (red circles)
INa reduction decreases the upstroke velocity of action potential phase 0, and, as a result, slows atrial and ventricular electrical conduction (Fig. 6c). This is often reflected by prolonged atrioventricular and ventricular conduction intervals (PR and QRS intervals, respectively) on the ECGs of BrS patients with an SCN5A mutation (Fig. 6a) [91]. During electrophysiological studies in such patients, electrical conduction is particularly delayed between the His bundle and the ventricles (HV interval prolongation), indicating the importance of INa for the initiation and propagation of action potentials in Purkinje fiber myocytes and the ventricular conduction system [72]. The right-precordial ST segment elevation is less well understood, and explained by two hypotheses. The first hypothesis focuses on the presence of transmural voltage gradients due to heterogeneity in action potential duration between the right ventricular epicardium and endocardium. Indeed, action potential durations are shorter in the epicardium, where the repolarizing transient outward potassium current (Ito) is more prominently expressed. INa reduction would further shorten epicardial action potential durations, and facilitate reentrant excitation waves between depolarized endocardium and prematurely repolarized epicardium [7]. The second hypothesis involves preferential conduction slowing in the right ventricular outflow tract. Regional differences in conduction velocity in the right ventricular epicardium would be aggravated by INa reduction, and trigger the occurrence of epicardial reentrant excitation waves [49]. This hypothesis is supported by the increased prevalence of mild (subclinical) structural abnormalities in the right ventricles of BrS patients (e.g., inflammation, fatty tissue replacement, and fibrosis) [24]. Regardless of the mechanism, since ST segment elevation is equally seen in SCN5A mutation carriers and non-carriers, other genetic variants (e.g., in genes encoding ion channels or regulatory proteins) and exogenous factors (e.g., electrolyte imbalances, hormones, body temperature) may contribute to the pathophysiology of BrS [4, 30, 46, 74].
Arrhythmic events in BrS occur more frequently in men, and at rest or during sleep. Experimental studies suggest that the effects of gender on the BrS phenotype may be due to intrinsic differences in ion channel expression between men and women (e.g., higher Ito densities in men) or due to differences in hormone levels (e.g., higher testosterone levels in men) [7, 74]. The latter is confirmed by the disappearance of typical BrS ECG changes after castration [46]. The occurrence of arrhythmias during sleep may be due to an increased vagal activity and/or decreased sympathetic activity. This is confirmed in BrS patients by right-precordial ST segment elevation following intracoronary injection of acetylcholine, and decreased levels of norepinephrine in the synaptic cleft on positron emission tomography [32, 54]. Moreover, the typical ECG changes and arrhythmias in BrS may also be triggered by fever [4]. Although the mechanism is not fully understood, some SCN5A mutations have been shown to alter the gating properties of cardiac sodium channels in a temperature-dependent manner, e.g., more slow inactivation at higher temperatures [6, 21]. Finally, the BrS ECG changes are reported to worsen during exercise. This may be partially attributed to an enhanced slow inactivation in mutant channels, leading to an accumulation of the mutant channels in the slow inactivation state at fast heart rates. However, other factors (e.g., autonomic nervous system, ion current imbalances) may also play a role [3].
So far, no mutation-specific therapy is available for BrS. Decreased sarcolemmal expression of mutant Nav1.5 proteins can be restored with cardiac sodium channel blocking drugs (e.g., mexiletine). Such drugs bind to mutant proteins, and act as molecular chaperones to rescue their trafficking from the ER to the sarcolemma [78]. However, it is questionable whether these drugs can be used as therapy, because (once expressed on the sarcolemma) the mutant proteins display arrhythmia-causing gating defects (e.g., Isus) [58]. Moreover, since sodium channel blocking drugs reduce INa, they may aggravate ECG changes or trigger arrhythmias in BrS, and should therefore be avoided [30, 59]. An implantable cardioverter defibrillator and adequate treatment of fever are currently the only effective therapies to prevent sudden death in BrS.
BrS and mutations in sodium channel regulatory proteins
INa reduction in BrS may also be due to mutations in genes encoding β-subunits or regulatory proteins of the cardiac sodium channel. A mutation in SCN1B was found in one family with BrS. When heterologously expressed, the mutation resulted in formation of truncated β1-subunits, which failed to interact with Nav1.5 proteins and to increase INa densities as normal β1-subunits did [89]. A missense mutation in SCN3B was found in one subject with BrS. The mutation reduced INa by disrupting the trafficking of Nav1.5 proteins from the ER to the sarcolemma, and by altering the gating properties (e.g., earlier and faster inactivation) [29]. Finally, a mutation in GPD1-L, encoding glycerol-3-phosphate dehydrogenase 1-like protein (GPD1-L), was found in a family with BrS [39]. Recently, GPD1-L was shown to inhibit the phosphorylation of Nav1.5 proteins at residues where phosphorylation would lead to INa reduction (probably by decreasing the sarcolemmal expression of Nav1.5 proteins). The mutant GPD1-L failed to inhibit the phosphorylation of Nav1.5 proteins, and resulted in reduced INa densities [79].
Progressive cardiac conduction disease
PCCD (also called Lev or Lenègre disease) manifests as progressive prolongation of the conduction parameters (P wave, PR, and QRS intervals), and right or left bundle branch block, without ST segment elevation or QT interval prolongation (Fig. 8). A complete atrioventricular block may develop and cause syncope or sudden death. The ECG changes indicate slowing of electrical conduction through the atria, atrioventricular node, His bundle, Purkinje fibers, and ventricles, accompanied by an age-related degenerative process, in which fibrosis affects only the cardiac conduction system. In heritable PCCD, conduction slowing may be attributed to loss-of-function mutations in SCN5A. The mutations are reported to reduce INa by decreasing the expression of channel proteins on the sarcolemma, expressing non-functional channels, or altering the gating properties (e.g., delayed activation, earlier inactivation, enhanced slow inactivation, or slower recovery from inactivation) [63, 67, 75, 93]. These loss-of-function mechanisms are also described for BrS-linked SCN5A mutations (Fig. 7). Moreover, a single SCN5A mutation may cause PCCD alone (isolated) or PCCD combined with BrS (overlap syndrome) [35, 73]. This suggests that, in addition to INa reduction, which is enough to cause conduction slowing in PCCD or BrS, other factors are required for the development of the typical right-precordial ST segment elevation in BrS.
Fig. 8.
