At least 20 core ribosome proteins are modified by O-GlcNAc. O-GlcNAcase is localized to the nucleolus and O-GlcNAc transferase is excluded from the nucleolus. Both enzymes associate with active polysomes. Overexpression of OGT disrupts ribosomal subunit homeostasis. Data suggest that O-GlcNAc regulates translation and ribosome biogenesis.
Abstract
Protein synthesis is globally regulated through posttranslational modifications of initiation and elongation factors. Recent high-throughput studies have identified translation factors and ribosomal proteins (RPs) as substrates for the O-GlcNAc modification. Here we determine the extent and abundance of O-GlcNAcylated proteins in translational preparations. O-GlcNAc is present on many proteins that form active polysomes. We identify twenty O-GlcNAcylated core RPs, of which eight are newly reported. We map sites of O-GlcNAc modification on four RPs (L6, L29, L32, and L36). RPS6, a component of the mammalian target of rapamycin (mTOR) signaling pathway, follows different dynamics of O-GlcNAcylation than nutrient-induced phosphorylation. We also show that both O-GlcNAc cycling enzymes OGT and OGAse strongly associate with cytosolic ribosomes. Immunofluorescence experiments demonstrate that OGAse is present uniformly throughout the nucleus, whereas OGT is excluded from the nucleolus. Moreover, nucleolar stress only alters OGAse nuclear staining, but not OGT staining. Lastly, adenovirus-mediated overexpression of OGT, but not of OGAse or GFP control, causes an accumulation of 60S subunits and 80S monosomes. Our results not only establish that O-GlcNAcylation extensively modifies RPs, but also suggest that O-GlcNAc play important roles in regulating translation and ribosome biogenesis.
INTRODUCTION
The ribosome is the central component of the translational apparatus. Its function is to decode the nucleotide sequence carried by the mRNA and convert it into an amino acid primary structure by the catalysis of peptide bonds (Marshall et al., 2008). In eukaryotes, ribosomes consist of two different subunits: a 40S small subunit and a 60S large subunit. These two subunits exist as separate pools in the cytosol and when translation is initiated they assemble into an active 80S monosome suitable for elongation of the polypeptide chain (Pestova et al., 2001; Acker and Lorsch, 2008). Ribosomal subunits contain both RNA and protein components. In mammals, the 40S subunit contains 33 different proteins and an 18S rRNA, whereas the 60S subunit is composed of 49 unique polypeptides and three rRNAs: a 5S, a 5.8S, and a 28S (Wool et al., 1995). In eukaryotes, ribosomal proteins (RPs) are assembled around the newly transcribed pre-rRNA within the nucleolus and then exported to the cytoplasm as mature subunits (Boisvert et al., 2007).
The small ribosomal subunit contains the decoding center in which aminoacyl-tRNAs base pair with the corresponding codons in the mRNA (Schluenzen et al., 2000; Wimberly et al., 2000). The peptidyl transferase center is located in the large subunit and rRNA is the sole enzyme responsible for this catalytic activity (Noller et al., 1992; Ban et al., 2000; Nissen et al., 2000; Harms et al., 2001). Ribosomal proteins are thought to have mainly a scaffolding/chaperone role in facilitating the processing and folding of rRNA during biogenesis and stabilizing the mature particle during protein synthesis (Dresios et al., 2006).
Translation factors are additional elements of the protein synthesis machinery that associate transiently with the ribosome (Proud, 2006). In general, they function by coupling ATP or GTP hydrolysis to the conformational rearrangements that occur during ribosomal motion (scanning and translocation; Marshall et al., 2008). It has been well established that global regulation of protein synthesis in eukaryotes is mainly achieved by posttranslational modification (PTM) of translation factors in response to environmental cues (Gebauer and Hentze, 2004). Mammalian ribosomal proteins also bear many different PTMs such as acetylation, methylation, ubiquitination, and phosphorylation among others (Odintsova et al., 2003; Yu et al., 2005). However, the impact of ribosomal protein modification on ribosome assembly, performance, or translation has not been studied. Recent studies suggest that ribosomal protein modification seems to play an important role in the extraribosomal functions of some individual riboproteins (Spence et al., 2000; Mazumder et al., 2003; Ruvinsky et al., 2005).
Several proteins of the translational machinery, including core ribosomal proteins, were identified as O-GlcNAc–modified in global glycoproteomic studies (Khidekel et al., 2007; Wang et al., 2007; Gurcel et al., 2008; Teo et al., 2010). O-GlcNAcylation is a PTM present in certain bacteria and in all metazoans, in which the O-linked monosaccharide β-N-acetylglucosamine cycles dynamically on serine or threonine residues of nuclear and cytoplasmic proteins (Hart et al., 2007). Two highly conserved enzymes catalyze the cycling of O-GlcNAc on proteins: the adding enzyme O-GlcNAc transferase (OGT; uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetyl-glucosaminyltransferase; Haltiwanger et al., 1992), and the removing enzyme β-N-acetylglucosaminidase O-GlcNAcase (OGAse), a neutral hexosaminidase (Dong and Hart, 1994; Gao et al., 2001). O-GlcNAc cycling plays an important role in many fundamental processes of cell physiology. OGT is essential for embryonic stem cell viability and is required for somatic function in several mammalian adult cell types. O-GlcNAcylation is a direct regulator of cellular growth and proliferation, protein turnover, and cellular stress, among others. The role of O-GlcNAc in cellular function is partly mediated by a complex interplay with phosphorylation that is substrate-dependent. O-GlcNAc and phosphate can compete for the same site on a protein. Alternatively, the two modifications can reciprocally occupy different sites (adjacent or distant) on a protein or coexist on another. In addition, each modification regulates the other's cycling enzymes. This cross-talk provides the cell with a mechanism to create great molecular diversity in response to stimuli and fine-tune protein interactions and functions (Hart et al., 2007; Wang et al., 2008b).
Little is known with respect to the functional consequences of the O-GlcNAc modification on translational proteins or on the process of protein synthesis itself. O-GlcNAc seems to play an important role in protein stabilization and cellular protection during stress. O-GlcNAcylation of the eIF2-associated factor p67 (which protects eIF2 from phosphorylation by heme-regulated kinases) prevents p67's degradation and stabilizes the protein allowing its association with eIF2. In contrast, non-O-GlcNAcylated p67 is rapidly degraded, resulting in phosphorylation of eIF2 and translation inhibition (Datta et al., 1988, 1989, 2001; Ray et al., 1992). In a recent study, the O-GlcNAcylation of ribosomal proteins was found to increase rapidly upon arsenite treatment, and OGT was required for stress granule formation during these conditions (Ohn et al., 2008).
In this report, we describe the incidence of the O-GlcNAc modification on purified translational preparations by direct methods. We identify several core ribosomal proteins as substrates for O-GlcNAcylation and map sites of modification on four of these proteins. In addition, we show that OGT and OGAse associate with different subpopulations of ribosomes and these interactions may play roles in ribosome biogenesis and translational regulation.
MATERIALS AND METHODS
Cell Culture and Treatments
HepG2 human hepatoma cells were maintained in minimum essential medium (Mediatech, Herndon, VA) supplemented with 10% vol/vol fetal bovine serum (Gemini Bio-Products, Woodland, CA) and penicillin/streptomycin. HeLa human adenocarcinoma cells and HEK293 human embryonic kidney cells were maintained in Dulbecco's modified Eagle's medium (Mediatech) (20 mM glucose) supplemented with 10% vol/vol fetal bovine serum and penicillin/streptomycin. Neuro2a murine neuroblastoma cells were maintained in Dulbecco's modified Eagle's medium (5 mM glucose) supplemented with 10% vol/vol fetal bovine serum and penicillin/streptomycin. All cell lines were maintained at 37°C in a humidified incubator with 5% CO2.
Glucose deprivation treatment of Neuro2a cells was performed exactly as previously described (Cheung and Hart, 2008). For immunofluorescence experiments nucleolar stress was induced by treating HeLa cells with actinomycin D (Sigma, St. Louis, MO; 0.5 μg/ml final concentration, 1 h), whereas control cells were treated with vehicle (DMSO, Sigma, 1:1000 vol/vol dilution).
Antibodies
For immunoblotting, the following commercially available primary antibodies were used at 1:5000 dilution: O-GlcNAc (CTD110.6, Covance Laboratories, Madison, WI), hemagglutinin (HA; Covance; HA.11), S6 ribosomal protein (2217, Cell Signaling Technology, Beverly, MA), phospho-S6 ribosomal protein (Ser235/236; 2211, Cell Signaling Technology), actin (Sigma), tubulin (Sigma), and fibrillarin (5821, 4566, Abcam, Cambridge, MA). Polyclonal antibodies for OGT (AL-28) and OGAse (345, 346) were raised in rabbit and chicken, respectively, and were also used at a 1:5000 dilution. Horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from GE Healthcare (Piscataway, NJ; rabbit: NA934V, mouse: NA931V) or Sigma (chicken: A 9046 and anti-mouse IgM: A 8786). For immunofluorescence studies, final dilutions were 1:1000 for OGT (AL-28), OGAse (345, 346), and tubulin antibodies, 1:100 vol/vol for S6 ribosomal protein (2317, Cell Signaling Technology) and 1:200 or 1:500 for fibrillarin (5821, 4566). Isotype controls for OGAse were performed with normal chicken IgY (sc-2718, Santa Cruz Biotechnology, Santa Cruz, CA). Fluorescently labeled secondary antibodies Alexa Fluor 647, 568, 546, and 488 were purchased from Invitrogen-Molecular Probes (Eugene, OR).
Plasmid- and Adenovirus-mediated Overexpression
Mammalian expression vectors encoding HA-tagged ribosomal protein S6 wild-type and phosphorylation mutants (HA-S6, HA-S6-Ser235/236S, and HA-S6-Ser235/236D) and HA (empty vector) were a kind gift of Dr. Philippe R. Roux (Université de Montréal, Montréal, QC, Canada; Roux et al., 2007). HEK 293 cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. Cells were infected at a multiplicity of infection of 25 for OGAse and 50 for OGT and GFP adenovirus 48 h before harvesting (Slawson et al., 2005).
