BiP availability represents a powerful tool for reporting global secretory protein misfolding levels and investigating the molecular events of ER stress in single cells, independent of traditional UPR markers.
Abstract
Accumulation of misfolded secretory proteins causes cellular stress and induces the endoplasmic reticulum (ER) stress pathway, the unfolded protein response (UPR). Although the UPR has been extensively studied, little is known about the molecular changes that distinguish the homeostatic and stressed ER. The increase in levels of misfolded proteins and formation of complexes with chaperones during ER stress are predicted to further crowd the already crowded ER lumen. Surprisingly, using live cell fluorescence microscopy and an inert ER reporter, we find the crowdedness of stressed ER, treated acutely with tunicamycin or DTT, either is comparable to homeostasis or significantly decreases in multiple cell types. In contrast, photobleaching experiments revealed a GFP-tagged variant of the ER chaperone BiP rapidly undergoes a reversible quantitative decrease in diffusion as misfolded proteins accumulate. BiP mobility is sensitive to exceptionally low levels of misfolded protein stressors and can detect intermediate states of BiP availability. Decreased BiP availability temporally correlates with UPR markers, but restoration of BiP availability correlates less well. Thus, BiP availability represents a novel and powerful tool for reporting global secretory protein misfolding levels and investigating the molecular events of ER stress in single cells, independent of traditional UPR markers.
INTRODUCTION
Maintenance of homeostasis is essential for cell viability. The importance of homeostatic regulation is evident from the array of cellular pathways evolved to detect and respond to cellular stresses including oxidative damage, starvation, and the accumulation of misfolded proteins. Studies of misfolded protein stress can often be divided into two broad categories: 1) investigation of specific misfolded proteins and 2) detection of activation of stress pathway components. Although such approaches have provided valuable insights, neither approach evaluates the global levels of protein misfolding or the biophysical changes in the cellular environment that distinguish stress and homeostasis. Rephrased, one can ask, what does misfolded protein stress “look” like at the molecular level in cells? The answer to this question will help define the extent of stress and impact the mechanisms by which the cell can restore homeostasis.
In the homeostatic endoplasmic reticulum (ER), a constant influx of nascent secretory proteins (∼0.1–1 million per minute per cell) presents a significant challenge for correct protein folding and quality control (QC; Alberts et al., 1994). Multiple families of ER chaperones directly assist secretory protein folding and QC (Kleizen and Braakman, 2004). The specialized folding environment of the ER can substantially remodel secretory proteins with posttranslational modifications, including disulfide bonds and N-linked glycosylation (Anken et al., 2005). When such modifications are inhibited or proteins contain misfolding mutations, ER chaperones bind and retain misfolded secretory proteins to help prevent proteins from aggregating into large insoluble complexes (Ma and Hendershot, 2004). Chaperones assist in refolding the proteins and can also help direct failed proteins to the ER-associated degradation (ERAD) machinery (Brodsky et al., 1999; Vembar and Brodsky, 2008).
When misfolded secretory protein levels exceed the capacity of the ER QC machinery, an adaptive stress pathway termed the unfolded protein response (UPR) can be induced (Rutkowski and Kaufman, 2004; Bernales et al., 2006; Ron and Walter, 2007). In metazoan cells, the UPR decreases the secretory protein burden, enhances protein-folding capacity of the ER, and promotes misfolded protein degradation (Meusser et al., 2005; Ron and Walter, 2007). However, what happens to ER luminal crowdedness (viscosity) leading up to and during misfolded protein stress remains unclear. For example, do chaperone-misfolded protein complexes obstruct the ER lumen?
During homeostasis, the luminal environment of the ER significantly impacts chaperone availability. The crowded environment (100 mg/ml protein) and the complex three-dimensional architecture of tubules and cisterna create a highly tortuos space. The combined effect decreases diffusion and hence the “availability” of molecules by up to threefold relative to that of the cytoplasm (Dayel et al., 1999; Siggia et al., 2000; Sbalzarini et al., 2005). The decreased mobility of molecules in the ER increases the challenge of chaperones to encounter unfolded nascent peptides before the peptides misfold. Chaperone availability will depend on chaperone concentration, substrate concentration, duration of association, and the mobilities (measured by diffusion coefficients) of the chaperones and substrates. Mobile chaperones must be available at sufficient concentrations and diffusion coefficients to encounter client proteins in the window of time needed to prevent off pathway folding. Some ER chaperones, including binding immunoglobulin protein (BiP), approach millimolar concentrations (Guth et al., 2004; Gilchrist et al., 2006). The mobility parameters of most chaperones or substrates have not been established.
In homeostatic cells, the ER lumen appears to be fluid. Live cell photobleaching experiments have revealed that small proteins, such as an ER-localized green fluorescent protein (ER-GFP), diffuse freely in the ER lumen (Dayel et al., 1999; Snapp et al., 2004, 2006). Some protein misfolding stresses including brefeldin A, hypertonicity, and low doses (1 mM) of dithiothreitol (DTT) have been acutely applied to the ER lumen and either have no significant effect or affect only modified luminal proteins, such as a glycosylated ER-GFP (Dayel et al., 1999; Nagaya et al., 2008). The molecular and environmental changes caused by acute inducers of global protein misfolding, such as tunicamycin (Tm) and high doses (5–10 mM) of DTT, have not been extensively investigated in the ER lumen. It remains unclear if the effects of distinct misfolded protein stresses on the ER share common biophysical features.
Misfolded protein accumulation correlates with UPR activation, but it has not been established whether the activation state of the UPR reflects the level of misfolded proteins in cells (Credle et al., 2005; Zhou et al., 2006; Kimata et al., 2007). Measurement of global levels of misfolded proteins in cells remains a major technical challenge. In addition, not all misfolded secretory proteins activate the UPR. For example, the α1-antitrypsin Z mutant aggregates in hepatocytes, but does not trigger the UPR (Hidvegi et al., 2005). Thus, measurement of the levels of ER-retained secretory proteins would not necessarily indicate ER stress. What distinguishes a UPR stress from “mere” misfolded protein accumulation may relate to the degree of BiP availability (Bertolotti et al., 2000; Credle et al., 2005; Oikawa et al., 2009).
To restore homeostasis and increase the folding capacity of the ER, the UPR increases expression of ER chaperones, especially the essential Hsp70 family member BiP (Yoshida et al., 1998; Travers et al., 2000; Lee et al., 2003). BiP plays a key role in secretory protein QC (Brodsky et al., 1995; Gething, 1999; Kabani et al., 2003; Hendershot, 2004; Kang et al., 2006). In metazoans, BiP regulates the UPR by binding the UPR stress sensors ATF6, PERK, and IRE1. Release from the sensors leads to activation of UPR signaling and effector pathways (Bertolotti et al., 2000; Shen et al., 2002; Kimata et al., 2004; Oikawa et al., 2009). During both homeostasis and ER stress, BiP binds exposed hydrophobic domains of nascent and misfolded proteins to prevent aggregation and helps retain incompletely folded proteins in the ER (Haas, 1994; Gething, 1999). BiP can bind secretory protein domains that fail to be posttranslationally modified with N-linked sugars or disulfide bonds (Machamer et al., 1990; de Silva et al., 1993; Lodish and Kong, 1993; Molinari and Helenius, 2000). After global misfolded protein stress induced by treatments such as DTT (10 mM for 30 min) and Tm (2 μg/ml for 8 h), a substantial fraction of BiP incorporates into high-molecular-weight and insoluble complexes (Marciniak et al., 2004; Kang et al., 2006). Taken together, BiP is a generalist chaperone that can bind a variety of substrates under an array of conditions and is therefore an excellent candidate for a chaperone whose occupancy reflects levels of incompletely folded or misfolded proteins. In this study, we investigated how BiP occupancy differs between homeostasis and stress in single live cells.
A related issue concerns what happens to the availability of ER chaperones as misfolded proteins accumulate. In the crowded ER lumen, protein folding efficiency depends on the availability of chaperones (Ellis, 2001). During stress, are chaperone-bound substrates partitioned within subdomains of the ER? Are complexes of misfolded proteins mobile? Immobilization of misfolded proteins would impact the mechanism of clearance to restore homeostasis. Degradative machinery must be able to encounter and interact with the immobilized proteins. Imaging studies in homeostatic cells indicate at least some ER chaperones, such as calreticulin and GRP94, can readily diffuse throughout the ER lumen (Snapp et al., 2006; Ostrovsky et al., 2009).
To better understand the physical differences between homeostasis and misfolded protein stress in the ER, we first asked how the overall ER lumen changes. Then, we directly examined the unfolded protein burden in live homeostatic and stressed cells.
MATERIALS AND METHODS
Drugs
DTT (Fisher Scientific, Pittsburgh, PA) was dissolved in distilled H2O in a 1 M stock and Tm (Calbiochem, La Jolla, CA) was dissolved in DMSO as a 5 mg/ml stock and used at the concentrations and times indicated.
