The study identifies Hsp90 as a molecular chaperone for KATP channels. Inhibition of Hsp90 function reduces, whereas overexpression of Hsp90 enhances, channel expression at the cell surface. Hsp90 facilitates channel biogenesis by targeting the SUR1 subunit. Up-regulation of Hsp90 also enhances expression of some SUR1 mutants with folding defects.
Abstract
The pancreatic β-cell ATP-sensitive potassium (KATP) channel is a multimeric protein complex composed of four inwardly rectifying potassium channel (Kir6.2) and four sulfonylurea receptor 1 (SUR1) subunits. KATP channels play a key role in glucose-stimulated insulin secretion by linking glucose metabolism to membrane excitability. Many SUR1 and Kir6.2 mutations reduce channel function by disrupting channel biogenesis and processing, resulting in insulin secretion disease. To better understand the mechanisms governing KATP channel biogenesis, a proteomics approach was used to identify chaperone proteins associated with KATP channels. We report that chaperone proteins heat-shock protein (Hsp)90, heat-shock cognate protein (Hsc)70, and Hsp40 are associated with β-cell KATP channels. Pharmacologic inhibition of Hsp90 function by geldanamycin reduces, whereas overexpression of Hsp90 increases surface expression of wild-type KATP channels. Coimmunoprecipitation data indicate that channel association with the Hsp90 complex is mediated through SUR1. Accordingly, manipulation of Hsp90 protein expression or function has significant effects on the biogenesis efficiency of SUR1, but not Kir6.2, expressed alone. Interestingly, overexpression of Hsp90 selectively improved surface expression of mutant channels harboring a subset of disease-causing SUR1 processing mutations. Our study demonstrates that Hsp90 regulates biogenesis efficiency of heteromeric KATP channels via SUR1, thereby affecting functional expression of the channel in β-cell membrane.
INTRODUCTION
ATP-sensitive potassium (KATP) channels in pancreatic β-cells, by virtue of their sensitivities to intracellular nucleotides ATP and ADP, serve as molecular linkers between cell metabolism and cell excitability, thus mediating glucose-regulated insulin secretion (Aguilar-Bryan and Bryan, 1999; Nichols, 2006). The β-cell KATP channel is an octameric complex of four inward rectifier potassium channel (Kir6.2) subunits and four sulfonylurea receptor 1 (SUR1) subunits (Aguilar-Bryan and Bryan, 1999; Nichols, 2006). Mutations in the genes ABCC8 encoding SUR1 or KCNJ11 encoding Kir6.2 that uncouple channel activity from glucose metabolism underlie congenital forms of hyperinsulinism and diabetes (Aguilar-Bryan and Bryan, 1999; Ashcroft, 2005; Flanagan et al., 2009). Studies characterizing disease-associated KATP channel mutations have indicated that many mutations, especially those identified in congenital hyperinsulinism, cause channel dysfunction by disrupting folding or assembly of channel proteins in the endoplasmic reticulum (ER) and thereby subsequent trafficking to the plasma membrane (Cartier et al., 2001; Taschenberger et al., 2002; Ashcroft, 2005; Lin et al., 2006; Yan et al., 2007). Despite the importance of channel biogenesis and surface expression in β-cell function, little is known about how these processes are governed at the molecular level.
Generation of KATP channels in the ER requires correct folding and coassembly of two different channel subunits with distinct structures. Whereas Kir6.2 is a relatively simple protein with only two-transmembrane helices and cytosolic N- and C-terminal domains, SUR1 contains 17 transmembrane helices divided into three transmembrane domains TMD0, TMD1, and TMD2, and two cytoplasmic nucleotide binding domains, NBD1 and NBD2 (Neagoe and Schwappach, 2005). We have shown previously that normal biogenesis of KATP channels is an inefficient process; only ∼20% newly synthesized channel subunits mature into functional channel complex, whereas the majority is degraded by the ubiquitin–proteasome pathway (Yan et al., 2004, 2005). With regard to channel assembly, studies have shown that a tripeptide ER retention/retrieval motif RKR present in both SUR1 and Kir6.2 prevents unassembled or partially assembled channel proteins from exiting the ER (Zerangue et al., 1999). Under physiological conditions where both subunits are coexpressed, assembly of SUR1 and Kir6.2 into octameric complexes masks the RKR signals to allow channel forward trafficking to the plasma membrane. Deletion or mutation of the RKR peptide to AAA also allows individual subunits to reach the cell surface by bypassing a critical ER quality-control checkpoint (Tucker et al., 1997; Zerangue et al., 1999).