Purification of Different Ribosomal Subpopulations and Isolation of Nucleoli
A total ribosome preparation was obtained from rat livers (PelFreez, Rogers, AR) or cells in culture as follows. Livers were thawed in ice-cold phosphate-buffered saline for 30 min and then homogenized in TMK buffer (50 mM; Tris-HCl, pH 7.5, 5 mM MgCl2, 100 mM KCl) supplemented with 1% Triton X-100 (Sigma), 1 μM O-GlcNAc-thiazoline (GT; Knapp et al., 1996; synthesized in house), 100 μg/ml cycloheximide (Sigma), 2 mM phenylmethylsulfonyl fluoride, and protease and phosphatase inhibitors with ∼15 strokes in a manual Potter-Elvehjem Dounce homogenizer (30 ml of buffer per liver). HepG2 or HeLa cell pellets were lysed in TMK buffer supplemented with 1% Triton X-100 (Sigma), 1 μM GT, 100 μg/ml cycloheximide, 2 mM phenylmethylsulfonyl fluoride, and protease and phosphatase inhibitors (∼3 × 108 cells/30 ml buffer) by pipetting up and down several times. After 15 min on ice, tissue homogenates or cell lysates were spun in a 70.1 Ti rotor (Beckman Coulter, Fullerton, CA) at 14,000 rpm for 30 min at 4°C. The resulting postmitochondrial fractions were loaded onto 1 M sucrose cushions prepared in TMK buffer supplemented with 100 μg/ml cycloheximide and 1 μM GT at a 1:1 ratio. Samples were subjected to ultracentrifugation at 41,000 rpm in SW41 Ti rotor for 3 h at 4°C. The resulting supernatants were saved as postribosomal fractions, and the ribosomal pellets were washed three times and resuspended with a pestle in 1 ml of TMK buffer. Only ribosomal preparations with OD260/280 > 1.8 were used for further experiments.
For salt-wash procedures, 100 μl of ribosomes were resuspended in TMK buffer containing 0.1, 0.25, 0.5, 0.75, or 1 M KCl, 100 μg/ml cycloheximide, and 1 μM GT and incubated for 30 min at 4°C with constant rotation. Then, ribosomes were pelleted through a sucrose cushion as described above.
For polysome profiles HepG2 cells were lysed as described above with the addition of 40 U/ml RNAse out (Promega, Madison, WI) and centrifuged at 6000 rpm for 15 min in a refrigerated centrifuge. Supernatants were loaded onto a continuous 15–60% sucrose gradient (10 ml) in TMK buffer supplemented with 1 μM GT and 100 μg/ml cycloheximide and subjected to ultracentrifugation at 38,000 rpm in SW41 Ti rotor for 2 h at 4°C. Gradients were fractionated using a Brandel fractionator and absorbance at A254 nm was continuously recorded.
The procedures for isolation of nucleoli were performed as previously described (Sullivan et al., 2001; Andersen et al., 2005).
Protein Extraction and Precipitation
Proteins from total ribosome preparations were extracted by adding 1/10 volume of 1 M MgCl2 and 2 volumes of acetic acid in rapid succession and incubating 45 min at 4°C with constant rotation. rRNA was precipitated by centrifugation at 14,000 rpm for 15 min and the proteins in the supernatant were precipitated with 10 volumes of cold acetone for 2 h at −20°C. After three washes with acetone the protein pellet was air-dried and resuspended in 1% SDS or 8 M urea and subjected to protein estimation by the bichoncinic acid method (Pierce, Rockford, IL).
Proteins from nucleoli were extracted with Trizol (Invitrogen) according to manufacturer's instructions before protein estimation analysis.
Proteins from sucrose gradient fractions (1 ml) were precipitated by adding 10 μl of 0.1% bovine serum albumin (BSA) and 100 μl of trichloroacetic acid (TCA) and incubating on ice for 1 h. Samples were spun at 12,000 rpm for 15 min. Pellets were recovered and washed twice with ethanol by incubation for at least 30 min on ice and centrifugation at 12,000 rpm for 5 min. Samples were air-dried and resuspended in Laemmli buffer for electrophoresis.
Protein Analysis
For immunoprecipitation experiments, cell pellets were lysed with 1% (vol/vol) Nonidet P-40 (Sigma) in phosphate-buffered saline supplemented with 1 μM GT, 2 mM phenylmethylsulfonyl fluoride, and protease and phosphatase inhibitors. Lysates were centrifuged at 14,000 rpm for 15 min to remove debris. Samples (1 mg/ml) were incubated overnight at 4°C with 1 μg of primary antibody. GammaBind G-Sepharose beads (GE Healthcare) were added and mixed for an additional 2 h at 4°C. The immunoprecipitates were washed, resuspended in Laemmli buffer, and subjected to immunoblot analysis.
For immunoblot analysis, samples were mixed with Laemmli buffer, boiled, separated on SDS-polyacrylamide gels (Bio-Rad, Richmond, CA) and transferred to polyvinylidene difluoride membrane (Millipore, Bedford, MA). Membranes were blocked for 1 h at room temperature (RT) in Tris-buffered saline with 0.1% (vol/vol) Tween-20 containing either 3% (wt/vol) BSA or 5% (wt/vol) nonfat dry milk. Membranes were then incubated overnight at 4°C with the appropriate antibody primary antibody and subsequently with the respective secondary antibody for 1 h at RT. Blots were developed using enhanced chemiluminescence (ECL; GE Healthcare) and exposed to Hyperfilm ECL (GE Healthcare). Blots were stripped in 200 mM glycine, pH 2.5, for 1 h at RT and reprobed using different antibodies.
For lectin blots, membranes were blocked as before and incubated with HRP-conjugated succinylated wheat germ agglutinin (sWGA; 2102-1, EY Laboratories, San Mateo, CA) for 1 h at RT. Membranes were washed in high-salt Tris-buffered saline (0.5 M NaCl) and developed as described for immunoblotting.
Galactosyltransferase Labeling
In the case of postmitochondrial and postribosomal fractions from rat pancreas, samples were denatured before labeling by incubating with 10 mM DTT and 0.5% (wt/vol) SDS and boiling for 10 min. For labeling, samples were diluted with 10 volumes of buffer containing 50 mM HEPES, pH 6.7, 50 mM NaCl, and 2% Triton X-100. Reactions were set up by mixing the sample with 10× galactosyltransferase labeling buffer (100 mM HEPES, pH 7.5, 100 mM galactose, and 50 mM MnCl2), 5′-AMP, 1 μCi UDP-[3H]Gal (American Radiolabeled Chemicals, St. Louis, MO; ART 0131), 1 U β-1,4-galactosyltransferase (Sigma, 48279) that had been previously autogalactosylated with cold UDP-Gal, and 1 U calf intestinal alkaline phosphatase (New England BioLabs, Beverly, MA) and incubating overnight at 4°C. Reactions were stopped by the addition of Laemmli buffer. Samples were boiled and separated by electrophoresis as described. Gels were stained with Coomassie brilliant blue G-250 (Bio-Rad) for total protein detection, and incubated in En3Hance solution (6NE9701, Perkin Elmer-Cetus, Boston, MA) for 1 h at 25°C upon destaining. The gels were then soaked in 1% (vol/vol) glycerol, dried, and exposed to film at −80°C. Where indicated, samples were treated with jack bean hexosaminidase (Sigma) for 18 h at 25°C before galactosyltransferase labeling.
Reverse-Phase HPLC
Proteins from total ribosomes were extracted in 3 vol of 6 M GuHCl followed by 4 vol of 0.2% trifluoroacetic acid for 1 h at 4°C with constant rotation. The insoluble rRNA was precipitated by centrifugation at 14,000 rpm for 15 min at 4°C. Protein extracts were separated in a Smart System (Pharmacia Biotech, Arlington Heights, IL). One milligram of sample was injected into a C8 column (Vydac, Hesperia, CA) and eluted at 25°C by a gradient of buffer A (0.1% trifluoroacetic acid in water) and buffer B (0.09% trifluoroacetic acid in 75% acetonitrile) for 90 min, with a flow rate of 100 μl/min. Detection was simultaneously monitored at 214, 260, and 280 nm. Approximately, 60 fractions of 150 μl were collected, dried in a speed-vac, resuspended in Laemmli buffer, and examined by one-dimensional gel electrophoresis.
Immunofluorescence Microscopy
HeLa cells were fixed, permeabilized, and stained as previously described (Slawson et al., 2008). Fluorescent images were obtained on the 3i Spinning Disk Confocal microscope using the Olympus Slidebook software (Melville, NY) at the Johns Hopkins University School of Medicine Core Microscopy Facility.
Site Mapping and Protein Identification
For site mapping, proteins extracted from total ribosome preparation were digested with Endoproteinase Lys-C (Roche, Indianapolis, IN) and tagged as described before (Wang et al., 2007). O-GlcNAc peptides were enriched by avidin chromatography and derivatized by BEMAD (Wells et al., 2002b). The derivatized peptides were separated by a reverse-phase C18 column (5 μm, 120 Å, YMC ODS-AQ, Waters, Milford, MA) connected to an Eksigent nano-LC system (Dublin, CA). The main HPLC gradient was 5–40% solvent B (A, 0.1% formic acid; B, 90% acetonitrile, 0.1% formic acid) in 60 min at a flow rate of 300 nl/min. Mass spectrometric analysis was performed by a linear trap quadrupole (LTQ)-Orbitrap (Thermo Scientific, Waltham, MA). Briefly, each survey scan (FT-MS, 60,000 resolution at m/z 400) of m/z 400-2000 was followed by CAD fragmentation of up to the five most intense precursor ions. Normalized collision energy was set at 35%. Dynamic exclusion was enabled with repeat count of 2 in 30 s and exclusion duration of 60 s. Database search was performed by using Mascot Daemon (version 2.2.0; Matrix Science, Boston, MA) against the Swiss-Prot database with the following parameters: enzymes, Lys-C; variable modifications, deamidated (NQ), DTT (ST); max missed cleavages, 2; peptide mass tolerance, 0.1 Da; and fragment mass tolerance, 0.8 Da.