Plasmid Constructions
Hamster BiP cDNA (from Amy Lee at the University of Southern California) was PCR amplified with the following primers: forward: 5′ GATCAGATCTACCATGAAGTTCCCTATG and reverse: 5′ GATCGGATCCCCTTCTGATGTATCCTC, to remove the COOH-terminal KDEL sequence and add BamHI and BglII sites for ease of subcloning into a monomeric GFP (Snapp et al., 2003b) or mCherry (Shaner et al., 2004) vector based on the Clontech N1-GFP backbone (Palo Alto, CA). Both fluorophores were modified to contain a KDEL sequence at the COOH terminus for localization of BiP to the ER.
ER-mKate2 was constructed by fusing the bovine prolactin signal sequence and the 10 amino acids after the signal cleavage site to the N1-mKate2 vector (MBL International, San Diego, CA). The construct was further modified by PCR to append a KDEL sequence to the COOH terminus of the mKate2 sequence, and the construct was reinserted into the N1-mKate2 vector.
All constructs were confirmed by sequencing. ER-GFP, ER-red fluorescent protein (RFP), and calreticulin-GFP have been previously described (Snapp et al., 2006). Murine immunoglobulin heavy chain GFP (IgHC-GFP) was provided by Linda Hendershot (St. Jude Children's Research Hospital). All constructs were transiently transfected for 16–48 h into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Cells
HepG2, Cos7, Madin-Darby canine kidney (MDCK), U2-OS, and HeLa cells were all grown in RPMI lacking phenol red plus l-glutamine, 10% heat -inactivated fetal bovine serum, and penicillin/streptomycin at 37°C in 5% CO2. Stable BiP-GFP MDCK cells were cultured with 450 μg/ml G418 (Invitrogen). Highly expressing cells transiently transfected for BiP-GFP exhibited intense immobile fluorescent accumulations on the nuclear envelope and adjacent to the nuclear envelope (Supplemental Figure 5A). For this reason, we limited imaging to low to moderate expressing cells that did not exhibit such structures. Using these criteria, we observed no significant quantitative differences in BiP-GFP mobility in transiently or stably transfected cells (Supplemental Figure 5B). For all imaging experiments, cells were grown in eight-well LabTek coverglass chambers (Nunc, Naperville, IL).
Immunofluorescence and Imaging of Live and Fixed Cells
Cells were imaged in phenol red–free RPMI supplemented with 10 mM HEPES, glutamine, and 10% fetal bovine serum. Live cells were imaged on a 37°C environmentally controlled chamber of a confocal microscope system (Duoscan; Carl Zeiss MicroImaging, Thornwood, NY) with a 63×/1.4 NA oil objective and a 489-nm 100-mW diode laser with a 500–550-nm bandpass filter for GFP and a 40-mW 561-nm diode laser with a 565-nm longpass filter for mCherry or mRFP. Composite figures were prepared using Photoshop CS4 and Illustrator CS4 software (Adobe Systems, San Jose, CA).
Photobleaching Analysis
Fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) were performed by photobleaching a small region of interest (ROI) and monitoring fluorescence recovery or loss over time, as described previously (Siggia et al., 2000; Snapp et al., 2003a). Fluorescence intensity plots and D measurements were calculated as described previously (Siggia et al., 2000; Snapp et al., 2003a). To create the fluorescence recovery curves, the fluorescence intensities were converted into a 0–100% scale and were plotted using Kaleidagraph 3.5 (Synergy Software, Reading, PA). p values were calculated using a Student's two-tailed t test in Excel (Microsoft, Redmond, WA) or Prism 5.0 (GraphPad Software, San Diego, CA). The relatively large spread of D values for ER proteins likely reflects differences in ER geometry between cells (Sbalzarini et al., 2005). For this reason, we use the more stringent significance cutoff of p ≤ 0.01 to define differences in distributions as statistically significant in FRAP analyses. Composite figures were prepared using Photoshop and Illustrator CS4 software (Adobe).
Immunoblots and Pulldowns
Total cell lysates for immunoblotting were prepared in 1% SDS, 0.1 M Tris, pH 8.0, using cells in six-well plates at 80–90% confluence. Proteins were separated using 12% Tris-tricine gels, transferred to nitrocellulose, probed with the indicated antibodies, and developed using enhanced chemiluminescent reagents from Pierce (Rockford, IL) and exposed to x-ray film. Antibodies used included anti-GFP (a gift from Ramanujan S. Hegde, National Institutes of Health), anti-phospho-eIF2α (Epitomics, Burlingame, CA), anti-tRFP (Evrogen, Moscow, Russia), horseradish peroxidase–labeled anti-rabbit and anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA), and anti-BiP (BD mouse 610979 or Sigma-Aldrich, St. Louis, MO; ET-21).
For pulldown analyses, cells in six-well plates were washed twice with 1× PBS and lysed with pulldown buffer (1% Triton X-100, 50 mM HEPES, pH 7.4, 100 mM NaCl) containing EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN) and 10 U apyrase or 50 U/ml and 15 mM glucose to deplete cellular ATP to lock BiP-GFP onto substrates. Lysates were clarified for 10 min at maximum speed in a microcentrifuge at 4°C and incubated for 2 h at 4°C with Affi-gel protein A beads (Bio-Rad, Richmond, CA). The beads were washed four times in pulldown buffer and once in distilled water, eluted with SDS-PAGE sample buffer, and analyzed on 12% Tris/glycine minigels, followed by blotting, staining, and development as for immunoblots.
XBP1 Splicing Assay
For detection of XBP1, total RNA was harvested using TRIZol (Invitrogen) according to manufacturer's protocol. Using XBP1-specific primers and Titan One-Tube PCR kit (Roche), unspliced and spliced variants of XBP1 were amplified according manufacturer's protocol, with annealing temperature at 52°C. The PCR products were run on 3% agarose gel and visualized with GelRed (Biotium, Hayward, CA). The gel image was processed using Adobe Photoshop CS4.
The following human XBP1 primers were used: XBP1F: 5′-CCTTGTAGTTGAGAACC-3′; XBP1R: 5′- CTGGGGAAGGGCATTTG-3′.
Statistics
To minimize cell-to-cell variables such as cell cycle stage or contact inhibition, we always selected flat, mononucleate, nonmitotic cells in cultures at between 40 and 70% confluence for analysis. We used a two-tailed Student's t test (Prism 5.0) to compare the different conditions. Variances of data sets were compared using an F-test (Prism) to establish whether to use equal or nonequal variance t tests. Significance was tested using α ≤ 0.01.
RESULTS
Rationale and Experimental Approach
In this study, we wanted to investigate the burden of acute misfolded protein stress on the ER, independent of UPR activation. First, we determined whether or not the viscosity of the ER lumen changes during misfolded protein stress. Second, we asked if the availability of the ER QC machinery, especially BiP, decreases during the acute accumulation of nascent misfolded proteins. Although BiP availability decreases in cellular fractionation experiments (Marciniak et al., 2004; Kang et al., 2006), it remains unclear if all cells are similarly stressed at the same rate and whether stress occurs homogenously throughout the ER or within subdomains. Monitoring BiP in single cells could help resolve these questions.
To explore luminal viscosity and chaperone availability, we expressed fluorescent probes in live cells and used photobleaching techniques to measure changes in probe mobility (Lippincott-Schwartz et al., 2001; Wouters et al., 2001). An inert probe, ER-GFP, reports on general parameters of the ER environment including its crowdedness and interconnectivity, two variables that directly impact ER QC machinery availability. The second probe is a GFP-tagged ER chaperone whose interactions with substrates could be tracked by changes in its diffusion. For these tools to accurately report changes, the ER environment must remain intact and the stressors must not alter our probes. Not all stressors meet these criteria. For example, calcium depletion by A21387 treatment can induce ER fragmentation (Subramanian and Meyer, 1997). For our study, we first identified doses of two commonly used pharmacologic stressors, Tm and DTT, which promote global misfolding of nascent secretory proteins that do not acutely alter or disrupt ER structure (Supplemental Figure 1). DTT predominantly affects denatured and nascent proteins (Tatu et al., 1993). Secretory proteins that lack disulfide bonds and post-ER compartments of the secretory pathway appear to remain functional in DTT (Lodish and Kong, 1993; Tatu et al., 1993). Neither of our probes, ER-GFP, ER-RFP nor BiP contain N-glycosylation consensus sequences nor disulfide bonds that would be affected by the drug treatments (Hendershot et al., 1996). Together, DTT and Tm satisfy our criteria that only newly synthesized proteins will misfold, leaving the preexisting QC machinery intact and capable of responding to the accumulation of misfolded protein under acute treatment conditions. Prolonged treatments (12+ h) represent adaptation by the cell and will be investigated in a future study.
Acute Misfolded Protein Accumulation and Luminal ER Viscosity
The viscosity of the ER lumen during homeostasis in living cells has been previously described through the use of an average sized protein, ER-GFP, which has no known interacting partners and rapidly samples the entire ER lumen (Dayel et al., 1999; Snapp et al., 2006). Functional GFP folds independently of chaperones and exhibits fluorescence only when properly folded (Cubitt et al., 1995). Using the photobleaching technique FRAP, the diffusion of ER-GFP fluorescence can report changes in the ability of an ER protein to sample its environment (Dayel et al., 1999; Snapp et al., 2006). Mobility or availability changes are reflected in the diffusion coefficient, D, which quantitates both changes in the environment and the size of a molecule or an associated complex (Snapp et al., 2003a). In the Stokes Einstein equation, D is inversely proportional to environmental crowdedness (viscosity) multiplied by the hydrodynamic radius (Rh; Einstein, 1905). A doubling of D indicates the molecule can now stochastically sample an area in half the time. Similarly, an increase in protein complex size by a factor of two would decrease D by half. Thus, even modest changes in D can reflect biologically significant changes. Within the ER, decreased ER-GFP mobility would indicate decreased access of nascent or misfolded proteins to ER chaperones. In unstressed cells, ER-GFP rapidly samples the ER, indicating small to moderate sized proteins are readily available for their substrates.