Molecular chaperones represent a heterogeneous group of conserved cellular proteins that assist the folding of newly translated proteins. Recent studies suggest that manipulation of molecular chaperones could be used to treat human disease caused by protein misfolding (Welch, 2004; Balch et al., 2008; Powers et al., 2009; Yang et al., 2009). A prominent example is the cystic fibrosis transmembrane conductance regulator (CFTR) mutant ΔF508, which accounts for most of the cystic fibrosis cases (Rowe et al., 2005). CFTR is structurally homologous to SUR1, both proteins belonging to the ATP-binding cassette (ABC) transporter protein family (Higgins, 1995). The molecular chaperone network monitoring CFTR biogenesis has been extensively studied and shown to involve the ER luminal chaperone calnexin and the cytosolic heat-shock protein (Hsp)40/70/90 chaperone complex (Loo et al., 1998; Wang et al., 2006). By contrast, virtually nothing is known about the molecular chaperones that participate in folding and degradation of KATP channels. In addition, there is a lack of understanding of how biogenesis of heteromeric protein complexes such as KATP channels is regulated. This lack of knowledge hinders progress on exploiting molecular chaperones for treatment of disease caused by misfolding of KATP channels.
To address these issues, we have taken a proteomics approach in which epitope-tagged SUR1 and Kir6.2 are expressed in the insulinoma cell line INS-1. Proteins associated with the channel subunits were isolated by affinity purification and identified by tandem mass spectrometry. Here, we show that KATP channel subunits are associated with components of the Hsp90 chaperone complex in the ER, including Hsp40, heat-shock complex (Hsc)70, and Hsp90. The Hsp90 chaperone network has been reported to affect the biogenesis/maturation efficiency of several membrane proteins, including the aforementioned ABC transporter CFTR and the voltage-gated potassium channel human ether-a-go-go-related gene (hERG; Kv11.1). However, other membrane proteins with similar complex topology such as the P-glycoprotein and the multidrug resistance-associated protein (MRP), also ABC transporters, and other Kv channels, including Kv1.5 and Kv2.1, are not affected by perturbation of Hsp90 function (Loo et al., 1998; Ficker et al., 2003). We therefore sought to determine whether the Hsp90 chaperone complex plays a role in KATP channel processing and maturation. We demonstrate that inhibition of Hsp90 reduces KATP channel expression at the cell surface, whereas overexpression of Hsp90 increases the number of KATP channels in the plasma membrane. Importantly, we found that the Hsp90 complex interacts preferentially with the SUR1 subunit. Moreover, although perturbation of Hsp90 significantly alters cell surface levels of a SUR1 expressed alone, it has little effect on those of a Kir6.2 expressed alone. Our results indicate that the Hsp90 chaperone regulates biogenesis efficiency of KATP channels by targeting the SUR1 but not the Kir6.2 subunit.
MATERIALS AND METHODS
Expression Constructs
Wild-type hamster SUR1 or FLAG epitope (DYKDDDDK)-tagged SUR1 (referred to as fSUR1) cDNAs were cloned in the pECE plasmid. Wild-type rat Kir6.2 or HA epitope (YPYDVPDYA)-tagged Kir6.2 cDNAs were cloned into pcDNA3, as described previously (Cartier et al., 2001). Construction of adenovirus carrying wild-type (WT) fSUR1 or WT Kir6.2 or hemagglutinin (HA)-tagged WT Kir6.2 cDNA was as described previously (Lin et al., 2008). The FLAG-tag for SUR1 was placed at the extracellular N terminus of the protein. The HA-tag for Kir6.2 was inserted between amino acid 100 and 101 in the extracellular domain; an extra nine amino acids, DLYAYMEKG, was added between amino acid 98 and 99 to aid accessibility of the epitope. We have shown in previous studies that these epitope tags do not affect the trafficking or function of the channels (Cartier et al., 2001; Lin et al., 2005). The fSUR1 recombinant adenovirus was made using a modified pShuttle plasmid (AdEasy kit; Stratagene, La Jolla, CA) containing a tetracycline-inducible promoter. Recombinant viruses were amplified in human embryonic kidney 293 cells and purified according to the manufacturer's instructions.