For protein identification, bands were excised and digested in-gel with Trypsin (Promega) or endoproteinase Lys-C. Peptides were extracted with 0.1% trifluoroacetic acid, 60% acetonitrile, and dried in a speed-vac. In the case of Hela cells, the extracted peptides were subjected to vMALDI-LTQ (1.0, Thermo Electron, Franklin, MA) analysis at the Johns Hopkins University Proteomics Core Facility and search was performed using Mascot against the National Center for Biotechnology Information database. In the case of rat liver, extracted peptides were analyzed by LTQ-Orbitrap (Thermo Scientific) as described for site mapping. Raw data were analyzed using Mascot (Matrix) as for site mapping, except no DTT(ST) was selected as variable modification. Only proteins with at least two significant and unique peptides are included in the results.
Statistical Analysis
Protein band densitometry from Western blots was measured using the software ImageJ (http://rsb.info.nih.gov/ij/). Statistical differences were determined by a one-tail, paired Student's t test and were considered significant when p = 0.07.
RESULTS
The O-GlcNAc Modification Is Part of the Translational Machinery
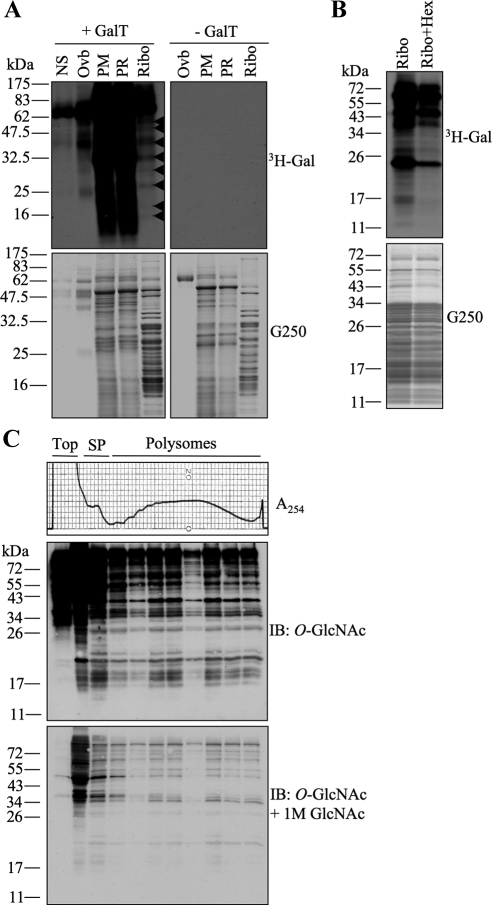
Several components of the translational machinery have been identified as O-GlcNAc modified proteins in different high-throughput studies using complex mixtures (i.e., whole-tissue homogenates or total-cell lysates) as a starting material (Khidekel et al., 2007; Wang et al., 2007; Gurcel et al., 2008; Teo et al., 2010). We set out to determine the extent and abundance of O-GlcNAcylation in subcellular fractions rich in translational components. We first selected rat pancreatic tissue based on its high amount of O-GlcNAc modified proteins (Akimoto et al., 1999; Hanover et al., 1999), fractionated it into postmitochondrial, postribosomal, and ribosomal preparations and performed galactosyltransferase reactions using UDP-[3H]Gal to label terminal O-GlcNAc moieties (Whelan and Hart, 2006). As expected, the postmitochondrial and postribosomal fractions contained many proteins that were strongly labeled (Figure 1A). Interestingly, the ribosome-rich fractions also showed labeling of several bands (Figure 1A, arrowheads), indicating that components associated with the translational machinery are modified with O-GlcNAc. Reactions excluding galactosyltransferase were included as controls (Figure 1A). To obtain the purest possible ribosomal preparations, we then fractionated rat liver tissue. Translational preparations showed intense labeling of many low-molecular-weight proteins (Figure 1B), coinciding with the typical ribosomal proteins masses. Pretreatment with commercial hexosaminidase for 18 h before labeling partially decreased the signal (Figure 1B). This enzyme removes terminal β-GlcNAc and O-GlcNAc and is commonly used as a specificity control for galactosyltransferase reactions (Whelan and Hart, 2006); however, its optimal low pH (4–5) often contributes to protein degradation during longer times of incubation.
Figure 1.
Preparations enriched in ribosomal components contain numerous O-GlcNAc proteins. (A) Postmitochondrial (PM), postribosomal (PR), and ribosomal (Ribo) preparations were obtained by subcellular fractionation of rat pancreas and subjected to galactosyltransferase labeling in the presence of UDP-[3H]Gal. Top, autoradiographs of the gels stained with G250 seen in the bottom panels. Many O-GlcNAcylated low-molecular-weight proteins (<50 kDa) are observed in the Ribo preparation (arrowheads). No substrate (NS) and positive ovalbumin (Ovb) controls are included. Reactions containing all the components except galactosyltransferase (−GalT) are shown in the top right panel. (B) Ribosomal preparations from rat liver were obtained and labeled as in A. Partial pretreatment with commercial hexosaminidase (Ribo+Hex) decreases labeling by galactosyltransferase. (C) Fractions from polysome profiles (top panel) obtained from HepG2 cells growing under normal conditions were TCA precipitated, separated by electrophoresis, and subjected to immunoblot (IB) analysis with O-GlcNAc antibody (middle). A competition control obtained by pre-incubating the antibody with 1 M GlcNAc shows specificity of the labeling (bottom). Sedimentation was from left to right. Data shown are representative of at least three independent experiments.
Ribosomal preparations purified under these conditions are translationally competent when incubated in vitro with exogenous mRNAs (Sugano et al., 1967; Schreier and Staehelin, 1973; Moldave and Sadnik, 1979; Ogata and Terao, 1979). To directly confirm that active ribosomes contain O-GlcNAc proteins, we isolated polysomes from HepG2 cells via continuous sucrose gradient centrifugation and analyzed the different fractions using an antibody that specifically recognizes O-GlcNAc–modified proteins (Comer et al., 2001). We found that both, subpolysomal fractions (represented by the 40S and 60S subunit peaks and the 80S monosome peak) as well as light and heavy polysomes (subunits actively engaged in translation of mRNA) showed many proteins modified with O-GlcNAc (Figure 1C, top). Competition experiments performed by preincubating 1 M GlcNAc with the antibody confirmed specificity (Figure 1C, bottom). These results show that several components of the translational machinery are indeed modified with O-GlcNAc, even under conditions of active protein synthesis.
Many Core Ribosomal Proteins are O-GlcNAcylated
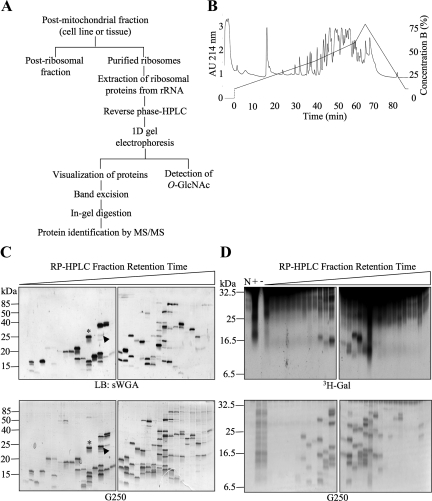
We determined the identity of some of the O-GlcNAc proteins isolated in our translational preparations. We initially focused our attention on low-molecular-weight species (55 kDa and below), which we hypothesized would represent O-GlcNAcylated core ribosomal proteins, and developed a strategy to purify endogenous individual ribosomal proteins (Figure 2A). First, we sedimented total ribosomes by subcellular fractionation and extracted ribosomal proteins from rRNA as described in Materials and Methods. Then, the complex mixture of proteins was separated by HPLC on a reverse-phase column yielding multiple peaks over a gradient of mobile organic phase (Figure 2B). The collected fractions were then resolved individually by one-dimensional gel electrophoresis. A general protein stain of the resulting gels showed that the HPLC peaks varied in complexity, some containing only one visible protein band, whereas others contained more than 10 visible species (Figures 2, C and D, and 3, A and B). When parallel gels were run, transferred, and blotted with the terminal O-GlcNAc–specific lectin sWGA, a large number of these proteins displayed strong reactivity (Figure 2C, top). The presence of O-GlcNAcylated proteins in the HPLC fractions was also confirmed by labeling with galactosyltransferase labeling (Figure 2D) and by immunoblotting with an O-GlcNAc–specific antibody (Figure 3, A and B).
Figure 2.
Extensive separation of ribosomes allows the detection of multiple individual O-GlcNAcylated proteins. (A) Strategy for separation and identification of ribosomal proteins modified with O-GlcNAc. (B) Representative chromatogram obtained after HPLC separation of a purified rat liver ribosomal fraction over a reverse-phase C8 column. (C) Reverse-phase HPLC fractions containing ribosomal protein peaks from HeLa cells were separated by one-dimensional gel electrophoresis and subjected to lectin blot (LB) with sWGA (top panels). Fractions were loaded in subsequent order of retention time (early to late from left to right). Comparison with the corresponding G250-stained gel (bottom panels) shows that many, but not all, proteins present in the fractions contain O-GlcNAc. Two examples (asterisk and arrowhead) show that the lectin signal for O-GlcNAc has different stoichiometries when compared with equivalent levels of total protein. (D) Fractions as in C were subjected to galactosyltransferase labeling in the presence of UDP-[3H]Gal (top panels). Bottom panels show G250 protein stain of the top panel. No substrate (N), total preparation (previous to RP-HPLC) with galactosyltransferase (+), and total preparation without galactosyltransferase (−) controls are included. Data shown are representative of at least three independent experiments.