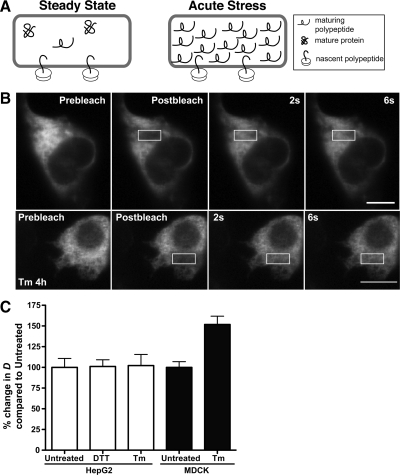
Misfolded proteins and misfolded protein-chaperone complexes accumulate in the ER with acute DTT and Tm stresses (Machamer et al., 1990; de Silva et al., 1993; Kuznetsov et al., 1997; Anken et al., 2005). We asked if misfolded proteins measurably increase ER crowdedness (Figure 1A). Transiently transfected HepG2 cells were analyzed by FRAP of a discrete ROI in a cell with intense laser light, followed by monitoring the movement of unbleached ER-GFP or ER-RFP into the ROI with low-intensity laser light (Figure 1B). FRAP analysis of ER-RFP in steady-state ER yielded a mean D value of 9.1 μm2/s (Figure 1C and Table 1), comparable to previously reported values (Dayel et al., 1999; Snapp et al., 2006). FRAP of Tm-treated cells revealed ER-RFP remains mobile even during acute misfolded protein stress (Figure 1B). Addition of Tm or DTT did not significantly alter mean D values (Figure 1C and Table 1). In contrast, Tm treatment increased ER-RFP mobility significantly in MDCK cells. Increased mobility indicates a decreased viscosity and most likely reflects an increase in ER volume. A recent study from Schuck et al. (2009) reports acute ER stress stimulates ER expansion in yeast. In mammalian cells, UPR stressors can also stimulate ER expansion, but data are not available for relatively short treatment times (Rutkowski et al., 2006). Potentially, properties of HepG2 and MDCK cells including ER geometry, the role of HepG2 hepatocyte–derived cells as professional secretory cells, tolerance for misfolded proteins, and cargo could account for the differences in Tm sensitivity. Regardless of the cell type differences, we can conclude an acute accumulation of misfolded secretory proteins does not generally impair mobility of an inert reporter in the ER lumen and may even lead to increased mobility (Figure 1C).
Figure 1.
Misfolded protein stress and the crowdedness (viscosity) of the ER lumen. (A) Proposed model of ER lumen crowdedness during steady state (left) and acute misfolded protein stress (right). At steady state, the lumen contains few immature polypeptides. Acute protein misfolding could fill the lumen with incompletely folded proteins, which could obstruct free diffusion of luminal ER proteins. (B) FRAP of HepG2 cell expressing ER-RFP, untreated or treated with 5 μg/ml Tm. Scale bar, 10 μm. (C) FRAP D values for HepG2 cells or MDCK cells transiently expressing ER-GFP or ER-RFP, untreated or treated with 5 mM DTT for 1 h or 5 μg/ml Tm for 4 h. The mean D values of the two treatments were compared with untreated cells, were converted to a percentage relative to untreated cells for ease of comparison, and are not significantly different for HepG2 cells, but are significant (p = 0.0002) for MDCK cells. D values are provided in Table 1.
Table 1.
UPR stress decreases BiP mobility
| Construct | DTT | Tm | n | D (μm2/s)a |
|---|---|---|---|---|
| HepG2 cells | ||||
| ER-GFP | Untreated | 18 | 6.6 ± 0.7 | |
| 1 h | 13 | 6.7 ± 0.5 | ||
| ER-RFP | Untreated | 10 | 9.1 ± 0.6 | |
| 4 h | 12 | 9.3 ± 1.2 | ||
| BiP-GFP | Untreated | 26 | 0.60 ± 0.06 | |
| 1 h | 13 | 0.33 ± 0.05b | ||
| MDCK cells | ||||
| ER-RFP | Untreated | 12 | 5.1 ± 0.4 | |
| 4h | 13 | 8.1 ± 0.6b | ||
| BiP-GFP | Untreated | 12 | 0.32 ± 0.03 | |
| 4 h | 13 | 0.16 ± 0.02b | ||
| Cos7 cells | ||||
| ER-RFP | Untreated | 15 | 10.4 ± 0.5 | |
| 1 h | 10 | 10.0 ± 0.9 | ||
| BiP-GFP | Untreated | 15 | 1.1 ± 0.1 | |
| 1 h | 10 | 0.6 ± 0.1 (p = 0.015) | ||
| ER-RFP | Untreated | 14 | 9.1 ± 0.5 | |
| 4 h | 13 | 10.9 ± 0.5 | ||
| BiP-GFP | Untreated | 14 | 1.2 ± 0.2 | |
| 4 h | 13 | 0.7 ± 0.1 (p = 0.0299) | ||
| U2-OS cells | ||||
| ER-RFP | Untreated | 15 | 6.6 ± 0.5 | |
| 4 h | 13 | 7.7 ± 1.1 | ||
| BiP-GFP | Untreated | 15 | 0.37 ± 0.03 | |
| 4 h | 13 | 0.17 ± 0.02b |
HepG2, MDCK, or U2-OS cells were transfected for 20–36 h with BiP-GFP, ER-GFP or ER-RFP treated with 5 mM DTT for 1 h or 5 μg/ml Tm for 4 h (10 mM DTT for 1 h for Cos7), and subjected to FRAP analysis.
a Values are mean ± SEM.
b p ≤ 0.01; two-tailed Student's t test comparing treated with untreated cells.
Previous studies have reported a range of ER-GFP mobilities depending on the type of perturbation of ER homeostasis. Altered osmolarity (Dayel et al., 1999; Nagaya et al., 2008) had little effect on ER-GFP mobility. Protein misfolding treatments had more pronounced effects. Treatments that disrupted the secretory pathway or dilated the ER (brefeldin A for 5 h or Tm 5 μg/ml for 18 h; Dayel et al., 1999; Rutkowski et al., 2006) increased ER-GFP mobility (Dayel et al., 1999; Nehls et al., 2000). Other stresses, such as ATP depletion, which globally disrupts cellular enzymes, profoundly decreases ER-GFP mobility (Nehls et al., 2000). Taken together with the findings in Figure 1, we conclude that misfolded proteins per se do not increase the viscosity of the ER lumen.
Generation and Characterization of a Functional BiP-GFP
Are there any measurable changes in live cells that can specifically report on levels of misfolded proteins? A recently reported, redox-sensitive fluorescent protein revealed an increase in the reducing potential of the normally oxidizing ER during a variety of misfolded protein stresses in yeast (Merksamer et al., 2008). This probe represents an important real time reporter of ER changes related to misfolded secretory protein stress. However, the redox probe ultimately does not directly measure levels of protein misfolding, but rather the environmental consequences of secretory protein misfolding. We sought to create a sensitive and robust probe to detect changes in misfolded secretory protein levels.
The abilities of BiP to bind misfolded proteins and incorporate into large complexes are ideal properties for a live cell diffusion-based reporter (Haas, 1994; Marciniak et al., 2004; Kang et al., 2006). Association with large complexes should either slow or even immobilize BiP within the ER lumen.
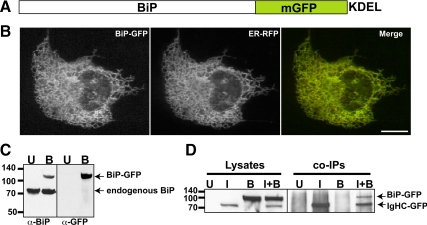
Therefore, we constructed a functional BiP-GFP (Figure 2A). Rational design of BiP-GFP (Figure 2A) mirrored the closely related functional GFP fusion with the essential BiP homolog Kar2p in haploid yeast (Huh et al., 2003). When expressed in cells, our construct correctly colocalizes in a tubular network pattern with ER-RFP (Figure 2B). In an immunoblot BiP-GFP migrates more slowly than wild-type BiP, consistent with the larger size of the BiP-GFP (Figure 2C). To confirm functionality of BiP-GFP, we tested whether the chaperone could interact with a classic BiP substrate, IgHC fused to EGFP. IgHC binds protein A affigel, and we exploited this feature for pulldown assays (Awad et al., 2008). Cells transfected with IgHC-GFP and BiP-GFP, separately or together, were lysed and incubated with protein A, and the bound proteins were probed in immunoblots with anti-GFP. Bound BiP-GFP was only detected when lysates also contained IgHC, consistent with substrate binding (Figure 2D).
Figure 2.