Virus Infection
INS-1 cells clone 832/13 (kindly provided by Dr. Christopher Newgard, Duke University, Durham, NC) were plated in 10-cm plates and cultured for 24 h in RPMI 1640 medium with 11.1 mM d-glucose (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES, 2 mM glutamine, 1 mM sodium pyruvate, and 50 μM β-mercaptoethanol (Hohmeier et al., 1997). For Kir6.2 expression, recombinant adenoviruses containing Kir6.2 or HA-Kir6.2 with desired titers were used as described previously (Lin et al., 2005, 2008). For fSUR1 expression, cells were coinfected with equal amounts of two recombinant adenoviruses, one adenovirus encoding a tetracycline-inhibited transactivator (tTA) and the other adenovirus a tTA-regulated gene expressing fSUR1 (Lin et al., 2008). Cells at ∼70% confluency were washed once with phosphate-buffered saline and then incubated for 1.5 h at 37°C in Opti-MEM containing low bovine serum and a mixture of viruses. The multiplicity of infection for each virus was determined empirically. After 90 min, 2× growth medium was added, and the cells were incubated at 37°C until reaching appropriate density for the various experiments.
Affinity Purification of KATP Channel Complexes
INS-1 (832/13) cells were transduced with adenoviruses carrying fSUR1, HA-Kir6.2, or fSUR1 and Kir6.2 as described above. Twenty-four hours after infection, the cells were lysed in 1 ml of lysis buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, and 1% Triton X-100, with Complete protease inhibitors [Roche Diagnostics, Indianapolis, IN]) on ice for 30 min. The cell lysate was centrifuged at 16,000 × g for 5 min at 4°C, and the supernatant was used for affinity purification by addition of 100 μl of FLAG- or HA-antibody conjugated agarose beads (Sigma-Aldrich, St. Louis, MO) overnight at 4°C. After washing three times with the lysis buffer, bound proteins were eluted by incubation with FLAG peptide (250 μg/ml, for fSUR1 sample) or HA peptide (10 μg/ml, for HA-Kir6.2 sample) at room temperature for 30 min.
Proteomics and Mass Spectrometry Analysis
Affinity-purified samples were concentrated to a final volume of 20 μl, mixed with Laemmli sample buffer, and electrophoresed briefly into 10% Bis-Tris gels (Invitrogen) at 200 V in 3-(N-morpholino)propanesulfonic acid buffer for 5 min. The gel was stained with Imperial Coomassie (Pierce Chemical, Rockville, IL), placed on a clean glass plate, and each sample was excised by cutting just below the dye front and removing the entire top of the lane including the bottom of the well. The excised gel pieces were then cut into 1- to 2-mm sections, washed twice by shaking in 0.5 ml of water for 15 min, washed twice again by shaking in 0.5 ml of 1:1 solution of acetonitrile:100 mM ammonium bicarbonate for 30 min, and then dried by vacuum centrifugation. After reduction and alkylation of proteins in the excised gel slices by incubation first in dithiothreitol (10 mM in 100 mM ammonium bicarbonate) and then in iodoacetamide (25 mM in 100 mM ammonium bicarbonate), proteins were digested by rehydrating the gel slice on ice in trypsin digestion solution containing 100 mM ammonium bicarbonate, and 10 μg/ml proteomics grade trypsin (Sigma-Aldrich). The excess trypsin solution was then removed and sufficient 100 mM ammonium bicarbonate was added to cover the gel slices; samples were incubated overnight at 37°C. The solution was then removed from the gel slices, additional peptides were extracted, the volume of the collected digests was decreased by vacuum centrifugation, and samples were transferred directly to autosampler vials for analysis.