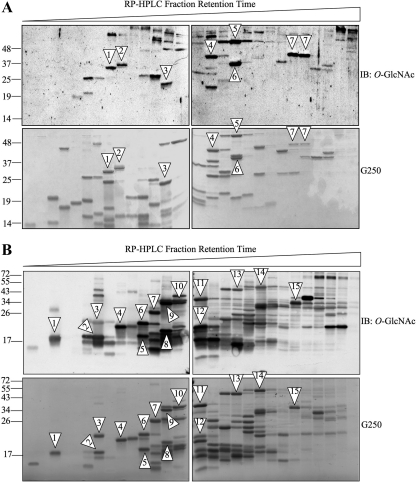
Figure 3.
Identification of ribosomal proteins modified with O-GlcNAc. (A) Reverse-phase HPLC fractions containing ribosomal protein peaks from HeLa cells were separated by one-dimensional gel electrophoresis and subjected to immunoblot (IB) analysis with O-GlcNAc-specific antibody (top panels). Fractions were loaded in subsequent order of retention time (early to late from left to right). Corresponding G250-stained gels are shown (bottom panels). Numbered arrowheads show protein bands selected for identification by MS/MS after careful comparison and gel line-up. Identified species are shown in Table 1. (B) Same as in A but with rat liver ribosomes. Data shown are representative of >5 independent experiments.
We proceeded to identify some of the modified proteins by carefully lining up the Coomassie-stained bands with the corresponding spot on the O-GlcNAc blots, for both HeLa cells and rat liver preparations (Figure 3, A and B). The criteria for selection included: good resolution on the gel and migration as a single band, strong signal on the O-GlcNAc blot, and ability to compete with free GlcNAc (not shown). Bands were sliced out of the gel, digested, and identified by tandem mass spectrometry (MS/MS). Proteins with at least two unique and significant peptides are listed in Table 1. All of the identified peptides were assigned to ribosomal proteins, corroborating the purity of the original preparation and the efficiency of our separation strategy. Interestingly, some of the identified proteins migrated differently than their predicted molecular mass. In HeLa cells RPL6 and RPSa each migrated at a molecular weight ∼10 kDa higher than their theoretical mass (calculated by the amino acid primary structure). It is important to note that RPL6 and RPSa are known substrates for acetylation and phosphorylation (Rush et al., 2005; Rikova et al., 2007; Yu et al., 2007; Wang et al., 2008a). Therefore, we hypothesize that the observed differences in molecular weights are due to the presence of other PTMs in addition to O-GlcNAcylation.
Table 1.
List of putative O-GlcNAcylated ribosomal proteins identified by MS/MS
| Source | Accesion no. | Name | MW (Da) | Coverage (%) | No. of peptides |
|---|---|---|---|---|---|
| HeLa adenocarcinoma (human) | |||||
| Band 1 | NP_150644 | RPL8 | 28,000 | 52 | 3 |
| Band 2 | NP_001001 | RPS6 | 28,600 | 30 | 2 |
| Band 3 | NP_002939 | RPL15 | 24,000 | 38 | 4 |
| Band 4 | NP_001019833 | RPL6 | 32,800 | 34 | 3 |
| Band 5 | NP_000959 | RPL4 | 47,600 | 31 | 4 |
| Band 6 | NP_000963 | RPL7a/RPS3a | 29,900 | 21 | 4 |
| Band 7 | NP_001012321 | RPSa | 31,700 | 36 | 5 |
| Rat liver | |||||
| Band 1 | P61928 | RPL37 | 11,071 | 23 | 2 |
| Band 2 | P83883 | RPL36a | 12,433 | 31 | 4 |
| Band 3 | P25886 | RPL29 | 17,315 | 24 | 3 |
| Band 4 | P12749 | RPL26 | 17,267 | 47 | 7 |
| Band 5 | P61354 | RPL27 | 15,788 | 24 | 3 |
| Band 6 | P20280 | RPL21 | 18,454 | 35 | 5 |
| Band 7 | P41123 | RPL13 | 24,294 | 16 | 3 |
| Band 8 | P62914 | RPS11 | 18,419 | 50 | 8 |
| Band 9 | P62755 | RPS6 | 28,663 | 18 | 5 |
| Band 10 | P62755 | RPS6 | 28,663 | 39 | 6 |
| Band 11 | P21533 | RPL6 | 33,541 | 16 | 6 |
| Band 12 | P24049 | RPL17 | 21,383 | 28 | 4 |
| Band 13 | P21531 | RPL3 | 46,107 | 17 | 5 |
| Band 14 | P50878 | RPL4 | 47,227 | 14 | 8 |
| Band 15 | P09895 | RPL5 | 34,437 | 28 | 9 |
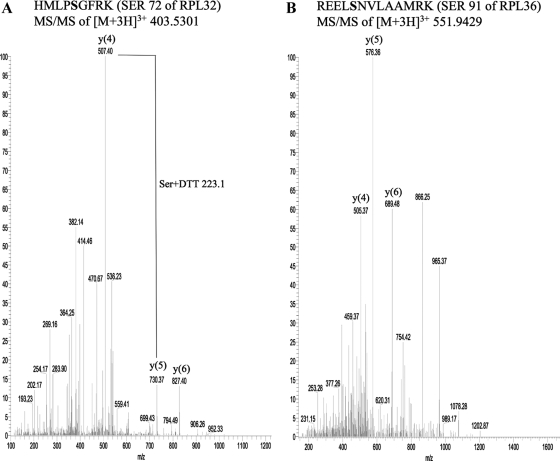
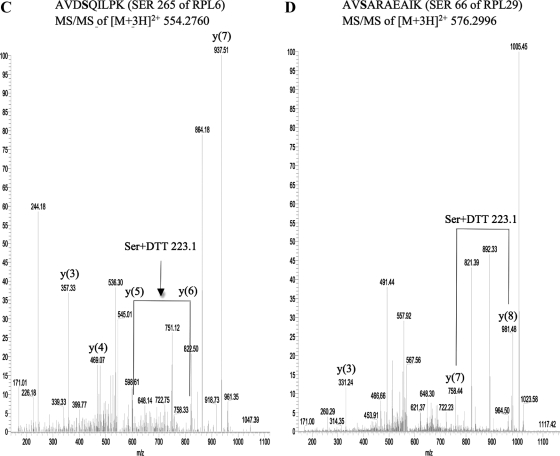
We decided to map the sites of O-GlcNAc modification on rat liver ribosomal proteins by employing a strategy we recently developed (Wang et al., 2007). Using ribosomal extracts we were able to identify sites of O-GlcNAcylation on four different ribosomal proteins (Figure 4, A–D). These sites are Ser 72 of RPL32 (Figure 4A), Ser 91 of RPL36 (Figure 4B), Ser 265 of RPL6 (Figure 4C), and Ser 66 of RPL29 (Figure 4D). The identification of L36, L6, and L29 as O-GlcNAcylated proteins by this method further validates our previous approach based on selection of single ribosomal protein bands that show immunoreactivity with the O-GlcNAc antibody.
Figure 4.
MS/MS of the enriched and derivatized peptides identified the sites of O-GlcNAc modification on four rat liver ribosomal proteins. (A) Serine 72 of RPL32. (B) Serine 91 of RPL36. (C) Serine 265 of RPL6. The m/z values of the y(5) and y(6) species are 599.48 and 822.5, respectively. (D) Serine 66 of RPL29.
O-GlcNAcylation of RPS6 Displays Different Dynamics than Phosphorylation in Response to Nutritional Changes
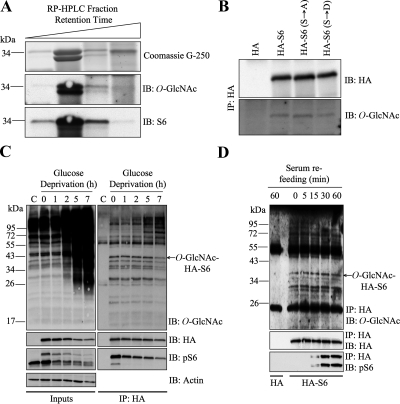
RPS6 phosphorylation upon activation of the mTOR signaling pathway has been widely studied (Ruvinsky and Meyuhas, 2006). Our screen showed that in both HeLa cells and rat liver RPS6 is O-GlcNAcylated. Interestingly, two protein bands that migrate at different molecular weights were identified as RPS6 by MS/MS, suggesting that both the naked and the phosphorylated forms of RPS6 are modified with O-GlcNAc (Figure 3, A and B, Table 1). We confirmed the identity of both bands by performing Western blot analysis with an antibody specific to RPS6 (Figure 5A). Because O-GlcNAcylation and phosphorylation are known to have a dynamic interplay in cells (Hart et al., 2007; Wang et al., 2007, 2008b; Copeland et al., 2008; Butkinaree et al., 2010), we determined if mutation of the phosphorylation sites on RPS6 (Krieg et al., 1988; Pende et al., 2004; Roux et al., 2007) abrogated O-GlcNAcylation. We transfected HEK293 cells with plasmids expressing HA, HA-S6 (wild type), HA-S6-S235/236A, and HA-S6-S235/236D. After immunoprecipitation and Western blot analysis, we were able to show that all three forms of the S6 protein are modified with O-GlcNAc to similar extents (Figure 5B).
Figure 5.
O-GlcNAcylation of RPS6 exhibits different dynamics than nutrient-induced phosphorylation. (A) Reverse-phase HPLC fractions containing rat liver RPS6 visible by G250 and identified by MS/MS were immunoblotted (IB) for O-GlcNAc and S6. (B) Lysates from HEK293 cells transfected with HA, HA-S6, HA-S6-S135/236A, or HA-S6-S235/236D were immunoprecipitated (IP) for HA and immunoblotted (IB) for HA and O-GlcNAc. (C) Lysates from Neuro-2a cells transfected with HA or HA-S6 were glucose-deprived for the indicated times, immunoprecipitated (IP) for HA, and immunoblotted (IB) for O-GlcNAc, HA, phospho-S6 (Ser240/244), and actin. Inputs are shown in the left panel. (D) HEK293 cells transfected with HA or HA-S6 were serum-starved overnight and stimulated with serum (10%) over the indicated times. Lysates were immunoprecipitated (IP) for HA and immunblotted (IB) for HA, O-GlcNAc, and phospho-S6 (Ser240/244). The vector-only control (HA) corresponding to 60-min serum stimulation is shown. Data shown are representative of at least three independent experiments.