Construction and Characterization of BiP-GFP. (A) Illustration of fusion of hamster BiP in-frame to monomeric GFP followed by a KDEL ER retrieval motif. (B) BiP-GFP colocalizes (yellow in merge panel) with ER-RFP in the ER of a cotransfected Cos7 cell. (C) BiP-GFP (top band) migrates slower than endogenous BiP (bottom band in both untransfected (lane U) and transfected (lane B) cells in an immunoblot of transiently transfected MDCK cell lysate (B lanes) stained with anti-BiP or anti-GFP. (D) BiP-GFP associates with IgHC fused to GFP and is pulled down when coexpressed in cells with IgG heavy chain. Results are displayed in an immunoblot probed with anti-GFP. In the left lanes (1–4), transiently transfected Cos7 cell lysates contain no band (untransfected, (U), IgHC-GFP (I), BiP-GFP (B) or both (I+B). Only I and I+B contain bands of IgHC-GFP, with BiP-GFP in the I+B lane, too. No material is present in U or B pulldown lanes, demonstrating the specificity of the interaction. Molecular-weight-marker positions are indicated to left of blots.
BiP Availability in the Homeostatic ER
First, we asked how available BiP is in the ER lumen during homeostasis with another photobleaching technique termed FLIP (Ellenberg et al., 1997). The technique resembles FRAP, except the ROI is photobleached multiple times, alternating intense bleaches with image acquisition at low laser power. If a molecule is mobile and localizes within a continuous compartment that passes through the ROI, fluorescence will be depleted from that compartment. The extent of fluorescence depletion will depend on the molecule's D value, the length of the FLIP experiment, and whether a subpopulation of the molecule is immobilized by binding or constrained in a discontinuous compartment. FLIP revealed that BiP-GFP fluorescence could be homogenously depleted from the entire ER (Figure 3A). Thus, during homeostasis, the majority of BiP is mobile throughout the ER and little, if any, BiP is sequestered within ER subcompartments. These data do not rule out the presence of small subpopulations of immobilized BiP, such as BiP bound to inactive ER translocation channels (Alder et al., 2005) or to UPR sensors (Rutkowski and Kaufman, 2004). This is because cellular autofluorescence often obscures signals from low densities of GFP (Niswender et al., 1995).
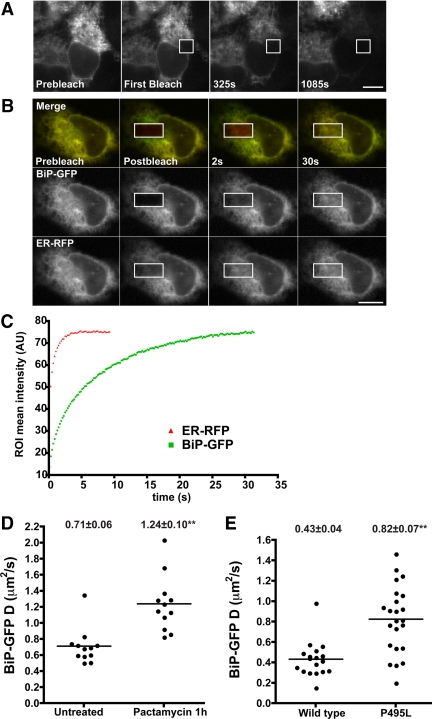
Figure 3.
Molecular availability of BiP-GFP in live cells. (A) Images of a transiently transfected Cos7 cell expressing BiP-GFP before (left panel) and various times during FLIP in the region outlined by the white box. Fluorescence in the bleached cell was depleted uniformly over 18 min. Fluorescence loss was specific, as adjacent cells remained fluorescent. (B) FRAP analysis of transiently transfected Cos7 cells expressing ER-RFP and BiP-GFP. Images were captured before (Prebleach), immediately after (Postbleach), and at times after photobleaching in the white box ROI. Both ER-GFP and BiP-GFP are highly mobile because unbleached fluorescent proteins rapidly diffuse into the bleached regions. Scale bar, 10 μm. (C) Plots of recovery rates reveal that the smaller protein, ER-RFP (red triangles), diffuses more rapidly than BiP-GFP (green squares). (D) MDCK cells stably transfected with BiP-GFP were treated with 0.2 μM pactamycin for 1 h and analyzed by FRAP. D values are plotted, and mean D values ± SEM are indicated above each column. (E) Transiently transfected Cos7 cells expressing BiP-GFP or the P495L mutant were analyzed by FRAP. D values are displayed as in D.
Next, we quantitated how available BiP-GFP is in homeostatic cells. FRAP and diffusional analysis of BiP-GFP could reveal whether the majority of BiP diffuses freely or is incorporated into complexes with substrates. We performed FRAP of BiP-GFP and compared its mean D value relative to ER-GFP (Table 1). The much larger BiP-GFP diffused significantly more slowly than the smaller ER-GFP. In a complimentary experiment, we coexpressed BiP-GFP with ER-RFP. These fluorescently labeled proteins can be imaged simultaneously and photobleached in the same ROI. Given the predicted size differences of BiP-GFP (Rh = 4.1 nm for the closely related DnaK plus Rh = 2.3 nm for GFP for a combined Rh of 6.4 nm; Shi et al., 1996; Hink et al., 2000) and ER-RFP (Rh = 2.3 nm for the closely related mCherry; Frey and Gorlich, 2009), ER-RFP would be predicted to recover much more rapidly. FRAP results in Figure 3, B and C, confirm this prediction, and eventually, BiP-GFP fluorescence intensity recovers (Figure 3C). Based on the Rh value (2.3 nm) and the D value of ER-GFP (6.6 μm2/s; Table 1) and using the Stokes-Einstein relationship, BiP-GFP D is predicted to be 2.4 μm2/s. However, BiP-GFP D is much lower, at 0.6 μm2/s (Table 1). Similarly, the analysis of D values in the Cos7 cells coexpressing BiP-GFP and ER-RFP predicts that the BiP-GFP D value should be 4 μm2/s, instead of the observed 1 μm2/s. The lower D values are consistent with the majority of BiP interacting with substrates. Two additional experiments in MDCK cells confirm this interpretation. First, depletion of newly synthesized BiP substrates by treating cells with the translational inhibitor pactamycin nearly doubles the mean D value of BiP-GFP (Figure 3D). Similarly, introduction of the P495L mutation, which impairs BiP interactions with substrates (Kabani et al., 2003) doubles the mobility of the mutant relative to BiP-GFP (Figure 3E). Both values more closely approximate the predicted D for BiP-GFP (1.9 μm2/s). The remaining differences may reflect association of BiP with cofactors (i.e., ERdj proteins) or other chaperones (Meunier et al., 2002; Weitzmann et al., 2007).
BiP-GFP as a Reporter of Misfolded Protein Stress
It has been impractical to directly measure the concentration of unfolded proteins in vivo. The main problems have been the lack of a tag to identify misfolded proteins and a method for detecting such a tag in live cells. Proteins can misfold in a number of ways ranging from the formation of insoluble aggregates to a subtle disturbance of functional conformation. Pharmacologic perturbations, such as Tm treatment, can inhibit posttranslational modifications that are essential for the folding of some proteins or well tolerated by other proteins. Also, Tm could indirectly promote protein misfolding. For example, a misfolded, normally glycosylated, QC protein could affect folding of a substrate protein, independent of whether the substrate is normally glycosylated. In another example, Gidalevitz et al. (2006) elegantly used temperature-sensitive mutant proteins to detect disruption of the cytoplasmic protein QC machinery. The mutants misfolded, changing distribution and functionality, when a separate unrelated polyglutamine protein misfolded. Therefore, a sensor with the capacity to detect a variety of types of misfolded proteins is needed to directly measure global levels of misfolded proteins within the ER. If it were possible to detect changes in levels of BiP-bound substrates, we should be able to measure changes in levels of misfolded secretory proteins.
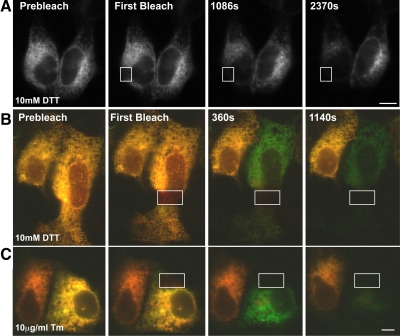
As BiP substrates include integral membrane proteins, nearly immobile translocon-bound proteins, and some large luminal proteins, increasing BiP substrate levels should decrease BiP diffusion and possibly immobilize or sequester BiP within ER subdomains (Suzuki et al., 1991; Kim et al., 1992; Nikonov et al., 2002; Daniels et al., 2003; Wang et al., 2005). To test this hypothesis, cells expressing BiP-GFP were treated with DTT and subjected to FLIP analysis (Figure 4A). BiP-GFP mobility substantially decreased, as evidenced by the much longer time required to deplete BiP fluorescence. We extended our observations to another cell type and stressor. Cos7 cells were transiently transfected with BiP-GFP and ER-RFP and treated with either DTT or Tm and then analyzed by FLIP (Figure 4, B and C). In both cases, ER-RFP remains mobile, and fluorescence was depleted rapidly and homogenously within treated cells. Continued FLIP eventually homogenously depleted the majority of BiP-GFP fluorescence. Thus, under conditions typically used to induce secretory protein misfolding in cells, BiP-GFP does not become trapped or sequestered in a matrix.