The peptide mixture was injected onto a 1 mm x 8 mm trap column (Michrom BioResources, Auburn, CA) at 20 μl/min in a mobile phase containing 0.1% formic acid. The trap cartridge was then placed in-line with a 0.5- × 250-mm column containing 5-μm Zorbax SB-C18 stationary phase (Agilent Technologies, Santa Clara, CA), and peptides were separated by a 2–30% acetonitrile gradient over 95 min at 10 μl/min by using a 1100 series capillary high-performance liquid chromotography (Agilent Technologies). Peptides were analyzed using a LTQ linear ion trap fitted with an Ion Max Source and 34-gauge metal needle kit (Thermo Fisher Scientific, Waltham, MA). Survey mass spectrometry (MS) scans were alternated with three data-dependant tandem MS (MS/MS) scans by using the dynamic exclusion feature of the software to increase the number of unique peptides analyzed. Peptide identification was performed by comparing observed MS/MS spectra to theoretical fragmentation spectra of peptides generated from a protein database using Sequest, version 27, revision 12 (Thermo Fisher Scientific). A Swiss-Prot database created in April 2008 containing hamster, mouse, and rat sequences was used (Swiss Institute of Bioinformatics, Geneva, Switzerland). Scaffold software (Proteome Software, Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide and protein identifications were accepted if they could be established at >95% and 95% probability, respectively (Keller et al., 2002; Nesvizhskii et al., 2003) and contained at least two identified peptides matched per protein entry.
Western Blotting and Immunoprecipitation Assay
INS-1 (832/13) cells were infected with SUR1 and Kir6.2 as described above. Twenty-four hours after infection, the cells were lysed in 1 ml of lysis buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, and 1% Triton X-100, with Complete protease inhibitors) on ice for 30 min. The cell lysate was centrifuged at 16,000 × g for 5 min at 4°C, and the supernatant was used for Western blot or immunoprecipitation. Immunoprecipitation was performed as described under Affinity Purification. Eluted proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membrane. The membrane was probed with appropriate primary antibodies including anti-FLAG (Sigma-Aldrich), anti-Hsp90α/β (Santa Cruz Biotechnology), anti-Hsp40 (Abcam, Cambridge, MA), and anti-Hsc70 (Abcam), followed by incubation with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and visualized by enhanced chemiluminescence (Super Signal West Femto; Pierce Chemical).
Chemiluminescence Assay for Surface Expression
COSm6 cells or INS-1 cells in 35-mm dishes were fixed with 2% paraformaldehyde for 20 min at room temperature 48 h after transfection or infection. Fixed cells were preblocked in phosphate-buffered saline (PBS) + 0.1% bovine serum albumin (BSA) for 1 h, incubated in M2 anti-FLAG antibody (10 μg/ml) for 1 h, washed 4 × 30 min in PBS + 0.1% BSA, incubated in horseradish peroxidase-conjugated anti-mouse secondary antibodies (1:1000 dilution; GE Healthcare) for 20 min, washed again 4 × 30 min in PBS + 0.1% BSA, and 2 × 5 min in PBS. Chemiluminescence signal was read in a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA) after 10-s incubation in Power Signal ELISA luminol solution (Pierce Chemical). The results of each experiment are the average of two dishes. Signals observed in untransfected COSm6 cells or uninfected INS-1 cells were subtracted as background for COSm6 or INS-1 cell experiments, respectively. Data points shown in figures are the average of three to 10 independent experiments as specified.
Metabolic Labeling and Immunoprecipitation
COSm6 cells grown on 35-mm dishes were transfected with fSUR1 and Kir6.2 for 24 h. The cells were incubated in methionine/cysteine-free DMEM supplemented with 5% dialyzed fetal bovine serum for 30 min before labeling with l-[35S]methionine (Tran35S-Label, 150–250 μCi/ml; MP Biomedicals, Solon, OH) for 60 min at 37°C. Labeled cultures were chased in regular medium supplemented with 10 mM methionine at 37°C. At the end of the chase, the cells were lysed in 500 μl of the lysis buffer described above. For immunoprecipitation, 500 μl of cell lysate was incubated with 100 μl of FLAG-antibody–conjugated agarose beads overnight at 4°C. The precipitate was washed three times in the lysis buffer, and the proteins were eluted with FLAG-peptide. The eluted proteins were separated by 8% SDS-PAGE, and the dried gels were analyzed using a Storm PhosphorImager (GE Healthcare).
86Rb+ Efflux Assay
INS-1 cells were plated onto 12-well plates and cultured for 2 d to confluence. Cells were incubated for 12 h in culture medium containing 86RbCl (1 μCi/ml) with or without 17-(allylamino)-17-demethoxygeldanamycin (17-AGG; 100 or 200 nM). Before measurement of 86Rb+ efflux, cells were incubated for 30 min at room temperature in Krebs-Ringer solution with metabolic inhibitors (2.5 μg/ml oligomycin and 1 mM 2-deoxy-d-glucose). At selected times, the solution was aspirated from the cells and replaced with fresh solution. At the end of a 40-min period, cells were lysed. The 86Rb+ in the aspirated solution and the cell lysate was counted. The percentage of efflux at each time was calculated as the cumulative counts in the aspirated solution divided by the total counts from the solutions and the cell lysate.