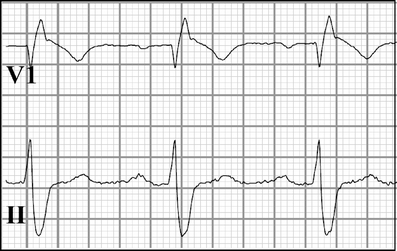
Prolonged conduction parameters (P wave, PR and QRS intervals), and right bundle branch block in an individual with progressive cardiac conduction disease
Whether the age-dependent fibrosis of the conduction system is a primary degenerative process in PCCD, or a physiologic process that is accelerated by INa reduction remains to be investigated. However, the latter is suggested by histological studies, in which fibrosis was found in cardiac biopsies of BrS patients [24], and animal studies in heterozygous SCN5A knockout mice (50% INa reduction), in which conduction defects and myocardial fibrosis that deteriorated progressively with age were observed [65]. So far, no molecular therapy for PCCD exists and the implantation of a pacemaker is the most effective treatment.
PCCD and mutations in sodium channel regulatory proteins
Two loss-of-function mutations in SCN1B have been identified in patients with PCCD who carried no mutation in SCN5A. One mutation resulted in truncated β1-subunits that failed to interact with Nav1.5 proteins, to increase INa densities and to alter the gating properties as normal β1-subunits did. The other mutation decreased the ability of normal β1-subunits to increase INa densities and to facilitate the activation of the co-expressed sodium channels [89]. Interestingly, normal and mutant β1-subunits were expressed to a higher degree in the Purkinje fibers than in the ventricles. This indicates that INa reduction in Purkinje fiber myocytes may underlie prolongation of PR and QRS intervals, and right or left bundle branch block in PCCD.
Dilated cardiomyopathy
DCM is characterized by ventricular dilatation and impaired systolic function, which may proceed into congestive heart failure. Although DCM is a final common pathway of various acquired diseases, up to 50% of cases are reported to be idiopathic (i.e., without any obvious aetiological trigger). Approximately 20% of idiopathic DCM cases display familial prevalence, and have been linked to mutations in various genes that encode proteins involved in the contractile apparatus and the cytoskeleton. Initially, linkage analysis in a large family with DCM mapped the disease locus to a region on the short arm of chromosome 3 (3p22-p25), which harbors the SCN5A gene [55]. Afterwards, a missense mutation in SCN5A was associated with the disease phenotype in this family [47]. Since then, several other missense and truncation mutations in SCN5A have been linked to DCM [25, 53, 56]. Remarkably, most of these mutations are found in patients who display multiple phenotypes, including DCM, sinoatrial node dysfunction, atrial flutter, atrial fibrillation, atrioventricular block, bundle branch block, ventricular tachycardia, and/or ventricular fibrillation. When expressed heterologously, DCM-linked SCN5A mutations usually do not disrupt the sarcolemmal expression of channel proteins, but cause diverse loss-of-function and gain-of-function changes in their gating properties (e.g., earlier or delayed activation, earlier inactivation, disrupted fast inactivation, and increased INaL or window current) [25, 53]. Although it remains unclear how such divergent gating changes lead to DCM, it is speculated that SCN5A mutations in DCM disrupt the interactions between cardiac sodium channels and intracellular (or extracellular) proteins that are essential for normal cardiomyocyte structure and architecture. However, it must be noted that mutations linked to DCM cases with conduction defects often cause INa reduction. This is supported by reduced SCN5A gene transcription and INa densities in a mouse model with DCM that also displayed prolonged conduction parameters [28]. No molecular therapy for DCM exists, and the standard therapy for congestive heart failure is applied.
Sick sinus syndrome
SSS encompasses various forms of arrhythmia that result from sinoatrial node dysfunction. Arrhythmias may be transient, are diagnosed electrocardiographically, and include sinus bradycardia, sinus arrest, and atrial tachycardia–bradycardia syndrome (i.e., atrial tachycardia alternating with episodes of sinus bradycardia; Fig. 9). Patients may suffer from syncope and require lifelong pacemaker therapy. Heritable SSS is associated with loss-of-function mutations in SCN5A, and often linked to compound heterozygous mutations in patients with severe symptoms at relatively young age [14]. INa reduction results from decreased sarcolemmal expression of mutant channel proteins, expression of non-functional channels, and altered gating properties (e.g., delayed activation, earlier inactivation; Fig. 7) [14, 37]. Not surprisingly, SSS may manifest concomitantly with other phenotypes that are linked to SCN5A loss-of-function mutations (i.e., BrS, PCCD) [37, 44, 73].
Fig. 9.
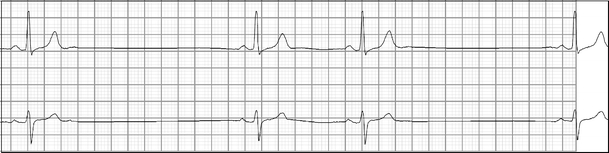
Sinus arrest in an individual with sick sinus syndrome. The patient also suffered from sinus bradycardia
Pacemaker cells in the sinoatrial node reach the voltage range for the sodium window current during action potential phase 4. The small inward window current contributes to the gradual depolarization of the sarcolemma of these cells, and allows spontaneous action potential generation, which underlies cardiac automaticity. In addition to their contribution to cardiac pacemaker activity, sodium channels also play an essential role in the propagation of action potentials from the central area of the sinoatrial node through its peripheral regions to the surrounding atrial muscle [37, 84]. Experimental studies in heterozygous SCN5A knockout mice suggest that the main mechanism of INa reduction to cause SSS indeed involves reduced automaticity, and conduction slowing or blocking of action potentials from the sinoatrial node to the surrounding atrial muscle [36]. Interestingly, LQT-3 patients with SCN5A gain-of-function mutations may also suffer from sinus bradycardia and sinus arrest. Computer simulation models showed that Isus in pacemaker cells may decrease sinus rate and induce sinus arrest by delaying the repolarization and prolonging action potential durations [83].
Atrial fibrillation
AFib is the most prevalent clinically relevant cardiac arrhythmia, and is characterized by a disorganized electrical activation of the atria. It usually affects elderly patients with structural heart disease. However, AFib may also occur in young patients with structurally normal hearts (i.e., lone AFib), and display a familial occurrence. SCN5A mutations were first linked to lone AFib in patients with DCM [56]. Next, a common polymorphism (H558R) in SCN5A was found more often in patients with lone AF than matched controls. Of note, H558R is known to reduce INa in heterologous systems [18]. Subsequently, an SCN5A loss-of-function mutation was identified in a family with lone AFib [23]. It was speculated that INa reduction may predispose to AFib by slowing the electrical conduction velocity. Conduction slowing is an essential determinant for maintaining reentrant excitation waves in the atria. This mechanism is supported by an increased prevalence of lone AFib, accompanied with prolonged atrial conduction intervals (e.g., P wave duration), in BrS patients [34]. Nevertheless, two SCN5A gain-of-function mutations have also been linked to lone AFib. Interestingly, both mutations were found to induce gain-of-function by delaying channel inactivation [38, 43]. In one mutation, larger INa densities corresponded with increased atrial excitability in mutation carriers [43]. The other mutation induced spontaneous action potential formation when expressed in atrial cardiomyocytes [38]. Thus, loss-of-function mutations in SCN5A predispose to AFib by facilitating the maintenance of reentrant excitation waves, while gain-of-function mutations may initiate AFib by increasing atrial excitability.