Figure 4.
A. Stably transfected MDCK cells expressing BiP-GFP and treated with 10 mM DTT for 1 h and then subjected to FLIP. Note the BiP-GFP fluorescence is homogenously depleted, but much more slowly than in Figure 3A. (B and C) BiP-GFP availability remains homogenous throughout cells during the accumulation of misfolded proteins in the ER. Cos7 cells transiently transfected with BiP-GFP and ER-RFP were treated for 1 h with 10 mM DTT or 10 μg/ml Tm for 5 h and then analyzed by FLIP. Red ER-RFP fluorescence in the bleached cell was depleted uniformly within 6 min, and green BiP-GFP fluorescence was virtually depleted by 19 min. Fluorescence loss was specific, as adjacent cells remained fluorescent. Scale bar, 10 μm.
Misfolded Protein Accumulation and BiP Availability
Next, we asked if BiP-GFP underwent a quantifiable change in availability during the accumulation of misfolded secretory proteins in the ER. DTT or Tm stresses were applied using cells transiently expressing BiP-GFP and ER-RFP, and cells were analyzed by FRAP (Figure 5). To incorporate the data for both BiP-GFP and the inert ER reporter, D values for each protein were combined and plotted on x-y axes. Proteins in untreated cells will produce values that cluster (outlined boxes on plots). On treatment, movement away from the cluster can be viewed for both parameters. If the chaperone-GFP alone changes in mobility, then the effect is chaperone-specific. If both proteins D values increase or decrease in the same direction, then a change in the ER environment could account for the paired shift.
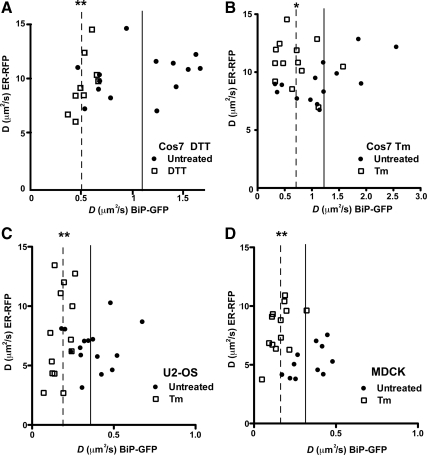
Figure 5.
Comparison of BiP-GFP and ER-RFP mobility in homeostatic and stressed single cells. Plot of D values for both ER-RFP and BiP-GFP in transiently transfected Cos7 (A and B) or U2-OS (C) or MDCK (D) cells expressing both proteins. Cells were either untreated (●) or treated with 10 mM DTT for 1 h or 5 μg/ml Tm for 4 h (□). Boxes have been drawn to highlight differences in distributions. Solid and dashed lines indicate mean BiP-GFP D values for untreated and treated cells, respectively. Means are statistically significant to * p ≤ 0.05 and ** p ≤ 0.01, according to Student's t test analyses.
Using this approach, and consistent with Figure 1, we observed no change in ER-RFP mobility upon treatment with DTT. In contrast, DTT-treated BiP-GFP quantitatively decreased in mobility (Figure 5A). Similar results were observed for Tm treatment (Figure 5B), though decreased BiP-GFP D values varied more broadly, but were still statistically significant compared with untreated cells (Table 1). The mobile fractions of BiP-GFP or ER-RFP did not significantly change with the treatments (83% for untreated vs. 77% for either Tm- or DTT-treated BiP-GFP and 97% for ER-RFP under all conditions). Other cell types, specifically U2-OS and MDCK cells, also displayed statistically significant decreases in BiP-GFP mobility after Tm treatment (Figure 5, C and D). Interestingly, ER-RFP mobility did not follow a general trend. The distribution of ER-RFP D values broadened for U2-OS, and D increased significantly in MDCK cells. Finally, DTT and Tm treatment significantly decreased BiP-GFP mobility in HepG2 and HeLa cells, respectively (Table 1 and Supplemental Figure 2). Together, the FRAP analyses reveal that two acute misfolded protein stresses significantly decrease BiP-GFP mobility in multiple cell types.
Do other ER chaperones also decrease in mobility as misfolded proteins accumulate? The lectin chaperone calreticulin-GFP is similar in size to BiP-GFP, but primarily binds monoglucoses on N-linked sugars (Ware et al., 1995; Rodan et al., 1996; Snapp et al., 2006). Unlike BiP, calreticulin-GFP dramatically increased in mobility after Tm treatment (Supplemental Figure 2A). The results with calreticulin-GFP have two important implications for our interpretation of BiP-GFP mobility. First, these data argue against a possible sieving effect on BiP-GFP mobility. If large protein-chaperone complexes form in the ER, they could permit passage of smaller proteins, such as GFP, but obstruct larger proteins. However, the ability of calreticulin-GFP to diffuse rapidly in the Tm-treated ER indicates proteins similar in size to BiP-GFP (85 and 100 kDa) do not experience barriers and decreased mobility (Supplemental Figure 2B). Second, the changes in chaperone mobility appear to be specific to chaperone substrate levels and/or size. The increase in calreticulin-GFP mobility is consistent with a decrease in substrate availability. The decrease in BiP mobility is consistent with biochemical data indicating a significant fraction of BiP becomes incorporated into large complexes because of increased levels of BiP substrates, decreased rates of release of BiP from substrates, and/or association of BiP with substrates bound to additional chaperones (Kuznetsov et al., 1997; Marciniak et al., 2004; Kang et al., 2006).
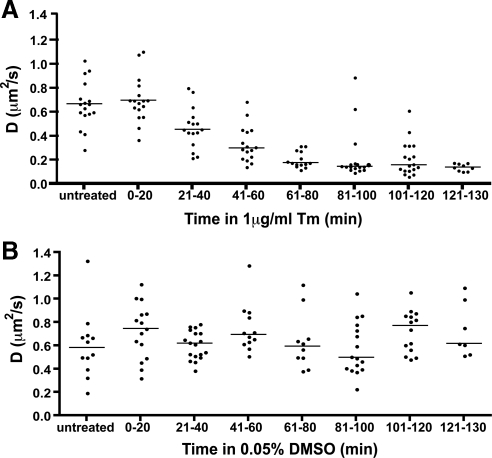
Next, we asked how the distribution of BiP availability temporally decreased in individual stressed cells. Stably transfected BiP-GFP–expressing cells were treated with Tm and assayed by FRAP, and D values were binned (Figure 6A). After 20 min, a significant decrease in BiP-GFP D occurs, continues to decrease through the first 80 min and then remains low. Thus, BiP-GFP mobility decreased up to sixfold in a temporally resolvable manner with increasing levels or sizes of misfolded proteins. Addition of DMSO carrier had no significant effect on BiP-GFP mobility (Figure 6B). One concern for interpreting the experiment is the level of BiP-GFP could be in substantial excess of the native BiP, artificially generating a BiP reserve. One would predict higher levels of BiP-GFP per cell would correlate with relatively higher D values. Arguing against this interpretation is a lack of correlation between intensity of BiP-GFP per cell and D values (Supplemental Figure 3).
Figure 6.
Decrease in BiP availability with misfolded protein stress. (A) D values of single MDCK cells stably expressing BiP-GFP analyzed by FRAP treated with Tm at the indicated times. D values are binned in 20-min intervals. Bars indicate median values. (B) The experiment in D was repeated with only 0.05% DMSO carrier. Bars indicate median values.
Sensitivity of BiP to Acute Misfolded Protein Stress
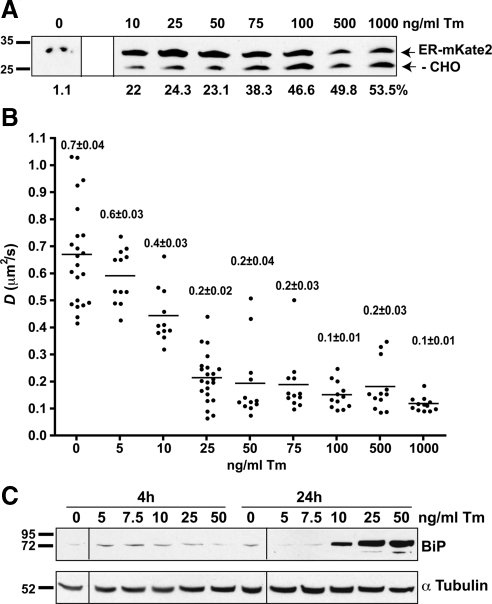
After demonstrating that BiP mobility decreases with conditions that increase the levels of misfolded proteins, we asked what levels of misfolded proteins are needed to decrease BiP mobility. First, we asked how much misfolded substrate can a stressor, such as Tm, generate. A reporter construct, ER-mKate2, which contains a single N-linked glycosylation consensus site, was transfected into MDCK cells, which were then treated with varying amounts of Tm for 4 h. Analysis of an immunoblot of whole cell lysates (Figure 7A) revealed that even very low doses (10 μg/ml) of Tm could produce detectable levels of nonglycosylated proteins. Higher doses more than doubled the amount of nonglycosylated protein.
Figure 7.