The mTOR pathway is known to integrate signals from nutrients and energy to regulate cell size and metabolism (Proud, 2007). RPS6 becomes phosphorylated in response to a wide range of nutritional cues, including growth factors and glucose-sensing hormones (Kimball et al., 2004; Proud, 2007; Roux et al., 2007). We decided to test if O-GlcNAcylation of RPS6 also responded to nutritional changes in the environment. We first performed glucose deprivation experiments, which are known to dramatically affect global protein O-GlcNAcylation (Cheung et al., 2008). Surprisingly, although phosphorylation of wild-type HA-S6 decreased rapidly and dramatically in response to glucose starvation, its O-GlcNAcylation remained unchanged (Figure 5C). Endogenous S6 coimmunoprecipitated with the HA-tagged S6 construct, indicating that either both forms of RPS6 interact at some point or that HA-S6 incorporates into polysomes and brings down ribosomes containing endogenous RPS6 (only one molecule of S6 is present per small subunit; Perry, 2007). Glucose deprivation increased the number of O-GlcNAcylated high-molecular-weight proteins that interact with HA-S6 at longer times (Figure 5C). We decided to also test the opposite scenario: O-GlcNAcylation of S6 under conditions of sudden nutrient abundance. We serum-starved HEK293 cells expressing HA-S6 wild type. Immunoprecipitation of HA-S6 and Western blot analysis showed that O-GlcNAcylation remained unchanged, whereas phosphorylation increased rapidly upon serum refeeding (Figure 5D). Our results suggest that the O-GlcNAc modification on RPS6 does not follow the same dynamics of phosphorylation, at least under our conditions of nutritional challenge.
O-GlcNAc Cycling Enzymes Associate with Different Populations of Ribosomes
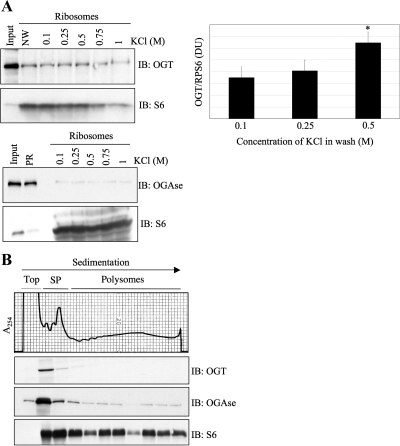
Ribosomal proteins are translated in the cytosol, imported into the nucleus, assembled around newly transcribed rRNA in the nucleolus, and exported back into the cytoplasm in the form of mature subunits (Andersen et al., 2005; Dresios et al., 2006; Boisvert et al., 2007). Because ribosomal proteins are substrates for O-GlcNAcylation, they must interact with OGT and/or OGAse in the cytoplasm, the nucleus/nucleolus, or both. To test this possibility, we purified total ribosomal preparations from rat liver and subjected them to washes with increasing concentrations of salt. After extraction from rRNA and Western blot analysis, we observed that both OGT and OGAse cosedimented with ribosomes, even after a salt wash of 1 M KCl (Figure 6A). Surprisingly, we find that OGT remains associated with the translational machinery even after the high-salt wash has stripped some of the core ribosomal protein S6, which significantly increases the ratio of OGT to ribosomes (Figure 6A, left and right top panels). Transiently associated translation factors are known to dissociate from the ribosome at salt concentrations of 0.5 M (Sugano et al., 1967; Schreier and Staehelin, 1973; Moldave and Sadnik, 1979; Ogata and Terao, 1979), suggesting that the O-GlcNAc cycling enzymes associate very strongly with ribosomes. Next, we fractionated actively translating polysomes from HepG2 cells growing under normal conditions and probed these fractions for OGT and OGAse. As shown (Figure 6B), we confirmed that both O-GlcNAc cycling enzymes copurify with actively translating ribosomes in the cytosol. Most of the endogenous OGT signal was observed in the subpolysomal populations, whereas OGAse was found in both, subpolysomal fractions and light and heavy polysomes (Figure 6B). When overexpressed, the majority of the OGT signal is still observed at the top of the gradient and associated with subpolysomal fractions; however, a significant amount of the protein is also found to cosediment with polysomes (see Figure 9B). The differential interaction of endogenous OGT and OGAse with various fractions of ribosomes observed here could be biologically significant, but there is the possibility that it could potentially be explained by limitations of the antibodies used.
Figure 6.
O-GlcNAc cycling enzymes associate with ribosomes. (A) Total ribosome preparation from rat liver was incubated with increasing concentrations of salt (KCl) as indicated and subjected to an additional round of purification through a sucrose cushion. Ribosomal proteins were extracted with acetic acid, acetone precipitated, separated by electrophoresis, and immunoblotted (IB) for OGT, OGAse, and S6 (top left panel and bottom panel). Densitometry analysis from four independent experiments shows the ratio of ribosome-associated OGT to RPS6 (top right panel), showing a significant difference between the 0.1 and the 0.5 M samples (P = 0.07). Bars in graph represent the values; error bars, SE. NW, no wash; DU, densitometry units; PR, postribosomal fraction. (B) Polysomal fractions from HepG2 cells growing under normal conditions were TCA precipitated, separated by electrophoresis, and subjected to immunoblot (IB) analysis with OGT, OGAse, and S6 antibodies. (SP) subpolysomal fractions. Data shown are representative of at least three independent experiments.
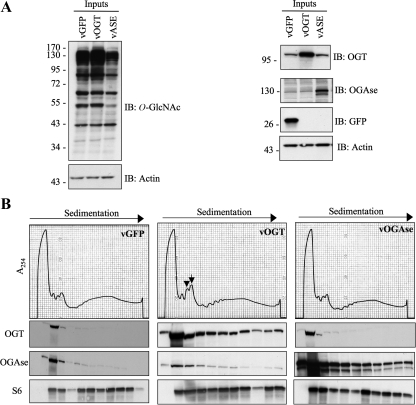
Figure 9.
Adenoviral-mediated overexpression of OGT causes accumulation of 60S subunits and 80S monosomes. (A) HepG2 cells were infected with adenovirus overexpressing either GFP control, OGT, or OGAse. Lysates were obtained after 48 h and immunoblotted (IB) for O-GlcNAc, OGT, OGAse, GFP, and actin to determine efficiency of overexpression. (B) HepG2 cells were infected as in A and polysome profiles were obtained 48 h after infection. Cells overexpressing OGT (vOGT) show an increase in the 60S (arrowhead) and the 80S (arrow) peaks as compared with vOGAse and vGFP control. Polysomal fractions were TCA precipitated, separated by electrophoresis, and subjected to immunoblot analysis with OGT, OGAse, and S6 antibodies. Data shown are representative of three independent experiments.
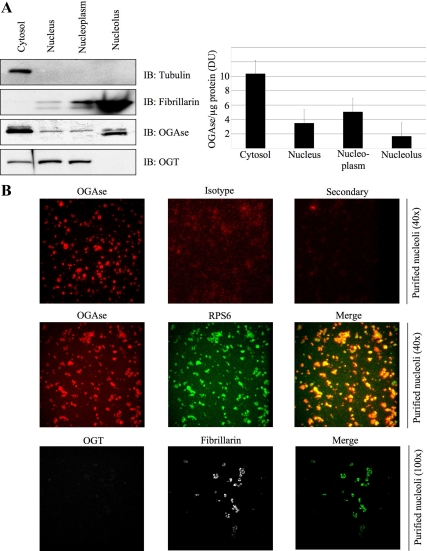
Next we wanted to test if the association of OGT and OGAse with ribosomes was exclusive to the cytosolic compartment. We isolated intact nuclei and nucleoli from HeLa cells (Andersen et al., 2005) and performed biochemical analysis for the presence of O-GlcNAc enzymes. Interestingly, whereas most of the OGAse was found in the cytoplasm, a fraction of this protein was also present in the nucleolar preparation (Figure 7A, left and right). Previous studies have reported predominant accumulation of OGAse in the cytosol; however, significant OGAse activity is localized to the nucleus as well (Gao et al., 2001; Wells et al., 2002a). Relative to its levels in the nucleus, OGT was absent from the nucleolus, and it accumulated in the nucleoplasmic fraction (Figure 7A, left and right). To further explore the cytolocalization of the O-GlcNAc cycling enzymes, we carried out immunofluorescence studies of the isolated nucleoli. This analysis showed no OGT in the structures labeled with fibrillarin as a nucleolar marker (Figure 7B, bottom). In contrast, endogenous OGAse localizes to nucleoli, surrounding regions labeled with RPS6 as a marker (Figure 7B, top and middle).
Figure 7.
OGAse is present in purified nucleoli. (A) HeLa cells were fractionated into different subcellular components in order to obtain intact nuclei and nucleoli. Equivalent amount of total proteins from the different fractions was separated by electrophoresis and immunoblotted (IB) for tubulin, fibrillarin, OGAse, and OGT (left). Densitometry analysis from four independent experiments shows the amount of OGAse per microgram of protein loaded in every fraction (right). Bars in graph represent the values; error bars, SE. (B) Nucleoli from A were spotted onto coverslips, fixed, permeabilized, and stained for immunofluorescence confocal microscopy with OGAse (red), RPS6 (green), OGT (red), and fibrillarin (green) antibodies. Isotype (red) and secondary (red) controls are included for the OGAse antibody. Magnifications are shown in parenthesis. All pictures in each panel were exposed for equal times and subjected to the same brightness/contrast adjustments. Data shown are representative of at least three independent experiments.