Although both gain-of-function mutations discussed above did not prolong QT intervals, another SCN5A gain-of-function mutation has recently been described in a family with LQT-3 and lone AFib. The mutation was shown to induce gain-of-function by causing Isus, suggesting that Isus may promote AFib by prolonging action potential duration and triggering EADs. Importantly, through inhibition of Isus, flecainide not only shortened the QT intervals in the affected family members, but also effectively restored the normal sinus rhythm [12]. In contrast, flecainide therapy in patients with AFib due to SCN5A loss-of-function mutations may be contraindicated, because flecainide also blocks the peak INa, and therefore may aggravate INa reduction in these patients.
AFib and mutations in sodium channel regulatory proteins
Recently, mutations in SCN1B and SCN2B (encoding the β1- and β2-subunits of the cardiac sodium channel, respectively) have been identified in patients with lone AFib. Interestingly, the reported patients with AFib and mutations in SCN1B or SCN2B often displayed an ECG pattern suspect for BrS. When heterologously co-expressed with SCN5A, the mutant β1-subunits failed to increase INa as normal β1-subunits did, and caused delayed channel activation. Mutant β2-subunits also induced loss-of-function, but only by delaying channel activation [88].
Sudden infant death syndrome
SIDS is diagnosed when an infant under the age of 1 year suddenly and unexpectedly dies, and when a detailed review of the clinical history, extensive examination of the death scene, and a complete medical autopsy fail to provide an explanation for the death. Although various exogenous factors are recognized to increase the risk for SIDS (e.g., tobacco and alcohol use by the mother, low socioeconomic status, prone sleeping), higher SIDS rates in some ethnicities suggest a role for genetic factors in the development of SIDS. Initially, this suggestion was reinforced when SIDS was linked to prolonged repolarization intervals as seen in LQTS [71]. This was followed by anecdotal reports linking SIDS to gain-of-function mutations in SCN5A and to mutations that were previously associated with LQT-3 [68, 90]. Subsequently, postmortem genetic testing in a population-based cohort indicated that gain-of-function mutations in SCN5A may be the most prevalent genetic cause of SIDS [8]. SCN5A mutations in SIDS commonly increase Isus, mostly in combination with altered gating properties that result in INa gain-of-function (e.g., earlier activation, delayed inactivation, or increased window current). Interestingly, some SIDS-linked SCN5A mutations display only Isus under acidic conditions, supporting the role of exogenous factors (here acidosis) in the development of SIDS [85]. Less frequently, SCN5A loss-of-function mutations have also been found in infants with SIDS [57]. However, it may be possible that in these patients SIDS represents a severe form of BrS that manifests during early childhood [62].
SIDS and mutations in sodium channel regulatory proteins
Recently, postmortem genetic testing in population-based cohorts identified mutations in CAV3 and GPD1-L as possible causative genetic factors in the development of SIDS [20, 80]. As mentioned earlier, mutations in CAV3 are linked to LQT-9, and mutations in GDPL-1 are found in patients with BrS [39, 81]. When co-expressed heterologously with SCN5A, SIDS-linked mutations in CAV3 or GDPL-1 exerted similar effects on the cardiac sodium channel as their equivalents in LQT-9 or BrS, respectively (i.e., increased Isus in the presence of mutant CAV3 proteins, and decreased peak INa in the presence of mutant GDPL-1 proteins). These data support the hypothesis that SIDS may be a severe form of LQTS or BrS during infancy.
Overlap syndromes
Overlap syndromes involve overlapping clinical symptoms or arrhythmias of various arrhythmogenic diseases (i.e., sodium channelopathies). The designation is also used when a mutation causes various arrhythmogenic phenotypes in different families or members of one family. Not surprisingly, SCN5A loss-of-function mutations have often been associated with overlapping phenotypes of BrS and PCCD or BrS and SSS [15, 35, 42, 44, 73, 83]. Since the underlying molecular mechanisms of these diseases implicate INa reduction, it is plausible that the clinical phenotype is determined by the degree of INa reduction, the nature of altered gating properties, the presence of exogenous factors (e.g., electrolyte imbalance, drugs, hormones, body temperature, subclinical cardiac structural changes) and co-inherited genetic variants (e.g., polymorphisms in SCN5A or genes encoding regulatory proteins).
More surprisingly, some SCN5A mutations cause symptoms of both LQT-3 (INa gain-of-function) and BrS (INa loss-of-function) in members of one family, or LQT-3 in one family and BrS in another [15, 26, 42]. Although exogenous and genetic factors discussed before may play a role, such mutations are believed to alter gating properties in a manner that results in both INa gain-of-function and loss-of-function. For example, the insertion of an aspartic acid residue at position 1795 of the Nav1.5 protein (1795insD) was the first mutation found in a multigenerational family with ECG signs of both LQT-3 and BrS [15]. While QT interval prolongation was found to be caused by an increased Isus, BrS was shown to be the result of delayed activation, earlier inactivation, and slower recovery from inactivation of the mutant channels [82]. Interestingly, while SCN5A mutations that are linked to LQT-3 often lead to increased Isus levels, mutations linked to overlap syndromes (LQT-3 and BrS with or without SSS) usually also induce earlier inactivation (i.e., inactivation at more negative membrane potentials) and enhanced tonic block by flecainide [42]. Finally, administration of sodium channel blocking drugs may induce typical ECG signs of BrS in patients with LQT-3, limiting the use of such drugs to inhibit Isus and restore delayed repolarization in these patients [61].