Patch-Clamp Recording
INS-1 cells were plated onto coverslips. Endogenous KATP currents were assessed using inside-out patch voltage-clamp recording after 12-h pretreatment with 17-AGG. Micropipettes were pulled from nonheparinized Kimble glass (Thermo Fisher Scientific) on a horizontal puller (Sutter Instrument, Novato, CA) with resistance typically ∼1–2 megaohms. The bath (intracellular) and pipette (extracellular) solution (K-INT) had the following composition: 140 mM KCl, 10 mM K-HEPES, and 1 mM K-EGTA, pH 7.3. ATP and ADP were added as the potassium salt. Recording was performed at room temperature and currents were measured at a membrane potential of −50 mV. In Figure 3, inward currents were shown as upward deflections. KATP current was calculated as the difference between current in ATP-free (channels open) and in 1 mM ATP (channels closed) solutions.
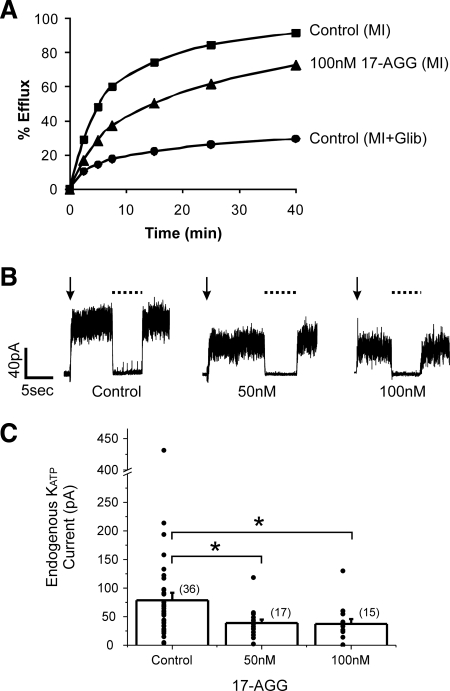
Figure 3.
The Hsp90 inhibitor 17-AGG diminishes endogenous INS-1 KATP channel activity. (A) Representative 86Rb+ efflux profile of INS-1 cells pretreated with metabolic inhibitors for 30 min to activate KATP channels. Treatment of cells with 100 nM 17-AGG for 12 h led to a decrease in the amount of efflux, reflecting reduced channel activity. (B) Representative current traces from inside-out patch-clamp recordings after 12-h treatment with 17-AGG. KATP current was calculated as the difference between current in ATP-free K-INT solution (channels open) and in K-INT with 1 mM ATP (channels closed, dashed lines). The downward arrow indicates the time of patch excision. (C) Bar graph showing average currents. The distribution of KATP currents are shown as circles for each condition, note the break in the y-axis. The total number of patches is given above each bar. The current density in cells treated with 50 or 100 nM 17-AGG was significantly lower than that of control cells (p = 0.03 and 0.05, respectively).
Statistics
Data are presented as mean ± SEM. Differences were tested using analysis of variance (ANOVA) when comparing three or more groups, and Dunnett's post hoc test was used to compare treated versus control group. When only two groups were compared, unpaired Student's t tests were used. Differences were assumed to be significant if p ≤ 0.05.
RESULTS
Identification of Molecular Chaperones Associated with KATP Channel Proteins
To identify proteins that interact with KATP channels in the early secretory pathway in β-cells, we overexpressed fSUR1 or HA-Kir6.2 in INS-1 cells (clone 823/13, a model β-cell line from rat; Hohmeier et al., 1997) by using recombinant adenoviruses (Lin et al., 2005, 2008). The rationale for the overexpression approach is that it will enhance the probability of capturing molecular chaperones that are associated with each subunit during biosynthesis and folding by increasing fraction of folding intermediates present. The epitope-tagged channel subunits were used to facilitate affinity purification, and we have shown previously that the FLAG- or HA-epitope on SUR1 or Kir6.2, respectively, do not affect channel expression or function (Cartier et al., 2001; Lin et al., 2005). Note that viral transduction was used because INS-1 cells exhibit low transfection efficiency using nonviral transfection methods (Lin et al., 2005, 2006; Yan et al., 2007). Proteins that interact with the channel subunits were captured using an affinity purification approach by incubation of the lysate with agarose beads conjugated with anti-FLAG or anti-HA antibody followed by elution using FLAG- or HA-peptide. The eluted proteins were identified by tandem mass spectrometry as described under Materials and Methods.