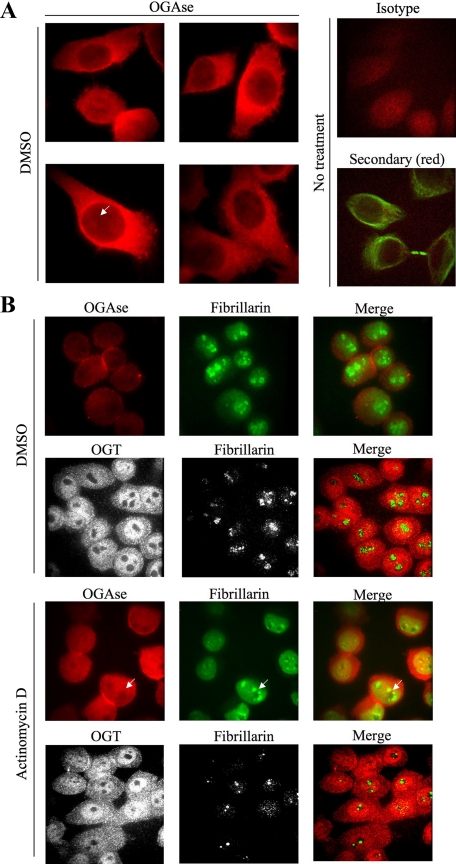
To confirm the differential localization of OGT and OGAse in the nucleolus, we performed immunofluorescence experiments on whole cells under conditions of nucleolar stress (Andersen et al., 2005; Lam et al., 2007; Figure 8). Control cells treated with vehicle (DMSO) show that OGAse localizes mostly to the cytosol, but is also present uniformly throughout the nucleus (Figure 8A). Nuclear staining of OGAse is apparent in the nucleoplasm and in subnuclear regions that presumably represent nucleoli (Figure 8A, white arrow). For OGT, most of the signal accumulated in the nucleoplasm, whereas less staining was present in the cytosolic compartment (Figure 8B, top middle panel). Despite its strong nucleoplasmic accumulation, OGT was almost completely excluded from nucleoli, as observed by the lack of colocalization with fibrillarin, which showed its typical granular staining (Figure 8B, top middle panel, left column, and green color in the merge). OGAse colocalization with fibrillarin in control cells is not apparent due to the strong signal generated by the typical granular staining of this nucleolar marker (Figure 8B, top left panel). Nucleolar stress induced by actinomycin D treatment clearly disrupted nucleolar structure as observed by diffusion of fibrillarin into the nucleoplasm, loss of granular staining in the nucleolus, and the appearance of dark regions (Figure 8B, bottom middle panel). This treatment did not affect the distribution or localization of OGT (Figure 8B, top middle panel and bottom panel). In contrast, nucleolar stress changed the distribution of OGAse in the nucleus, which showed a nonuniform staining and the appearance of dark regions that colocalized with the empty nucleolar spaces generated by the diffusion of fibrillarin into the nucleoplasm (Figure 8B, bottom middle panel and white arrows). Concomitantly, colocalization of fibrillarin and OGAse under nucleolar stress increased in the nucleoplasm (Figure 8B, bottom middle panel).
Figure 8.
Nucleolar stress disrupts staining of OGAse in the nucleus of cells. (A) HeLa cells were grown on coverslips, fixed, permeabilized, and stained for immunofluorescence confocal microscopy with OGAse (red) antibody. Nuclear OGAse staining shows a gradient from nucleoplasm to nucleoli (white arrow). Isotype (red) and secondary (red, tubulin shown in green) controls are included. Magnification, ×100. All pictures were exposed for equal times (50 ms) and subjected to the same brightness/contrast adjustments. (B) HeLa cells were subjected to treatments as indicated, processed as in A, and double-stained with OGAse (red) and fibrillarin (green) or OGT (red) and fibrillarin (green) antibodies. OGAse staining in the nucleus follows the same pattern as disrupted fibrillarin (white arrows). Magnifications, ×63 (OGT) and ×100 (OGAse). All pictures in each panel were exposed for equal times and subjected to the same brightness/contrast adjustments. Data shown are representative of at least three independent experiments.
Overall, our results indicate that OGT and OGAse strongly and differentially associate with ribosomes at different stages of their maturation process (translating ribosomes and assembling ribosomes).
Overexpression of OGT Causes Accumulation of 60S Subunits and 80S Monosomes
Polysome profiles not only provide information about the initiation and elongation status of actively translating ribosomes (monosome to polysome ratio), but also reflect imbalances in subunit populations. We tested whether overexpression of either O-GlcNAc cycling enzyme would affect polysome distribution. We infected HepG2 cells with either adenoviral OGT (vOGT), adenoviral OGAse (vOGAse), or adenoviral GFP (vGFP), the latter as a control for infection and overexpression. Forty-eight hours after infection, we obtained whole cell lysates and polysome profiles. Immunoblots showed that infection was efficient as observed by the increased levels of OGT, OGAse, or GFP and increase and decrease in O-GlcNAc levels, respectively (compared with vGFP control; Figure 9A). Analysis of polysome profiles from vGFP control and OGAse-overexpressing cells showed two smaller peaks for 40S and 60S subunits and a relatively taller peak for 80S monosomes, as well as polysomes (Figure 9B, left and right panels). However, overexpression of OGT caused a dramatic increase in the size of the 60S peak (Figure 9B, middle panel, arrowhead) and the 80S peak (Figure 9B, middle panel, arrow) when compared with vGFP and vOGAse. Immunoblot analysis showed that when overexpressed, both OGT and OGAse increase their association with polysomes. Taken together, our data suggest that O-GlcNAc cycling may play a role in ribosomal subunit homeostasis.
DISCUSSION
The ribosome is the most complex universal ribonucleoprotein machine (Dresios et al., 2006; Marshall et al., 2008). In this work, we have added a new level of complexity, the presence of a single β-N-acetylglucosamine monosaccharide attached in O-linkage to its core ribosomal proteins. We have identified ribosomal proteins Sa, S6, and S11 from the small subunit and L3, L4, L5, L6, L8, L13, L15, L17, L21, L26, L27, L29, L32, L36, L36a, and L37 from the large subunit as being modified with O-GlcNAc under conditions of normal cellular growth and active translation. In addition, either or both S3 and L7a are also O-GlcNAcylated; however, these two proteins migrate together during reverse-phase separation and gel electrophoresis, making it difficult to single out the O-GlcNAc immunoreactive band. O-GlcNAcylation of ribosomal proteins L8, L17, and L21 has been previously reported using a Click-iT chemistry tagging methodology in total MCF-7 cell extracts (Gurcel et al., 2008). O-GlcNAcylation of S3 and L13a has also been reported by metabolic labeling with UDP-GlcNAz (Ohn et al., 2008). This latter report also suggests that ribosomal proteins S11, L6, and L36a-like are modified by O-GlcNAc after their immunoprecipitation with an O-GlcNAc antibody (Ohn et al., 2008). More recently, the generation of a highly specific mAb against O-GlcNAcylated epitopes allowed the immunopurification of several ribosomal proteins (Teo et al., 2010). Our direct experimental approach of purifying preparations enriched in components of the translational machinery and applying several levels of biochemical separation has allowed us to directly identify 20 O-GlcNAcylated ribosomal proteins of which eight are newly reported. Along with previous observations, our findings bring the O-GlcNAc-ribosomal proteome to 34 proteins out of approximately 80 that comprise the mammalian ribosome.
Site mapping constitutes a more definitive approach of assigning PTMs to any given protein. In this work, we identified the amino acid residues to which O-GlcNAc is covalently attached in four of the mammalian ribosomal proteins. We report that Ser 265 of L6, Ser 66 of L29, Ser 72 of L32, and Ser 91 of L36 are modified by O-GlcNAc. Of these four proteins, only the sequence containing the O-GlcNAcylated residue in L32 is included within the known structure of a mammalian 80S complex recently solved at 8.7 Å by electron cryomicroscopy and single-particle methods (Chandramouli et al., 2008). Using the software PyMOL (DeLano Scientific, South San Francisco, CA), we have located the Serine 72 residue of the L32 polypeptide on a kink that is exposed to the solvent, but also is in close proximity with rRNA (not shown). This interesting location makes the O-GlcNAcylated residue of L32 available for dynamic modification under changing conditions and suggests that O-GlcNAc may play a structural role on the ribosome, helping to stabilize the entire mature complex and perhaps contributing to its ribozyme activity.
Assigning individual functions to ribosomal proteins has been a very difficult task for more than 50 years. Of the proteins identified in our analyses, studies in yeast have shown that S6, S11, L4, L5, and L8 are essential for cell viability, whereas L29 is a nonessential ribosomal protein (Dresios et al., 2006). S6, L8, and L29 are unique to eukaryotes, whereas L4, L5, and S11 have homologues in archaea (Dresios et al., 2006). O-GlcNAcylation of ribosomal proteins could impact the role of these components as a whole complex and thus have implications on overall ribosome performance. In addition, O-GlcNAc may have an influence on extra-ribosomal functions of individual proteins. In fact, the occurrence of other PTMs on ribosomal proteins has been functionally linked to activities independent of their roles as components of the ribosome. For example, ribosomal protein L26 binds to the 5′ untranslated region of the p53 mRNA and controls p53 translation and induction upon damage to DNA (Takagi et al., 2005). Ribosomal protein L6, also known as TaxREB107, contains a DNA-binding motif and may be involved in transactivation of transcription of human T-cell leukemia virus type I (Morita et al., 1993). Another intriguing ribosomal protein, Sa, is the monomeric precursor of the 67-kDa laminin receptor dimer. Sa is a multifunctional protein that interacts with alphaviruses and prions at the cell surface and is also involved in metastasis of solid tumors. Interestingly, Sa is required to be acylated in order to become the cell surface laminin receptor (Wewer et al., 1986; Rieger et al., 1997; Buto et al., 1998; Rieger et al., 1999; Gauczynski et al., 2001; Kim et al., 2005). One common property to all of these proteins is that they exist as separate cytosolic pools dissociated from the ribosome. It is possible that O-GlcNAcylation functions as a biochemical tag to help maintain these separate populations of the same protein species.