Conclusion
Cardiac sodium channelopathies that are described in this review emphasize the importance of INa for normal cardiac electrical activity. The association of most channelopathies to mutations not only in SCN5A but also in genes encoding the β-subunits or regulatory proteins indicates that, for normal functioning of the cardiac sodium channel, the contribution of various channel subunits is required. Most of our understanding of the molecular mechanisms of cardiac sodium channelopathies originates from experimental studies in heterologous expression system and transgenic mouse models. Although these models have greatly increased our knowledge of the structure and function of the cardiac sodium channel in normal hearts, and the nature and role of INa dysfunction in diseased hearts, they have by and large failed to explain the mechanism of different phenotypes or diseases caused by one single mutation in SCN5A. Since the native environment of cardiomyocytes is absent, the effects of intracellular and extracellular molecules on the function of cardiac sodium channels are greatly lost in heterologous expression models. Although this shortcoming is remedied in transgenic mouse models, the absence of environmental factors and co-inherited genetic variants (e.g., polymorphisms, mutations in non-coding genetic regions) in mouse models remains a limitation. These limitations compel careful interpretation of the experimental data and their translation into the patient phenotypes, and make it clear that future research is needed to design more appropriate expression systems (e.g., cardiomyocytes derived from induced pluripotent stem cells). Future research is also needed to discover novel gene-specific and mutation-specific pharmacological therapies in the management of cardiac sodium channelopathies. Currently, molecular therapy for cardiac sodium channelopathies due to INa loss-of-function is lacking, and the efficiency of sodium channel blocking drugs to restore QT intervals in patients with LQT-3 seems to be limited to mutations that induce earlier inactivation of the sodium channels.
Acknowledgments
Conflict of interest
The authors have declared that no conflict of interest exists.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Grant support
Dr. Tan was supported by the Netherlands Organization for Scientific Research (NWO, grant ZonMW Vici 918.86.616).
References
- 1.Abriel H, Kass RS. Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins. Trends Cardiovasc Med. 2005;15:35–40. doi: 10.1016/j.tcm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Albert CM, Nam EG, Rimm EB, Jin HW, Hajjar RJ, Hunter DJ, MacRae CA, Ellinor PT. Cardiac sodium channel gene variants and sudden cardiac death in women. Circulation. 2008;117:16–23. doi: 10.1161/CIRCULATIONAHA.107.736330. [DOI] [PubMed] [Google Scholar]
- 3.Amin AS, de Groot EAA, Ruijter JM, Wilde AA, Tan HL. Exercise-induced ECG changes in Brugada syndrome. Circ Arrhythmia Electrophysiol. 2009;2:531–539. doi: 10.1161/CIRCEP.109.862441. [DOI] [PubMed] [Google Scholar]
- 4.Amin AS, Meregalli PG, Bardai A, Wilde AA, Tan HL. Fever increases the risk for cardiac arrest in the Brugada syndrome. Ann Intern Med. 2008;149:216–218. doi: 10.7326/0003-4819-149-3-200808050-00020. [DOI] [PubMed] [Google Scholar]
- 5.Amin AS, Tan HL, Wilde AAM. Cardiac ion channels in health and disease. Heart Rhythm. 2009 doi: 10.1016/j.hrthm.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Amin AS, Verkerk AO, Bhuiyan ZA, Wilde AA, Tan HL. Novel Brugada syndrome-causing mutation in ion-conducting pore of cardiac Na+ channel does not affect ion selectivity properties. Acta Physiol Scand. 2005;185:291–301. doi: 10.1111/j.1365-201X.2005.01496.x. [DOI] [PubMed] [Google Scholar]
- 7.Antzelevitch C. Cellular basis for the repolarization waves of the ECG. Ann N Y Acad Sci. 2006;1080:268–2681. doi: 10.1196/annals.1380.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnestad M, Crotti L, Rognum TO, Insolia R, Pedrazzini M, Ferrandi C, Vege A, Wang DW, Rhodes TE, George AL, Jr, Schwartz PJ. Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation. 2007;115:361–367. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 9.Balser JR. The cardiac sodium channel: gating function and molecular pharmacology. J Mol Cell Cardiol. 2001;33:599–613. doi: 10.1006/jmcc.2000.1346. [DOI] [PubMed] [Google Scholar]
- 10.Bankston JR, Kass RS. Molecular determinants of local anesthetic action of beta-blocking drugs: implications for therapeutic management of long QT syndrome variant 3. J Mol Cell Cardiol. 2009 doi: 10.1016/j.yjmcc.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benhorin J, Taub R, Goldmit M, Kerem B, Kass RS, Windman I, Medina A. Effects of flecainide in patients with new SCN5A mutation: mutation-specific therapy for long-QT syndrome? Circulation. 2000;101:1698–1706. doi: 10.1161/01.cir.101.14.1698. [DOI] [PubMed] [Google Scholar]
- 12.Benito B, Brugada R, Perich RM, Lizotte E, Cinca J, Mont L, Berruezo A, Tolosana JM, Freixa X, Brugada P, Brugada J. A mutation in the sodium channel is responsible for the association of long QT syndrome and familial atrial fibrillation. Heart Rhythm. 2008;5:1434–1440. doi: 10.1016/j.hrthm.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Bennett PB, Yazawa K, Makita N, George AL., Jr Molecular mechanism for an inherited cardiac arrhythmia. Nature. 1995;376:683–685. doi: 10.1038/376683a0. [DOI] [PubMed] [Google Scholar]