Sensitivity of BiP-GFP to changes in misfolded protein levels in live cells. (A) Immunoblot of lysates of MDCK cells transiently transfected with ER-mKate2 and treated with indicated amounts of Tm for 4 h. Blot was probed with anti-tRFP. The normally glycosylated (ER-mKate2) and nonglycosylated (−CHO) forms are indicated by arrows at right of blot. Molecular weight marker positions are indicated to left of blots. The percent of the −CHO band intensity relative to glycosylated proteins is indicated beneath each lane. (B) Plot of D values of single MDCK cells (●) stably expressing BiP-GFP, untreated or treated with the indicated amount of Tm for 4 h. Bars indicate mean values. All treated D means for Tm concentrations of 10 ng/ml or greater are significantly lower than the untreated mean (p ≤ 0.001). All treated means are significantly higher than those of 1000 ng/ml (p ≤ 0.05); n ≥ 11. Values indicate mean ± SEM. (C) Immunoblot of nontransfected MDCK cells treated with indicated amounts of Tm for 4 or 24 h, lysed, and immunoblotted with anti-BiP (top blot) and reprobed with anti-α tubulin (bottom blot). No significant increase in BiP is observed in cells treated with less than 10 ng/ml Tm. Note that A and C blots have each been spliced from single blot images obtained at a single exposure. Discontinuous parts of the images have been marked with lines.
Applying the range of Tm concentrations to MDCK cells stably transfected with BiP-GFP and analyzed by FRAP produced a surprising result. Treatment with 1000 ng/ml Tm robustly decreased BiP-GFP mobility (Figure 7B). On the basis of the immunoblot results in Figure 7A, one might predict changes in BiP-GFP mobility would correlate with Tm dose and detectable nonglycosylated protein levels. Instead, doses as low as 25 ng/ml Tm decreased BiP-GFP mobility to a degree comparable to 1000 ng/ml. Furthermore, despite similar levels of nonglycosylated protein for 10–50-ng/ml treatments, 10 ng/ml produced a significantly higher BiP-GFP mobility.
Two nonexclusive hypotheses could explain these data. First, glycoproteins appear to exhibit differing degrees of susceptibility to Tm (Rutkowski et al., 2006). Thus, ER-mKate2 may not accurately report levels of misfolded protein in the ER. Although one might identify a more susceptible glycoprotein reporter than ER-mKate2, an important problem is highlighted. Would a more sensitive reporter more accurately report the global extent of protein misfolding? This point is directly relevant to the second hypothesis. Tm could indirectly promote misfolding of nonglycosylated proteins, either by preventing folding of newly synthesized unglycosylated QC proteins and depriving the cell of their activity or by depleting the existing pool of QC components and thus promoting misfolding of nonglycosylated proteins. Taken together, we postulate BiP-GFP mobility directly reports the global extent of protein misfolding. In this case, one does not have to monitor select glycoproteins or evaluate all secretory proteins by proteomic approaches to infer the levels of misfolded ER proteins.
The BiP-GFP assay does not require the UPR be activated, though our results in Figure 7C and Supplemental Figure 4 suggest a correlation between BiP-GFP mobility and the degree of UPR activation. BiP is critical for regulating activation of the UPR in mammalian cells (Bertolotti et al., 2000; Oikawa et al., 2009). Mean BiP-GFP availability in cells at 4 h correlated well with the degree of UPR activation achieved by 24 h, as measured by an increase in BiP in untransfected cells. Up-regulation of BiP levels is a relatively slow process compared with BiP-GFP mobility changes. In the final experiment, we directly compare early UPR marker activation and BiP-GFP mobility.
Reversibility of Misfolded Protein Stress on BiP Mobility
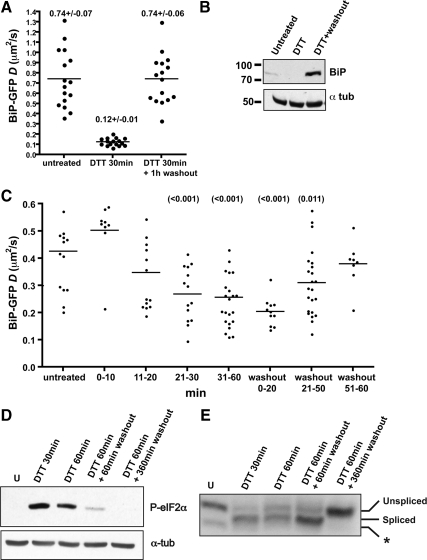
BiP association with nascent proteins is necessarily reversible (Bole et al., 1986; Knittler and Haas, 1992) to allow nascent proteins to complete folding. In contrast, BiP has been reported to bind irreversibly to some unfoldable substrates (Hendershot, 1990; Vanhove et al., 2001). At least some misfolded or aggregated proteins refold and releases from BiP (de Silva et al., 1990; Kim et al., 1992; Tatu et al., 1993). Whether or not the global pool of misfolded proteins rapidly refolds and release from BiP after DTT removal is unclear. We investigated this question by performing FRAP on BiP-GFP–expressing cells untreated, treated with DTT for 30–60 min, or followed by washing out the DTT for 1 h (Figure 8, A and C). As previously observed, DTT treatment rapidly decreased BiP-GFP mobility, consistent with accumulation of misfolded BiP substrates in the ER. However, washout led to a complete recovery of BiP-GFP mobility within 1 h (Figure 8A). BiP-GFP must reversibly bind and release most misfolded substrates in cells. Therefore, BiP-GFP can be considered a dynamic sensor of misfolded protein stress, similar to FRET biosensors that reversibly change conformation depending on the presence or absence of substrate (Bunt and Wouters, 2004). At any given time, the mobility of BiP-GFP represents the combined mobilities of unbound and substrate-bound chaperones. Conditions that alter the ratio of these two populations will be reflected in increases or decreases in BiP-GFP mobility. The DTT washout data suggest the levels of BiP substrates can be depleted quickly in cells after washout. The data do not distinguish between whether the misfolded proteins were degraded, aggregated, or correctly folded. ERAD rates for misfolded proteins do not appear to be fast enough to account for turnover of the misfolded proteins in 1 h. For example, mutant tyrosinase and Null Hong Kong α1-antitrypsin are degraded with 2–3-h halftimes (Svedine et al., 2004; Christianson et al., 2008). In light of the ability of proteins such as VSVG (vesicular stomatitis virus glycoprotein) to rapidly refold (15 min) after DTT washout (Lodish and Kong, 1993; Tatu et al., 1993), we favor the interpretation that the general pool of BiP substrates correctly refolds after DTT washout.
Figure 8.
Decreased BiP-GFP mobility is reversible. (A) Scatter plot of D values of stably transfected MDCK cells expressing BiP-GFP that are untreated, incubated with 10 mM DTT for 30–60 min, or incubated with 10 mM DTT for 30 min, followed by a 1-h washout. Values indicate mean D values ± SEM; n = 16 for each group. (B) Immunoblot of nontransfected cell lysates that were untreated, treated with 10 mM DTT for 30 min, or treated and then washed out for 6 h. Blots were stained with either anti-BiP or anti-α-tubulin (tub), as indicated. (C) Time course of DTT treatment and washout effects on U2-OS cells transiently transfected with BiP-GFP for 16 h. Cells were treated for the indicated time periods with 5 mM DTT and analyzed by FRAP. Bars indicate mean D values. Values in parentheses indicate p values in comparison to D values of the untreated cells using a Student's t test. (D and E) Analysis of UPR markers for U2-OS cells treated with 5 mM DTT for 30 min or 1 h and then washed out with DTT-free media for 1 or 6 h. (D) Immunoblot of the early UPR reporter phosphorylated eIF2α. Samples were lysed, separated by SDS-PAGE, and immunoblotted with anti-P-eIF2α antibody. Equal loading of samples was confirmed by reprobing with anti-α-tubulin. (E) XBP1 splicing in DTT-treated U2-OS cells as described in Materials and Methods. Asterisk indicates a nonspecific band.
Finally, we asked how UPR activity compares to BiP-GFP availability. As with Tm treatment in Figure 7, up-regulation of BiP levels occurs several hours downstream of an ER stress event (Figure 8B). In contrast, early markers, XBP1 splicing by Ire1 and eIF2α phosphorylation by PERK, are rapidly activated in cells and can be detected at the same time that BiP-GFP mobility significantly decreases (30 min; Figures 8, D and E). Similarly, most eIF2α is dephosphorylated by the time BiP-GFP mobility returns to unstressed D values, but spliced XBP1 levels remain persistent for some time after stress resolution.
Together, our results help establish the temporal relationship between misfolded protein stress and UPR status. Events most proximal to activation of UPR sensors can be activated exceptionally rapidly. However, attenuation of different aspects of the UPR does not appear directly related to stress levels. Levels of spliced XBP1 and BiP remained high or became elevated after resolution of the misfolded protein stress. Similarly, Wu et al. and Mori et al. (Yoshida et al., 2006; Wu et al., 2007) demonstrated several markers of UPR increase or are activated for several hours after a DTT washout. Persistent UPR markers and effectors represent the establishment of a new cellular state or “memory” reflecting the exposure to stressor and improved resistance to future stressors (Rutkowski and Kaufman, 2007; Burrill and Silver, 2010).