Here we also report that the widely studied ribosomal protein S6, a constituent of the mTOR pathway, is modified with O-GlcNAc. In response to mitogens and growth factors, S6 becomes phosphorylated by various kinases at five distinct serine residues clustered in its C-terminus (Ruvinsky and Meyuhas, 2006; Roux et al., 2007). It was postulated that the physiological consequence of these phosphorylation events was to control the translation of 5′ tract of oligopyrimidine (TOP)-mRNAs, which encode numerous components of the translational apparatus (Ruvinsky and Meyuhas, 2006). However, a breakthrough study showed that knockin mutations at all five phosphorylatable residues of RPS6 do not affect translation of TOP-mRNAs, but rather increase global protein synthesis, accelerate cell division, and result in smaller cell size (Ruvinsky et al., 2005). More interestingly, these mutations have a profound deleterious effect on glucose homeostasis in several tissues (Ruvinsky et al., 2005). It has been well established that deregulation of glucose metabolism increases protein GlcNAcylation of several key components of the insulin signaling pathway (Dias and Hart, 2007). The sites of O-GlcNAc modification on RPS6 have yet to be determined; however, we hypothesize that the O-GlcNAcylated form of RPS6 may play a role in regulating glucose metabolism in cells.
In mammalian cells, ribosomal protein S6 is also required for biogenesis of the small ribosomal subunit and maintenance of the size of the free 40S cytosolic pool (Volarevic et al., 2000; Pachler et al., 2004). L29, another ribosomal protein found to be O-GlcNAcylated at Ser 66 in our study, is also important for the assembly of large ribosomal subunits (DeLabre et al., 2002). In this work, we present evidence that the O-GlcNAc modification may in fact play a role in ribosome biogenesis. We showed that OGAse colocalized in the nucleolus with fibrillarin, a ribosome-assembly processing factor (Tollervey et al., 1993), whereas OGT was excluded from the nucleolus. OGAse nuclear distribution was sensitive to nucleolar disruption treatments that caused redistribution of fibrillarin to the nucleoplasm, suggesting that OGAse may transiently associate with nucleolar resident proteins. OGAse has been shown to directly interact with ATP-dependent RNA helicase A, a protein that resides in the nuclear/nucleolar compartment and participates in the biogenesis of the eukaryotic 60S subunit (Stelzl et al., 2005). Moreover, increasing the levels of OGT caused an accumulation of the 60S peak, pointing at a role for O-GlcNAc cycling in large subunit assembly. The process of subunit biogenesis is different for small and large subunits. Although 40S particles are entirely assembled in the nucleolus, the late stages of 60S particle assembly take place in the nucleoplasm and in the cytoplasm (Boisvert et al., 2007). We hypothesize that the role of the O-GlcNAc–cycling enzymes on ribosome assembly is dependent on their ability to differentially localize to the nucleus/nucleolus. In addition, the mechanisms by which OGT is kept predominantly in the nucleoplasm and is excluded from the nucleolus are very intriguing.
Our results also show that adenoviral-mediated OGT overexpression caused an accumulation of 80S monosomes, which is suggestive of a role for OGT in keeping an active translational apparatus during viral infection. A role for OGT during cellular stress has been previously documented by our laboratory (Zachara et al., 2004). OGT activity and global O-GlcNAcylation of proteins increase drastically and dynamically under a wide variety of stress, and these events are required for recovery and survival of the cells. A recent study shows that OGT directly participates in formation of stress granules under conditions of arsenite treatment and which OGT correlates with a general increase in the O-GlcNAcylation of ribosomal proteins (Ohn et al., 2008). This study also shows that O-GlcNAc is present in proteins that form the stress granules (Ohn et al., 2008). These data support our hypothesis that O-GlcNAc may serve as a biochemical tag for assigning ribosomal proteins new functions or subcellular locations under changing environmental conditions.
With this work, we have opened a new area of study on the possible role of O-GlcNAcylation and its cycling enzymes in the translational control of gene expression. Future research should be dedicated to establishing the functions of OGT, OGAse and O-GlcNAcylated ribosomal proteins in ribosome structure and performance, extraribosomal functions of ribosomal proteins, biogenesis of 40S and 60S particles, and control of polysomal populations and translation.
ACKNOWLEDGMENTS
We greatly appreciate the gift of mammalian expression plasmids encoding RPS6, RPS6-S235/236A, and RPS6-S235/236D from Dr. Philippe P. Roux (Université de Montréal, Montréal, QC, Canada). We thank Dr. Robert N. Cole and Dr. Tatiana Boronina of the Johns Hopkins School of Medicine Mass Spectrometry and Proteomics Facility and Dr. Robert J. Cotter for advice with mass spectrometry. We graciously thank Dr. Jon R. Lorsch and members of his laboratory and Dr. Michael G. Acker for support with polysome fractionation. We especially thank Dr. Natasha E. Zachara for assistance with reverse-phase HPLC. Finally, we thank members of the Hart laboratory and in particular Dr. Chad Slawson, Dr. Kaoru Sakabe, and Dr. Win D. Cheung for invaluable technical assistance and helpful discussions.
Abbreviations used:
- BEMAD
β-elimination followed by Michael addition with dithiothreitol
- DMSO
dimethyl sulfoxide
- eIF2
eukaryotic initiation factor 2
- GFP
green fluorescent protein
- GT
O-GlcNAc-thiazoline
- HA
hemagglutinin
- LTQ
linear trap quadrupole
- MALDI
matrix-assisted laser desorption ionization
- mTOR
mammalian target of rapamycin
- OGAse
O-linked β-N-acetylglucosaminidase (EC 3.2.1.52)
- O-GlcNAc
O-linked β-N-acetylglucosamine
- OGT
O-linked β-N-acetylglucosaminyltransferase (EC 2.4.1.94)
- PTM
posttranslational modification
- RP
ribosomal protein
- TOP
tract of oligopyrimidine
- UDP-Gal
uridine diphosphate galactose.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-11-0941) on April 21, 2010.
REFERENCES
- Acker M. G., Lorsch J. R. Mechanism of ribosomal subunit joining during eukaryotic translation initiation. Biochem. Soc. Trans. 2008;36:653–657. doi: 10.1042/BST0360653. [DOI] [PubMed] [Google Scholar]
- Akimoto Y., Kreppel L. K., Hirano H., Hart G. W. Localization of the O-linked N-acetylglucosamine transferase in rat pancreas. Diabetes. 1999;48:2407–2413. doi: 10.2337/diabetes.48.12.2407. [DOI] [PubMed] [Google Scholar]
- Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Boisvert F. M., van Koningsbruggen S., Navascues J., Lamond A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Butkinaree C., Park K., Hart G. W. O-linked β-N-acetylglucosamine (O-GlcNAc): extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buto S., Tagliabue E., Ardini E., Magnifico A., Ghirelli C., van den Brule F., Castronovo V., Colnaghi M. I., Sobel M. E., Menard S. Formation of the 67-kDa laminin receptor by acylation of the precursor. J. Cell. Biochem. 1998;69:244–251. doi: 10.1002/(sici)1097-4644(19980601)69:3<244::aid-jcb2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Chandramouli P., Topf M., Menetret J. F., Eswar N., Cannone J. J., Gutell R. R., Sali A., Akey C. W. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W. D., Hart G. W. AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J. Biol. Chem. 2008;283:13009–13020. doi: 10.1074/jbc.M801222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W. D., Sakabe K., Housley M. P., Dias W. B., Hart G. W. O-GlcNAc transferase substrate specificity is regulated by MYPT1 and other interacting proteins. J. Biol. Chem. 2008;283:33935–33941. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer F. I., Vosseller K., Wells L., Accavitti M. A., Hart G. W. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal. Biochem. 2001;293:169–177. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- Copeland R. J., Bullen J. W., Hart G. W. Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am. J. Physiol. Endocrinol. Metab. 2008;295:E17–E28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta B., Chakrabarti D., Roy A. L., Gupta N. K. Roles of a 67-kDa polypeptide in reversal of protein synthesis inhibition in heme-deficient reticulocyte lysate. Proc. Natl. Acad. Sci. USA. 1988;85:3324–3328. doi: 10.1073/pnas.85.10.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta B., Ray M. K., Chakrabarti D., Wylie D. E., Gupta N. K. Glycosylation of eukaryotic peptide chain initiation factor 2 (eIF-2)-associated 67-kDa polypeptide (p67) and its possible role in the inhibition of eIF-2 kinase-catalyzed phosphorylation of the eIF-2 alpha-subunit. J. Biol. Chem. 1989;264:20620–20624. [PubMed] [Google Scholar]
- Datta R., Choudhury P., Bhattacharya M., Soto Leon F., Zhou Y., Datta B. Protection of translation initiation factor eIF2 phosphorylation correlates with eIF2-associated glycoprotein p67 levels and requires the lysine-rich domain I of p67. Biochimie. 2001;83:919–931. doi: 10.1016/s0300-9084(01)01344-x. [DOI] [PubMed] [Google Scholar]
- DeLabre M. L., Kessl J., Karamanou S., Trumpower B. L. RPL29 codes for a non-essential protein of the 60S ribosomal subunit in Saccharomyces cerevisiae and exhibits synthetic lethality with mutations in genes for proteins required for subunit coupling. Biochim. Biophys. Acta. 2002;1574:255–261. doi: 10.1016/s0167-4781(01)00372-4. [DOI] [PubMed] [Google Scholar]
- Dias W. B., Hart G. W. O-GlcNAc modification in diabetes and Alzheimer's disease. Mol. Biosyst. 2007;3:766–772. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- Dong D. L., Hart G. W. Purification and characterization of an O-GlcNAc selective N-acetyl-β-d-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- Dresios J., Panopoulos P., Synetos D. Eukaryotic ribosomal proteins lacking a eubacterial counterpart: important players in ribosomal function. Mol. Microbiol. 2006;59:1651–1663. doi: 10.1111/j.1365-2958.2006.05054.x. [DOI] [PubMed] [Google Scholar]