- 14.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, Rhodes TH, George AL., Jr Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J Clin Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bezzina C, Veldkamp MW, van Den Berg MP, Postma AV, Rook MB, Viersma JW, van Langen IM, Tan-Sindhunata G, Bink-Boelkens MT, van Der Hout AH, Mannens MM, Wilde AA. A single Na+ channel mutation causing both long-QT and Brugada syndromes. Circ Res. 1999;85:1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 16.Brugada P, Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol. 1992;20:1391–1396. doi: 10.1016/0735-1097(92)90253-J. [DOI] [PubMed] [Google Scholar]
- 17.Charpentier F, Bourgé A, Mérot J. Mouse models of SCN5A-related cardiac arrhythmias. Prog Biophys Mol Biol. 2008;98:230–237. doi: 10.1016/j.pbiomolbio.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Chen LY, Ballew JD, Herron KJ, Rodeheffer RJ, Olson TM. A common polymorphism in SCN5A is associated with lone atrial fibrillation. Clin Pharmacol Ther. 2007;81:35–41. doi: 10.1038/sj.clpt.6100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clancy CE, Tateyama M, Liu H, Wehrens XH, Kass RS. Non-equilibrium gating in cardiac Na+ channels: an original mechanism of arrhythmia. Circulation. 2003;107:2233–2237. doi: 10.1161/01.CIR.0000069273.51375.BD. [DOI] [PubMed] [Google Scholar]
- 20.Cronk LB, Ye B, Kaku T, Tester DJ, Vatta M, Makielski JC, Ackerman MJ. Novel mechanism for sudden infant death syndrome: persistent late sodium current secondary to mutations in caveolin-3. Heart Rhythm. 2007;4:161–166. doi: 10.1016/j.hrthm.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumaine R, Towbin JA, Brugada P, Vatta M, Nesterenko DV, Nesterenko VV, Brugada J, Brugada R, Antzelevitch C. Ionic mechanisms responsible for the electrocardiographic phenotype of the Brugada syndrome are temperature dependent. Circ Res. 1999;85:803–809. doi: 10.1161/01.res.85.9.803. [DOI] [PubMed] [Google Scholar]
- 22.Dun W, Boyden PA. The Purkinje cell; 2008 style. J Mol Cell Cardiol. 2008;45:617–624. doi: 10.1016/j.yjmcc.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellinor PT, Nam EG, Shea MA, Milan DJ, Ruskin JN, MacRae CA. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 2008;5:99–105. doi: 10.1016/j.hrthm.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Frustaci A, Priori SG, Pieroni M, Chimenti C, Napolitano C, Rivolta I, Sanna T, Bellocci F, Russo MA. Cardiac histological substrate in patients with clinical phenotype of Brugada syndrome. Circulation. 2005;112:3680–3687. doi: 10.1161/CIRCULATIONAHA.105.520999. [DOI] [PubMed] [Google Scholar]
- 25.Ge J, Sun A, Paajanen V, Wang S, Su C, Yang Z, Li Y, Wang S, Jia J, Wang K, Zou Y, Gao L, Wang K, Fan Z. Molecular and clinical characterization of a novel SCN5A mutation associated with atrioventricular block and dilated cardiomyopathy. Circ Arrhythmia Electrophysiol. 2008;1:83–92. doi: 10.1161/CIRCEP.107.750752. [DOI] [PubMed] [Google Scholar]
- 26.Grant AO, Carboni MP, Neplioueva V, Starmer CF, Memmi M, Napolitano C, Priori S. Long QT syndrome, Brugada syndrome, and conduction system disease are linked to a single sodium channel mutation. J Clin Invest. 2002;110:1201–1209. doi: 10.1172/JCI15570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haïssaguerre M, Extramiana F, Hocini M, Cauchemez B, Jaïs P, Cabrera JA, Farré J, Leenhardt A, Sanders P, Scavée C, Hsu LF, Weerasooriya R, Shah DC, Frank R, Maury P, Delay M, Garrigue S, Clémenty J. Mapping and ablation of ventricular fibrillation associated with long-QT and Brugada syndromes. Circulation. 2003;108:925–928. doi: 10.1161/01.CIR.0000088781.99943.95. [DOI] [PubMed] [Google Scholar]
- 28.Hesse M, Kondo CS, Clark RB, Su L, Allen FL, Geary-Joo CT, Kunnathu S, Severson DL, Nygren A, Giles WR, Cross JC. Dilated cardiomyopathy is associated with reduced expression of the cardiac sodium channel Scn5a. Cardiovasc Res. 2007;75:498–509. doi: 10.1016/j.cardiores.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 29.Hu D, Barajas-Martinez H, Burashnikov E, Springer M, Wu Y, Varro A, Pfeiffer R, Koopmann TT, Cordeiro JM, Guerchicoff A, Pollevick GD, Antzelevitch C. A mutation in the β3 subunit of the cardiac sodium channel associated with Brugada ECG phenotype. Circ Cardiovasc Genet. 2009;2:270–278. doi: 10.1161/CIRCGENETICS.108.829192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Junttila MJ, Gonzalez M, Lizotte E, Benito B, Vernooy K, Sarkozy A, Huikuri HV, Brugada P, Brugada J, Brugada R. Induced Brugada-type electrocardiogram, a sign for imminent malignant arrhythmias. Circulation. 2008;117:1890–1893. doi: 10.1161/CIRCULATIONAHA.107.746495. [DOI] [PubMed] [Google Scholar]
- 31.Kapplinger JD, Tester DJ, Salisbury BA, Carr JL, Harris-Kerr C, Pollevick GD, Wilde AA, Ackerman MJ. Spectrum and prevalence of mutations from the first 2, 500 consecutive unrelated patients referred for the FAMILION® long QT syndrome genetic test. Heart Rhythm. 2009;6:1297–1303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kies P, Wichter T, Schäfers M, Paul M, Schäfers KP, Eckardt L, Stegger L, Schulze-Bahr E, Rimoldi O, Breithardt G, Schober O, Camici PG. Abnormal myocardial presynaptic norepinephrine recycling in patients with Brugada syndrome. Circulation. 2004;110:3017–3022. doi: 10.1161/01.CIR.0000146920.35020.44. [DOI] [PubMed] [Google Scholar]
- 33.Ko SH, Lenkowski PW, Lee HC, Mounsey JP, Patel MK. Modulation of Nav1.5 by β1-and β3-subunit co-expression in mammalian cells. Pflügers Arch. 2005;449:403–412. doi: 10.1007/s00424-004-1348-4. [DOI] [PubMed] [Google Scholar]
- 34.Kusano KF, Taniyama M, Nakamura K, Miura D, Banba K, Nagase S, Morita H, Nishii N, Watanabe A, Tada T, Murakami M, Miyaji K, Hiramatsu S, Nakagawa K, Tanaka M, Miura A, Kimura H, Fuke S, Sumita W, Sakuragi S, Urakawa S, Iwasaki J, Ohe T. Atrial fibrillation in patients with Brugada syndrome relationships of gene mutation, electrophysiology, and clinical backgrounds. J Am Coll Cardiol. 2008;51:1169–1175. doi: 10.1016/j.jacc.2007.10.060. [DOI] [PubMed] [Google Scholar]