Conceptually, the UPR must represent a distinct state from the accumulated misfolded secretory protein levels detected by BiP. Markers of the UPR indicate that misfolded proteins did accumulate in a cell, but the downstream effectors of the UPR do not necessarily indicate the presence or the levels of misfolded protein. In contrast, BiP-GFP mobility directly reports on levels of misfolded secretory protein. Taken together, BiP-GFP represents an important new tool for studying the cell biology of secretory protein misfolding.
DISCUSSION
Although numerous assays are available to detect changes in the active state of UPR sensors or levels of UPR effectors, these assays only report whether a response has been initiated, not whether homeostasis has been restored. In this study, we sought to develop new UPR-independent measures of the levels and biophysical effects of misfolded secretory proteins in the ER.
First, we investigated how the ER luminal environment changes between homeostasis and the accumulation of misfolded proteins. No significant changes were observed in luminal crowdedness. Then, we examined how the availability of the key ER chaperone BiP changes spatially and temporally in response to misfolded proteins. We have presented evidence that BiP-GFP availability sensitively reports levels of global secretory protein misfolding, whether protein misfolding is induced with DTT or TM (Figures 4–8). Both inducers of ER stress similarly decrease BiP-GFP availability. Misfolded protein levels could be detected rapidly in single cells (Figures 7A and 8, A and C). Thus, the BiP-GFP assay now provides a sensitive assay of misfolded secretory protein levels in live cells and will open new avenues of investigation.
The mobility of BiP-GFP will depend on a few key parameters. First, levels of BiP may differ significantly between tissue culture fibroblasts and professional secretory cells, such as a plasma B-cell or a pancreatic beta cell. BiP-GFP mobility represents the combined mobilities of the populations of free chaperones and chaperone-substrate complexes. The substrates will vary in size, the number of BiP occupancy sites, and the number of other chaperone occupancy sites and subenvironments (i.e., lumen or membrane). The dynamics of BiP binding and releasing from substrates, as well as availability of cofactors, such as ATP, GRP170, and ERdj proteins, will also influence BiP-GFP mobility (Weitzmann et al., 2007). Thus, although accumulation of misfolded secretory proteins should produce the same general trend in decreased BiP-GFP mobility, the rate and degree of change are likely to vary significantly in different cell types.
Homeostasis and Misfolded Protein Stress
These photobleaching assays have provided a novel insight into the molecular organizational strategies evolved by the ER to maintain and restore homeostasis. Our probes reveal a highly dynamic ER environment and chaperones that remain mobile, albeit with lower D values, as misfolded substrates acutely accumulate. Maintaining chaperone mobility and luminal fluidity could conceivably aid in the restoration of homeostasis. BiP-GFP mobility in cells containing misfolded secretory proteins is consistent with previous reports of the mobility of misfolded secretory proteins, such as GFP-fused to temperature-sensitive VSVG protein or CFTRΔ508-GFP (Nehls et al., 2000; Haggie et al., 2002). Together, our data suggest chaperone-misfolded protein complexes can readily diffuse to ER clearance sites, such as the ERAD machinery. In addition, new chaperones could enter and redistribute throughout the ER, unimpeded, to restore the balance of available QC machinery. In the future, it will be important to examine how the luminal environment changes with prolonged stress and UPR-modulated adaptation (Rutkowski et al., 2006; Schuck et al., 2009).
Supplementary Material
ACKNOWLEDGMENTS
We thank Zuzana Cabartova and Lindsey Costantini for constructing fluorescent protein fusions. We thank the Einstein Analytical Imaging Facility for use of the Zeiss Duoscan. ELS was an Ellison Medical Foundation New Scholar in Aging and is supported by National Institute on Aging Grant R21 1R21AG032544–01 and National Institute of General Medical Sciences (NIGMS) Grant R01GM086530–01. D.E.A. was supported by the National Institutes of Health (NIH) Training Program in Cellular and Molecular Biology and Genetics Grant T32 GM007491 and a National Research Service Award Ruth L. Kirschstein Award Number F31GM089090 from the NIGMS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH. C.W.L. is supported by the NIH Medical Scientist Training Program T32GM07288.
Abbreviations used:
- Crt
calreticulin
- D
diffusion coefficient
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum–associated degradation
- FLIP
fluorescence loss in photobleaching
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- IgHC
immunoglobulin heavy chain
- RFP
red fluorescent protein
- Rh
hydrodynamic radius
- ROI
region of interest
- Tm
tunicamycin
- UPR
unfolded protein response.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-12-1066) on April 21, 2010.
REFERENCES
- Alberts B., Bray D., Lewis J., Raff M., Roberts K., Watson J. D. Molecular Biology of the Cell. New York: Garland Publishing; 1994. [Google Scholar]
- Alder N. N., Shen Y., Brodsky J. L., Hendershot L. M., Johnson A. E. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 2005;168:389–399. doi: 10.1083/jcb.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anken E., Braakman I., Craig E. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev. Biochem. Mol. Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- Awad W., Estrada I., Shen Y., Hendershot L. M. BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc. Natl. Acad. Sci. USA. 2008;105:1164–1169. doi: 10.1073/pnas.0702132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S., Papa F. R., Walter P. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Bole D. G., Hendershot L. M., Kearney J. F. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J. Cell Biol. 1986;102:1558–1566. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L., Goeckeler J., Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky J. L., Werner E. D., Dubas M. E., Goeckeler J. L., Kruse K. B., McCracken A. A. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Bunt G., Wouters F. S. Visualization of molecular activities inside living cells with fluorescent labels. Int. Rev. Cytol. 2004;237:205–277. doi: 10.1016/S0074-7696(04)37005-1. [DOI] [PubMed] [Google Scholar]
- Burrill D. R., Silver P. A. Making cellular memories. Cell. 2010;140:13–18. doi: 10.1016/j.cell.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credle J. J., Finer-Moore J. S., Papa F. R., Stroud R. M., Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubitt A. B., Heim R., Adams S. R., Boyd A. E., Gross L. A., Tsien R. Y. Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- Daniels R., Kurowski B., Johnson A. E., Hebert D. N. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol. Cell. 2003;11:79–90. doi: 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- Dayel M. J., Hom E. F., Verkman A. S. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys. J. 1999;76:2843–2851. doi: 10.1016/S0006-3495(99)77438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva A., Braakman I., Helenius A. Posttranslational folding of vesicular stomatitis virus G protein in the ER: involvement of noncovalent and covalent complexes. J. Cell Biol. 1993;120:647–655. doi: 10.1083/jcb.120.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva A. M., Balch W. E., Helenius A. Quality control in the endoplasmic reticulum: folding and misfolding of vesicular stomatitis virus G protein in cells and in vitro. J. Cell Biol. 1990;111:857–866. doi: 10.1083/jcb.111.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einstein A. Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Ann. Phys. 1905;17:549–560. [Google Scholar]
- Ellenberg J., Siggia E. D., Moreira J. E., Smith C. L., Presley J. F., Worman H. J., Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Frey S., Gorlich D. FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. EMBO J. 2009;28:2554–2567. doi: 10.1038/emboj.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. J. Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K. H., Brignull H. R., Morimoto R. I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Gilchrist A., et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Guth S., Volzing C., Muller A., Jung M., Zimmermann R. Protein transport into canine pancreatic microsomes: a quantitative approach. Eur. J. Biochem. 2004;271:3200–3207. doi: 10.1111/j.1432-1033.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- Haas I. G. BiP (GRP78), an essential hsp70 resident protein in the endoplasmic reticulum. Experientia. 1994;50:1012–1020. doi: 10.1007/BF01923455. [DOI] [PubMed] [Google Scholar]
- Haggie P. M., Stanton B. A., Verkman A. S. Diffusional mobility of the cystic fibrosis transmembrane conductance regulator mutant, delta F508-CFTR, in the endoplasmic reticulum measured by photobleaching of GFP-CFTR chimeras. J. Biol. Chem. 2002;277:16419–16425. doi: 10.1074/jbc.M112361200. [DOI] [PubMed] [Google Scholar]
- Hendershot L., Wei J., Gaut J., Melnick J., Aviel S., Argon Y. Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc. Natl. Acad. Sci. USA. 1996;93:5269–5274. doi: 10.1073/pnas.93.11.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot L. M. Immunoglobulin heavy chain and binding protein complexes are dissociated in vivo by light chain addition. J. Cell Biol. 1990;111:829–837. doi: 10.1083/jcb.111.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot L. M. The ER function BiP is a master regulator of ER function. Mt. Sinai J. Med. 2004;71:289–297. [PubMed] [Google Scholar]