- Gao Y., Wells L., Comer F. I., Parker G. J., Hart G. W. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Gauczynski S., et al. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J. 2001;20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F., Hentze M. W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcel C., Vercoutter-Edouart A. S., Fonbonne C., Mortuaire M., Salvador A., Michalski J. C., Lemoine J. Identification of new O-GlcNAc modified proteins using a click-chemistry-based tagging. Anal. Bioanal. Chem. 2008;390:2089–2097. doi: 10.1007/s00216-008-1950-y. [DOI] [PubMed] [Google Scholar]
- Haltiwanger R. S., Blomberg M. A., Hart G. W. Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase. J. Biol. Chem. 1992;267:9005–9013. [PubMed] [Google Scholar]
- Hanover J. A., Lai Z., Lee G., Lubas W. A., Sato S. M. Elevated O-linked N-acetylglucosamine metabolism in pancreatic beta-cells. Arch. Biochem. Biophys. 1999;362:38–45. doi: 10.1006/abbi.1998.1016. [DOI] [PubMed] [Google Scholar]
- Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Hart G. W., Housley M. P., Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- Khidekel N., Ficarro S. B., Clark P. M., Bryan M. C., Swaney D. L., Rexach J. E., Sun Y. E., Coon J. J., Peters E. C., Hsieh-Wilson L. C. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- Kim K. J., Chung J. W., Kim K. S. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J. Biol. Chem. 2005;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- Kimball S. R., Siegfried B. A., Jefferson L. S. Glucagon represses signaling through the mammalian target of rapamycin in rat liver by activating AMP-activated protein kinase. J. Biol. Chem. 2004;279:54103–54109. doi: 10.1074/jbc.M410755200. [DOI] [PubMed] [Google Scholar]
- Knapp V., Gao, Kirk, Lou, Withers NAG-thiazoline, an N-Acetyl-β-hexosaminidase inhibitor that implicates acetamido participation. J. Am. Chem. Soc. 1996;118:6804–6805. [Google Scholar]
- Krieg J., Hofsteenge J., Thomas G. Identification of the 40 S ribosomal protein S6 phosphorylation sites induced by cycloheximide. J. Biol. Chem. 1988;263:11473–11477. [PubMed] [Google Scholar]
- Lam Y. W., Lamond A. I., Mann M., Andersen J. S. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R. A., Aitken C. E., Dorywalska M., Puglisi J. D. Translation at the single-molecule level. Annu. Rev. Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- Mazumder B., Sampath P., Seshadri V., Maitra R. K., DiCorleto P. E., Fox P. L. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Moldave K., Sadnik I. Preparation of derived and native ribosomal subunits from rat liver. Methods Enzymol. 1979;59:402–410. doi: 10.1016/0076-6879(79)59101-0. [DOI] [PubMed] [Google Scholar]
- Morita T., Sato T., Nyunoya H., Tsujimoto A., Takahara J., Irino S., Shimotohno K. Isolation of a cDNA clone encoding DNA-binding protein (TAXREB107) that binds specifically to domain C of the tax-responsive enhancer element in the long terminal repeat of human T-cell leukemia virus type I. AIDS Res. Hum. Retrovir. 1993;9:115–121. doi: 10.1089/aid.1993.9.115. [DOI] [PubMed] [Google Scholar]
- Nissen P., Hansen J., Ban N., Moore P. B., Steitz T. A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Hoffarth V., Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Odintsova T. I., Muller E. C., Ivanov A. V., Egorov T. A., Bienert R., Vladimirov S. N., Kostka S., Otto A., Wittmann-Liebold B., Karpova G. G. Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J Protein Chem. 2003;22:249–258. doi: 10.1023/a:1025068419698. [DOI] [PubMed] [Google Scholar]
- Ogata K., Terao K. Analytical methods for ribosomal proteins of rat liver 40 S and 60 S subunits by “three-dimensional” acrylamide gel electrophoresis. Methods Enzymol. 1979;59:502–515. doi: 10.1016/0076-6879(79)59110-1. [DOI] [PubMed] [Google Scholar]
- Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachler K., et al. Functional interaction in establishment of ribosomal integrity between small subunit protein rpS6 and translational regulator rpL10/Grc5p. FEMS Yeast Res. 2004;5:271–280. doi: 10.1016/j.femsyr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Pende M., Um S. H., Mieulet V., Sticker M., Goss V. L., Mestan J., Mueller M., Fumagalli S., Kozma S. C., Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P. Balanced production of ribosomal proteins. Gene. 2007;401:1–3. doi: 10.1016/j.gene.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T. V., Kolupaeva V. G., Lomakin I. B., Pilipenko E. V., Shatsky I. N., Agol V. I., Hellen C. U. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud C. G. Regulation of protein synthesis by insulin. Biochem. Soc. Trans. 2006;34:213–216. doi: 10.1042/BST20060213. [DOI] [PubMed] [Google Scholar]
- Proud C. G. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem. J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- Ray M. K., Datta B., Chakraborty A., Chattopadhyay A., Meza-Keuthen S., Gupta N. K. The eukaryotic initiation factor 2-associated 67-kDa polypeptide (p67) plays a critical role in regulation of protein synthesis initiation in animal cells. Proc. Natl. Acad. Sci. USA. 1992;89:539–543. doi: 10.1073/pnas.89.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger R., Edenhofer F., Lasmezas C. I., Weiss S. The human 37-kDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nat. Med. 1997;3:1383–1388. doi: 10.1038/nm1297-1383. [DOI] [PubMed] [Google Scholar]
- Rieger R., Lasmezas C. I., Weiss S. Role of the 37 kDa laminin receptor precursor in the life cycle of prions. Transfus. Clin. Biol. 1999;6:7–16. doi: 10.1016/S1246-7820(99)80006-8. [DOI] [PubMed] [Google Scholar]
- Rikova K., et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Roux P. P., Shahbazian D., Vu H., Holz M. K., Cohen M. S., Taunton J., Sonenberg N., Blenis J. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I., Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I., Sharon N., Lerer T., Cohen H., Stolovich-Rain M., Nir T., Dor Y., Zisman P., Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F., et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Schreier M. H., Staehelin T. Initiation of mammalian protein synthesis: the importance of ribosome and initiation factor quality for the efficiency of in vitro systems. J. Mol. Biol. 1973;73:329–349. doi: 10.1016/0022-2836(73)90346-x. [DOI] [PubMed] [Google Scholar]
- Slawson C., Lakshmanan T., Knapp S., Hart G. W. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol. Biol. Cell. 2008;19:4130–4140. doi: 10.1091/mbc.E07-11-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., Hart G. W. Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- Spence J., Gali R. R., Dittmar G., Sherman F., Karin M., Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- Stelzl U., et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Sugano H., Watanabe I., Ogata K. Stabilizing effect of ribonuclease inhibitor on structure of polysomes and some properties of four classes of ribosomal particles in rat liver cytoplasm. J. Biochem. 1967;61:778–786. doi: 10.1093/oxfordjournals.jbchem.a128613. [DOI] [PubMed] [Google Scholar]
- Sullivan G. J., Bridger J. M., Cuthbert A. P., Newbold R. F., Bickmore W. A., McStay B. Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J. 2001;20:2867–2874. doi: 10.1093/emboj/20.11.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Absalon M. J., McLure K. G., Kastan M. B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Teo C. F., Ingale S., Wolfert M. A., Elsayed G. A., Not L. G., Chatham J. C., Wells L., Boons G. J. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat. Chem. Biol. 2010;6:338–343. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E. C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Volarevic S., Stewart M. J., Ledermann B., Zilberman F., Terracciano L., Montini E., Grompe M., Kozma S. C., Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- Wang B., Malik R., Nigg E. A., Korner R. Evaluation of the low-specificity protease elastase for large-scale phosphoproteome analysis. Anal. Chem. 2008a;80:9526–9533. doi: 10.1021/ac801708p. [DOI] [PubMed] [Google Scholar]
- Wang Z., Gucek M., Hart G. W. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. USA. 2008b;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Pandey A., Hart G. W. Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol. Cell Proteomics. 2007;6:1365–1379. doi: 10.1074/mcp.M600453-MCP200. [DOI] [PubMed] [Google Scholar]
- Wells L., Gao Y., Mahoney J. A., Vosseller K., Chen C., Rosen A., Hart G. W. Dynamic O-glycosylation of nuclear and cytosolic proteins: further characterization of the nucleocytoplasmic β-N-acetylglucosaminidase, O-GlcNAcase. J. Biol. Chem. 2002a;277:1755–1761. doi: 10.1074/jbc.m109656200. [DOI] [PubMed] [Google Scholar]
- Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., Hart G. W. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell Proteomics. 2002b;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- Wewer U. M., et al. Altered levels of laminin receptor mRNA in various human carcinoma cells that have different abilities to bind laminin. Proc. Natl. Acad. Sci. USA. 1986;83:7137–7141. doi: 10.1073/pnas.83.19.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S. A., Hart G. W. Identification of O-GlcNAc sites on proteins. Methods Enzymol. 2006;415:113–133. doi: 10.1016/S0076-6879(06)15008-9. [DOI] [PubMed] [Google Scholar]
- Wimberly B. T., Brodersen D. E., Clemons W. M., Jr, Morgan-Warren R. J., Carter A. P., Vonrhein C., Hartsch T., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wool I. G., Chan Y. L., Gluck A. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 1995;73:933–947. doi: 10.1139/o95-101. [DOI] [PubMed] [Google Scholar]
- Yu L. R., Zhu Z., Chan K. C., Issaq H. J., Dimitrov D. S., Veenstra T. D. Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J. Proteome Res. 2007;6:4150–4162. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]
- Yu Y., Ji H., Doudna J. A., Leary J. A. Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci. 2005;14:1438–1446. doi: 10.1110/ps.041293005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachara N. E., O'Donnell N., Cheung W. D., Mercer J. J., Marth J. D., Hart G. W. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]