- 35.Kyndt F, Probst V, Potet F, Demolombe S, Chevallier JC, Baro I, Moisan JP, Boisseau P, Schott JJ, Escande D, Le Marec H. Novel SCN5A mutation leading either to isolated cardiac conduction defect or Brugada syndrome in a large French family. Circulation. 2001;104:3081–3086. doi: 10.1161/hc5001.100834. [DOI] [PubMed] [Google Scholar]
- 36.Lei M, Goddard C, Liu J, Léoni AL, Royer A, Fung SS, Xiao G, Ma A, Zhang H, Charpentier F, Vandenberg JI, Colledge WH, Grace AA, Huang CL. Sinus node dysfunction following targeted disruption of the murine cardiac sodium channel gene Scn5a. J Physiol. 2005;567(Pt 2):387–400. doi: 10.1113/jphysiol.2005.083188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei M, Huang CL, Zhang Y. Genetic Na+ channelopathies and sinus node dysfunction. Prog Biophys Mol Biol. 2008;98:171–178. doi: 10.1016/j.pbiomolbio.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Li Q, Huang H, Liu G, Lam K, Rutberg J, Green MS, Birnie DH, Lemery R, Chahine M, Gollob MH. Gain-of-function mutation of Nav1.5 in atrial fibrillation enhances cellular excitability and lowers the threshold for action potential firing. Biochem Biophys Res Commun. 2009;380:132–137. doi: 10.1016/j.bbrc.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 39.London B, Michalec M, Mehdi H, Zhu X, Kerchner L, Sanyal S, Viswanathan PC, Pfahnl AE, Shang LL, Madhusudanan M, Baty CJ, Lagana S, Aleong R, Gutmann R, Ackerman MJ, McNamara DM, Weiss R, Dudley SC., Jr Mutation in glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) decreases cardiac Na+ current and causes inherited arrhythmias. Circulation. 2007;116:2260–2268. doi: 10.1161/CIRCULATIONAHA.107.703330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupoglazoff JM, Cheav T, Baroudi G, Berthet M, Denjoy I, Cauchemez B, Extramiana F, Chahine M, Guicheney P. Homozygous SCN5A mutation in long-QT syndrome with functional two-to-one atrioventricular block. Circ Res. 2001;89:E16–E21. doi: 10.1161/hh1401.095087. [DOI] [PubMed] [Google Scholar]
- 41.Makielski JC, Farley AL. Na+ current in human ventricle: implications for sodium loading and homeostasis. J Cardiovasc Electrophysiol. 2006;17:S15–S20. doi: 10.1111/j.1540-8167.2006.00380.x. [DOI] [PubMed] [Google Scholar]
- 42.Makita N. Phenotypic overlap of cardiac sodium channelopathies: individual-specific or mutation-specific? Circ J. 2009;73:810–817. doi: 10.1253/circj.CJ-09-0014. [DOI] [PubMed] [Google Scholar]
- 43.Makiyama T, Akao M, Shizuta S, Doi T, Nishiyama K, Oka Y, Ohno S, Nishio Y, Tsuji K, Itoh H, Kimura T, Kita T, Horie M. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J Am Coll Cardiol. 2008;52:1326–1334. doi: 10.1016/j.jacc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Makiyama T, Akao M, Tsuji K, Doi T, Ohno S, Takenaka K, Kobori A, Ninomiya T, Yoshida H, Takano M, Makita N, Yanagisawa F, Higashi Y, Takeyama Y, Kita T, Horie M. High risk for bradyarrhythmic complications in patients with Brugada syndrome caused by SCN5A gene mutations. J Am Coll Cardiol. 2005;46:2100–2106. doi: 10.1016/j.jacc.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 45.Mangoni ME, Nargeot J. Genesis and regulation of the heart automaticity. Physiol Rev. 2008;88:919–982. doi: 10.1152/physrev.00018.2007. [DOI] [PubMed] [Google Scholar]
- 46.Matsuo K, Akahoshi M, Seto S, Yano K. Disappearance of the Brugada-type electrocardiogram after surgical castration: a role for testosterone and an explanation for the male preponderance. Pacing Clin Electrophysiol. 2003;26:1551–1553. doi: 10.1046/j.1460-9592.2003.t01-1-00227.x. [DOI] [PubMed] [Google Scholar]
- 47.McNair WP, Ku L, Taylor MR, Fain PR, Dao D, Wolfel E, Mestroni L, Familial Cardiomyopathy Registry Research Group SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110:2163–2167. doi: 10.1161/01.CIR.0000144458.58660.BB. [DOI] [PubMed] [Google Scholar]
- 48.Medeiros-Domingo A, Kaku T, Tester DJ, Iturralde-Torres P, Itty A, Ye B, Valdivia C, Ueda K, Canizales-Quinteros S, Tusié-Luna MT, Makielski JC, Ackerman MJ. SCN4B-encoded sodium channel β4 subunit in congenital long-QT syndrome. Circulation. 2007;116:134–142. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meregalli PG, Wilde AA, Tan HL. Pathophysiological mechanisms of Brugada syndrome: depolarization disorder, repolarization disorder, or more? Cardiovasc Res. 2005;67:367–378. doi: 10.1016/j.cardiores.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Morita H, Wu J, Zipes DP. The QT syndromes: long and short. Lancet. 2008;372:750–763. doi: 10.1016/S0140-6736(08)61307-0. [DOI] [PubMed] [Google Scholar]
- 51.Moss AJ, Zareba W, Schwarz KQ, Rosero S, McNitt S, Robinson JL. Ranolazine shortens repolarization in patients with sustained inward sodium current due to type-3 long-QT syndrome. J Cardiovasc Electrophysiol. 2008;19:1289–1293. doi: 10.1111/j.1540-8167.2008.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Napolitano C, Priori SG, Schwartz PJ, Bloise R, Ronchetti E, Nastoli J, Bottelli G, Cerrone M, Leonardi S. Genetic testing in the long QT syndrome: development and validation of an efficient approach to genotyping in clinical practice. JAMA. 2005;294:2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen TP, Wang DW, Rhodes TH, George AL., Jr Divergent biophysical defects caused by mutant sodium channels in dilated cardiomyopathy with arrhythmia. Circ Res. 2008;102:364–371. doi: 10.1161/CIRCRESAHA.107.164673. [DOI] [PubMed] [Google Scholar]
- 54.Noda T, Shimizu W, Taguchi A, Satomi K, Suyama K, Kurita T, Aihara N, Kamakura S. ST-segment elevation and ventricular fibrillation without coronary spasm by intracoronary injection of acetylcholine and/or ergonovine maleate in patients with Brugada syndrome. J Am Coll Cardiol. 2002;40:1841–1847. doi: 10.1016/S0735-1097(02)02494-4. [DOI] [PubMed] [Google Scholar]