- Hidvegi T., Schmidt B. Z., Hale P., Perlmutter D. H. Accumulation of mutant alpha1-antitrypsin Z in the endoplasmic reticulum activates caspases-4 and -12, NFkappaB, and BAP31 but not the unfolded protein response. J. Biol. Chem. 2005;280:39002–39015. doi: 10.1074/jbc.M508652200. [DOI] [PubMed] [Google Scholar]
- Hink M. A., Griep R. A., Borst J. W., van Hoek A., Eppink M. H., Schots A., Visser A. J. Structural dynamics of green fluorescent protein alone and fused with a single chain Fv protein. J. Biol. Chem. 2000;275:17556–17560. doi: 10.1074/jbc.M001348200. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P. S., Bole D., Arvan P. Transient aggregation of nascent thyroglobulin in the endoplasmic reticulum: relationship to the molecular chaperone, BiP. J. Cell Biol. 1992;118:541–549. doi: 10.1083/jcb.118.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y., Ishiwata-Kimata Y., Ito T., Hirata A., Suzuki T., Oikawa D., Takeuchi M., Kohno K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J. Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y., Oikawa D., Shimizu Y., Ishiwata-Kimata Y., Kohno K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J. Cell Biol. 2004;167:445–456. doi: 10.1083/jcb.200405153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B., Braakman I. Protein folding and quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 2004;16:343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Knittler M. R., Haas I. G. Interaction of BiP with newly synthesized immunoglobulin light chain molecules: cycles of sequential binding and release. EMBO J. 1992;11:1573–1581. doi: 10.1002/j.1460-2075.1992.tb05202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov G., Chen L. B., Nigam S. K. Multiple molecular chaperones complex with misfolded large oligomeric glycoproteins in the endoplasmic reticulum. J. Biol. Chem. 1997;272:3057–3063. doi: 10.1074/jbc.272.5.3057. [DOI] [PubMed] [Google Scholar]
- Lee A. H., Iwakoshi N. N., Glimcher L. H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Snapp E., Kenworthy A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Lodish H. F., Kong N. The secretory pathway is normal in dithiothreitol-treated cells, but disulfide-bonded proteins are reduced and reversibly retained in the endoplasmic reticulum. J. Biol. Chem. 1993;268:20598–20605. [PubMed] [Google Scholar]
- Ma Y., Hendershot L. M. ER chaperone functions during normal and stress conditions. J. Chem. Neuroanat. 2004;28:51–65. doi: 10.1016/j.jchemneu.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Machamer C. E., Doms R. W., Bole D. G., Helenius A., Rose J. K. Heavy chain binding protein recognizes incompletely disulfide-bonded forms of vesicular stomatitis virus G protein. J. Biol. Chem. 1990;265:6879–6883. [PubMed] [Google Scholar]
- Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merksamer P. I., Trusina A., Papa F. R. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–947. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier L., Usherwood Y. K., Chung K. T., Hendershot L. M. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Molinari M., Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- Nagaya H., Tamura T., Higa-Nishiyama A., Ohashi K., Takeuchi M., Hashimoto H., Hatsuzawa K., Kinjo M., Okada T., Wada I. Regulated motion of glycoproteins revealed by direct visualization of a single cargo in the endoplasmic reticulum. J. Cell Biol. 2008;180:129–143. doi: 10.1083/jcb.200704078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls S., Snapp E. L., Cole N. B., Zaal K. J., Kenworthy A. K., Roberts T. H., Ellenberg J., Presley J. F., Siggia E., Lippincott-Schwartz J. Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- Nikonov A. V., Snapp E., Lippincott-Schwartz J., Kreibich G. Active translocon complexes labeled with GFP-Dad1 diffuse slowly as large polysome arrays in the endoplasmic reticulum. J. Cell Biol. 2002;158:497–506. doi: 10.1083/jcb.200201116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender K. D., Blackman S. M., Rohde L., Magnuson M. A., Piston D. W. Quantitative imaging of green fluorescent protein in cultured cells: comparison of microscopic techniques, use in fusion proteins and detection limits. J. Microsc. 1995;180:109–116. doi: 10.1111/j.1365-2818.1995.tb03665.x. [DOI] [PubMed] [Google Scholar]
- Oikawa D., Kimata Y., Kohno K., Iwawaki T. Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp. Cell Res. 2009;315:2496–2504. doi: 10.1016/j.yexcr.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Ostrovsky O., Makarewich C. A., Snapp E. L., Argon Y. An essential role for ATP binding and hydrolysis in the chaperone activity of GRP94 in cells. Proc. Natl. Acad. Sci. USA. 2009;106:11600–11605. doi: 10.1073/pnas.0902626106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodan A. R., Simons J. F., Trombetta E. S., Helenius A. N-linked oligosaccharides are necessary and sufficient for association of glycosylated forms of bovine RNase with calnexin and calreticulin. EMBO J. 1996;15:6921–6930. [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rutkowski D. T., Arnold S. M., Miller C. N., Wu J., Li J., Gunnison K. M., Mori K., Sadighi Akha A. A., Raden D., Kaufman R. J. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski D. T., Kaufman R. J. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Rutkowski D. T., Kaufman R. J. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem. Sci. 2007;32:469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Sbalzarini I. F., Mezzacasa A., Helenius A., Koumoutsakos P. Effects of organelle shape on fluorescence recovery after photobleaching. Biophys. J. 2005;89:1482–1492. doi: 10.1529/biophysj.104.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S., Prinz W. A., Thorn K. S., Voss C., Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shen J., Chen X., Hendershot L., Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shi L., Kataoka M., Fink A. L. Conformational characterization of DnaK and its complexes by small-angle X-ray scattering. Biochemistry. 1996;35:3297–3308. doi: 10.1021/bi951984l. [DOI] [PubMed] [Google Scholar]
- Siggia E. D., Lippincott-Schwartz J., Bekiranov S. Diffusion in inhomogeneous media: theory and simulations applied to whole cell photobleach recovery. Biophys. J. 2000;79:1761–1770. doi: 10.1016/S0006-3495(00)76428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E., Altan-Bonnet N., Lippincott-Schwartz J. Measuring protein mobility by photobleaching GFP-chimeras in living cells. In: Bonifacino J. S., Dasso M., Harford J., Lippincott-Schwartz J., Yamada K., editors. Current Protocols in Cell Biology. New York: John Wiley & Sons; 2003a. Unit 21.21. [DOI] [PubMed] [Google Scholar]
- Snapp E. L., Hegde R. S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 2003b;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E. L., Iida T., Frescas D., Lippincott-Schwartz J., Lilly M. A. The fusome mediates intercellular endoplasmic reticulum connectivity in Drosophila ovarian cysts. Mol. Biol. Cell. 2004;15:4512–4521. doi: 10.1091/mbc.E04-06-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E. L., Sharma A., Lippincott-Schwartz J., Hegde R. S. Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc. Natl. Acad. Sci. USA. 2006;103:6536–6541. doi: 10.1073/pnas.0510657103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K., Meyer T. Calcium-induced restructuring of nuclear envelope and endoplasmic reticulum calcium stores. Cell. 1997;89:963–971. doi: 10.1016/s0092-8674(00)80281-0. [DOI] [PubMed] [Google Scholar]
- Suzuki C. K., Bonifacino J. S., Lin A. Y., Davis M. M., Klausner R. D. Regulating the retention of T-cell receptor α chain variants within the endoplasmic reticulum: Ca2+-dependent association with BiP. J. Cell Biol. 1991;114:189–205. doi: 10.1083/jcb.114.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedine S., Wang T., Halaban R., Hebert D. N. Carbohydrates act as sorting determinants in ER-associated degradation of tyrosinase. J. Cell Sci. 2004;117:2937–2949. doi: 10.1242/jcs.01154. [DOI] [PubMed] [Google Scholar]
- Tatu U., Braakman I., Helenius A. Membrane glycoprotein folding, oligomerization and intracellular transport: effects of dithiothreitol in living cells. EMBO J. 1993;12:2151–2157. doi: 10.1002/j.1460-2075.1993.tb05863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Vanhove M., Usherwood Y. K., Hendershot L. M. Unassembled Ig heavy chains do not cycle from BiP in vivo but require light chains to trigger their release. Immunity. 2001;15:105–114. doi: 10.1016/s1074-7613(01)00163-7. [DOI] [PubMed] [Google Scholar]
- Vembar S. S., Brodsky J. L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Daniels R., Hebert D. N. The cotranslational maturation of the type I membrane glycoprotein tyrosinase: the heat shock protein 70 system hands off to the lectin-based chaperone system. Mol. Biol. Cell. 2005;16:3740–3752. doi: 10.1091/mbc.E05-05-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware F. E., Vassilakos A., Peterson P. A., Jackson M. R., Lehrman M. A., Williams D. B. The molecular chaperone calnexin binds Glc1Man9GlcNAc2 oligosaccharide as an initial step in recognizing unfolded glycoproteins. J. Biol. Chem. 1995;270:4697–4704. doi: 10.1074/jbc.270.9.4697. [DOI] [PubMed] [Google Scholar]
- Weitzmann A., Baldes C., Dudek J., Zimmermann R. The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum—a quantitative approach. FEBS J. 2007;274:5175–5187. doi: 10.1111/j.1742-4658.2007.06039.x. [DOI] [PubMed] [Google Scholar]
- Wouters F. S., Verveer P. J., Bastiaens P. I. Imaging biochemistry inside cells. Trends Cell Biol. 2001;11:203–211. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- Wu J., Rutkowski D. T., Dubois M., Swathirajan J., Saunders T., Wang J., Song B., Yau G. D., Kaufman R. J. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Haze K., Yanagi H., Yura T., Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Oku M., Suzuki M., Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu C. Y., Back S. H., Clark R. L., Peisach D., Xu Z., Kaufman R. J. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.