- 55.Olson TM, Keating MT. Mapping a cardiomyopathy locus to chromosome 3p22–p25. J Clin Invest. 1996;97:528–532. doi: 10.1172/JCI118445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olson TM, Michels VV, Ballew JD, Reyna SP, Karst ML, Herron KJ, Horton SC, Rodeheffer RJ, Anderson JL. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otagiri T, Kijima K, Osawa M, Ishii K, Makita N, Matoba R, Umetsu K, Hayasaka K. Cardiac ion channel gene mutations in sudden infant death syndrome. Pediatr Res. 2008;64:482–487. doi: 10.1203/PDR.0b013e3181841eca. [DOI] [PubMed] [Google Scholar]
- 58.Pfahnl AE, Viswanathan PC, Weiss R, Shang LL, Sanyal S, Shusterman V, Kornblit C, London B, Dudley SC., Jr A sodium channel pore mutation causing Brugada syndrome. Heart Rhythm. 2007;4:46–53. doi: 10.1016/j.hrthm.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Postema PG, Wolpert C, Amin AS, Probst V, Borggrefe M, Roden DM, Priori SG, Tan HL, Hiraoka M, Brugada J, Wilde AA. Drugs and Brugada syndrome patients: review of the literature, recommendations, and an up-to-date website (www.brugadadrugs.org) Heart Rhythm. 2009;6:1335–1341. doi: 10.1016/j.hrthm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Potet F, Chagot B, Anghelescu M, Viswanathan PC, Stepanovic SZ, Kupershmidt S, Chazin WJ, Balser JR. Functional interactions between distinct sodium channel cytoplasmic domains through the action of calmodulin. J Biol Chem. 2009;284:8846–8854. doi: 10.1074/jbc.M806871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Priori SG, Napolitano C, Schwartz PJ, Bloise R, Crotti L, Ronchetti E. The elusive link between LQT3 and Brugada syndrome: the role of flecainide challenge. Circulation. 2000;102:945–947. doi: 10.1161/01.cir.102.9.945. [DOI] [PubMed] [Google Scholar]
- 62.Probst V, Denjoy I, Meregalli PG, Amirault JC, Sacher F, Mansourati J, Babuty D, Villain E, Victor J, Schott JJ, Lupoglazoff JM, Mabo P, Veltmann C, Jesel L, Chevalier P, Clur SA, Haissaguerre M, Wolpert C, Le Marec H, Wilde AA. Clinical aspects and prognosis of Brugada syndrome in children. Circulation. 2007;115:2042–2048. doi: 10.1161/CIRCULATIONAHA.106.664219. [DOI] [PubMed] [Google Scholar]
- 63.Probst V, Kyndt F, Potet F, Trochu JN, Mialet G, Demolombe S, Schott JJ, Baró I, Escande D, Le Marec H. Haploinsufficiency in combination with aging causes SCN5A-linked hereditary Lenègre disease. J Am Coll Cardiol. 2003;41:643–652. doi: 10.1016/S0735-1097(02)02864-4. [DOI] [PubMed] [Google Scholar]
- 64.Rivolta I, Abriel H, Tateyama M, Liu H, Memmi M, Vardas P, Napolitano C, Priori SG, Kass RS. Inherited Brugada and long QT-3 syndrome mutations of a single residue of the cardiac sodium channel confer distinct channel and clinical phenotypes. J Biol Chem. 2001;276:30623–30630. doi: 10.1074/jbc.M104471200. [DOI] [PubMed] [Google Scholar]
- 65.Royer A, van Veen TA, Le Bouter S, Marionneau C, Griol-Charhbili V, Léoni AL, Steenman M, van Rijen HV, Demolombe S, Goddard CA, Richer C, Escoubet B, Jarry-Guichard T, Colledge WH, Gros D, de Bakker JM, Grace AA, Escande D, Charpentier F. Mouse model of SCN5A-linked hereditary Lenègre's disease: age-related conduction slowing and myocardial fibrosis. Circulation. 2005;111:1738–1746. doi: 10.1161/01.CIR.0000160853.19867.61. [DOI] [PubMed] [Google Scholar]
- 66.Ruan Y, Liu N, Bloise R, Napolitano C, Priori SG. Gating properties of SCN5A mutations and the response to mexiletine in long-QT syndrome type 3 patients. Circulation. 2007;116:1137–1144. doi: 10.1161/CIRCULATIONAHA.107.707877. [DOI] [PubMed] [Google Scholar]
- 67.Schott JJ, Alshinawi C, Kyndt F, Probst V, Hoorntje TM, Hulsbeek M, Wilde AA, Escande D, Mannens MM, Le Marec H. Cardiac conduction defects associate with mutations in SCN5A. Nat Genet. 1999;23:20–21. doi: 10.1038/12618. [DOI] [PubMed] [Google Scholar]
- 68.Schwartz PJ, Priori SG, Dumaine R, Napolitano C, Antzelevitch C, Stramba-Badiale M, Richard TA, Berti MR, Bloise R. A molecular link between the sudden infant death syndrome and the long-QT syndrome. N Engl J Med. 2000;343:262–267. doi: 10.1056/NEJM200007273430405. [DOI] [PubMed] [Google Scholar]
- 69.Schwartz PJ, Priori SG, Locati EH, Napolitano C, Cantù F, Towbin JA, Keating MT, Hammoude H, Brown AM, Chen LS. Long QT syndrome patients with mutations of the SCN5A and HERG genes have differential responses to Na+ channel blockade and to increases in heart rate. Implications for gene-specific therapy. Circulation. 1995;92:3381–3386. doi: 10.1161/01.cir.92.12.3381. [DOI] [PubMed] [Google Scholar]
- 70.Schwartz PJ, Priori SG, Spazzolini C, Moss AJ, Vincent GM, Napolitano C, Denjoy I, Guicheney P, Breithardt G, Keating MT, Towbin JA, Beggs AH, Brink P, Wilde AA, Toivonen L, Zareba W, Robinson JL, Timothy KW, Corfield V, Wattanasirichaigoon D, Corbett C, Haverkamp W, Schulze-Bahr E, Lehmann MH, Schwartz K, Coumel P, Bloise R. Genotype-phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation. 2001;103:89–95. doi: 10.1161/01.cir.103.1.89. [DOI] [PubMed] [Google Scholar]
- 71.Schwartz PJ, Stramba-Badiale M, Segantini A, Austoni P, Bosi G, Giorgetti R, Grancini F, Marni ED, Perticone F, Rosti D, Salice P. Prolongation of the QT interval and the sudden infant death syndrome. N Engl J Med. 1998;338:1709–1714. doi: 10.1056/NEJM199806113382401. [DOI] [PubMed] [Google Scholar]
- 72.Smits JP, Eckardt L, Probst V, Bezzina CR, Schott JJ, Remme CA, Haverkamp W, Breithardt G, Escande D, Schulze-Bahr E, LeMarec H, Wilde AA. Genotype-phenotype relationship in Brugada syndrome: electrocardiographic features differentiate SCN5A-related patients from non-SCN5A-related patients. J Am Coll Cardiol. 2002;40:350–356. doi: 10.1016/S0735-1097(02)01962-9. [DOI] [PubMed] [Google Scholar]
- 73.Smits JP, Koopmann TT, Wilders R, Veldkamp MW, Opthof T, Bhuiyan ZA, Mannens MM, Balser JR, Tan HL, Bezzina CR, Wilde AA. A mutation in the human cardiac sodium channel (E161K) contributes to sick sinus syndrome, conduction disease and Brugada syndrome in two families. J Mol Cell Cardiol. 2005;38:969–981. doi: 10.1016/j.yjmcc.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 74.Shimizu W, Matsuo K, Kokubo Y, Satomi K, Kurita T, Noda T, Nagaya N, Suyama K, Aihara N, Kamakura S, Inamoto N, Akahoshi M, Tomoike H. Sex hormone and gender difference—role of testosterone on male predominance in Brugada syndrome. J Cardiovasc Electrophysiol. 2007;18:415–421. doi: 10.1111/j.1540-8167.2006.00743.x. [DOI] [PubMed] [Google Scholar]
- 75.Tan HL, Bink-Boelkens MT, Bezzina CR, Viswanathan PC, Beaufort-Krol GC, van Tintelen PJ, van den Berg MP, Wilde AA, Balser JR. A sodium-channel mutation causes isolated cardiac conduction disease. Nature. 2001;409:1043–1047. doi: 10.1038/35059090. [DOI] [PubMed] [Google Scholar]
- 76.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 77.Ueda K, Valdivia C, Medeiros-Domingo A, Tester DJ, Vatta M, Farrugia G, Ackerman MJ, Makielski JC. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc Natl Acad Sci USA. 2008;105:9355–9360. doi: 10.1073/pnas.0801294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valdivia CR, Tester DJ, Rok BA, Porter CB, Munger TM, Jahangir A, Makielski JC, Ackerman MJ. A trafficking defective, Brugada syndrome-causing SCN5A mutation rescued by drugs. Cardiovasc Res. 2004;62:53–62. doi: 10.1016/j.cardiores.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 79.Valdivia CR, Ueda K, Ackerman MJ, Makielski JC. GPD1L links redox state to cardiac excitability by PKC-dependent phosphorylation of the sodium channel SCN5A. Am J Physiol, Heart Circ Physiol. 2009;297:H1446–H1452. doi: 10.1152/ajpheart.00513.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Norstrand DW, Valdivia CR, Tester DJ, Ueda K, London B, Makielski JC, Ackerman MJ. Molecular and functional characterization of novel glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) mutations in sudden infant death syndrome. Circulation. 2007;116:2253–2259. doi: 10.1161/CIRCULATIONAHA.107.704627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, Tester DJ, Balijepalli RC, Foell JD, Li Z, Kamp TJ, Towbin JA. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114:2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 82.Veldkamp MW, Viswanathan PC, Bezzina C, Baartscheer A, Wilde AA, Balser JR. Two distinct congenital arrhythmias evoked by a multidysfunctional Na+ channel. Circ Res. 2000;86:E91–E97. doi: 10.1161/01.res.86.9.e91. [DOI] [PubMed] [Google Scholar]
- 83.Veldkamp MW, Wilders R, Baartscheer A, Zegers JG, Bezzina CR, Wilde AA. Contribution of sodium channel mutations to bradycardia and sinus node dysfunction in LQT3 families. Circ Res. 2003;92:976–983. doi: 10.1161/01.RES.0000069689.09869.A8. [DOI] [PubMed] [Google Scholar]
- 84.Verkerk AO, Wilders R, van Borren MM, Tan HL. Is sodium current present in human sinoatrial node cells? Int J Biol Sci. 2009;5:201–204. doi: 10.7150/ijbs.5.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang DW, Desai RR, Crotti L, Arnestad M, Insolia R, Pedrazzini M, Ferrandi C, Vege A, Rognum T, Schwartz PJ, George AL., Jr Cardiac sodium channel dysfunction in sudden infant death syndrome. Circulation. 2007;115:368–376. doi: 10.1161/CIRCULATIONAHA.106.646513. [DOI] [PubMed] [Google Scholar]
- 86.Wang DW, Yazawa K, George AL, Jr, Bennett PB. Characterization of human cardiac Na+ channel mutations in the congenital long QT syndrome. Proc Natl Acad Sci USA. 1996;93:13200–13205. doi: 10.1073/pnas.93.23.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 88.Watanabe H, Darbar D, Kaiser DW, Jiramongkolchai K, Chopra S, Donahue BS, Kannankeril PJ, Roden DM. Mutations in sodium channel β1- and β2-subunits associated with atrial fibrillation. Circ Arrhythmia Electrophysiol. 2009;2:268–275. doi: 10.1161/CIRCEP.108.779181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Watanabe H, Koopmann TT, Le Scouarnec S, Yang T, Ingram CR, Schott JJ, Demolombe S, Probst V, Anselme F, Escande D, Wiesfeld AC, Pfeufer A, Kääb S, Wichmann HE, Hasdemir C, Aizawa Y, Wilde AA, Roden DM, Bezzina CR. Sodium channel β1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest. 2008;118:2260–2268. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wedekind H, Smits JP, Schulze-Bahr E, Arnold R, Veldkamp MW, Bajanowski T, Borggrefe M, Brinkmann B, Warnecke I, Funke H, Bhuiyan ZA, Wilde AA, Breithardt G, Haverkamp W. De novo mutation in the SCN5A gene associated with early onset of sudden infant death. Circulation. 2001;104:1158–1164. doi: 10.1161/hc3501.095361. [DOI] [PubMed] [Google Scholar]
- 91.Wilde AA, Antzelevitch C, Borggrefe M, Brugada J, Brugada R, Brugada P, Corrado D, Hauer RN, Kass RS, Nademanee K, Priori SG, Towbin JA, Study Group on the Molecular Basis of Arrhythmias of the European Society of Cardiology Proposed diagnostic criteria for the Brugada syndrome: consensus report. Circulation. 2002;106:2514–2519. doi: 10.1161/01.CIR.0000034169.45752.4A. [DOI] [PubMed] [Google Scholar]
- 92.Yan GX, Wu Y, Liu T, Wang J, Marinchak RA, Kowey PR. Phase 2 early afterdepolarization as a trigger of polymorphic ventricular tachycardia in acquired long-QT syndrome: direct evidence from intracellular recordings in the intact left ventricular wall. Circulation. 2001;103:2851–2856. doi: 10.1161/01.cir.103.23.2851. [DOI] [PubMed] [Google Scholar]
- 93.Zimmer T, Surber R. SCN5A channelopathies—an update on mutations and mechanisms. Prog Biophys Mol Biol. 2008;98:120–136. doi: 10.1016/j.pbiomolbio.2008.10.005. [DOI] [PubMed] [Google Scholar]