In total, 10 experiments were conducted: three in INS-1 cells transduced with viruses for fSUR1 overepxression only (fSUR1 virus plus tTA virus; see Materials and Methods), two in cells transduced with the virus for HA-Kir6.2 only, and five in cells transduced with viruses for overexpression of fSUR1 and untagged Kir6.2. In all experiments, uninfected INS-1 cells were subjected to the same experimental procedure to serve as a negative control. Results of three representing experiments from the three different conditions are shown in Table 1. The chaperone proteins consistently copurified with the channel subunits are Hsp40, Hsc70, and Hsp90α/β. Calnexin and Hsp organizing protein (HOP) were also present in a subset of experiments in which fSUR1 was used as a bait (Table 1). In yet another proteomics experiment using COS cells transfected with fSUR1 and Kir6.2 cDNAs, Hsp40, Hsc70, and Hsp90α/β were also found in the fSUR1 immunoprecipitate (data not shown).
Table 1.
KATP channel subunits-associated chaperone proteinsa
| Accession no. (NCBI) | Protein name | Sequence coverage (%) | Unique spectra | Total spectra |
|---|---|---|---|---|
| fSUR1 only | ||||
| Q09427 | SUR1 | 46 | 143 | 588 |
| Q61743 | Kir6.2 | 11 | 4 | 4 |
| Q76G10 | Hsp40 | 26 | 7 | 9 |
| P19378 | Hsc70 | 57 | 43 | 72 |
| P46633 | Hsp90α | 29 | 20 | 26 |
| P11499 | Hsp90β | 39 | 30 | 43 |
| O54981 | HOP | 16 | 6 | 6 |
| Q8K3H8 | Calnexin | 13 | 7 | 10 |
| fSUR1 + Kir6.2 | ||||
| Q09427 | SUR1 | 48 | 126 | 377 |
| Q61743 | Kir6.2 | 21 | 10 | 18 |
| Q76G10 | Hsp40 | 37 | 9 | 12 |
| P19378 | Hsc70 | 61 | 33 | 57 |
| P46633 | Hsp90α | 26 | 19 | 25 |
| P11499 | Hsp90β | 34 | 24 | 35 |
| O54981 | HOP | 7 | 3 | 3 |
| Q8K3H8 | Calnexin | 13 | 7 | 8 |
| HA-Kir6.2 only | ||||
| Q09427 | SUR1 | 8 | 7 | 9 |
| Q61743 | Kir6.2 | 30 | 22 | 100 |
| Q76G10 | Hsp40 | 29 | 6 | 10 |
| P19378 | Hsc70 | 57 | 42 | 95 |
| P46633 | Hsp90α | 17 | 9 | 13 |
| P11499 | Hsp90β | 23 | 13 | 18 |
| O54981 | HOP | 0 | 0 | 0 |
| Q8K3H8 | Calnexin | 0 | 0 | 0 |
a In total, 10 proteomics experiments were performed: three in INS-1 cells infected with fSUR1 virus only, five in INS-1 cells infected with fSUR1 and Kir6.2 viruses, and two in INS-1 cells infected with HA-Kir6.2 virus only. The fSUR1 or HA-Kir6.2 protein complexes in INS-1 cell lysates were affinity purified using FLAG- or HA-antibody–conjugated agarose beads and subjected to mass spectrometry analyses as described in Materials and Methods. Results from three proteomic experiments representing each condition are shown. Note Hsp40, Hsc70, and Hsp90α and -β are present in all experiments. The protein HOP is found in only a subset of experiments overexpressing fSUR1 alone or fSUR1 plus Kir6.2 (6 of 8 experiments), so is the protein calnexin (5 of 8 experiments); neither proteins showed up in cells overexpressing HA-Kir6.2 only. NCBI, National Center for Biotechnology Information.
Differential Association of KATP Channel Subunits with the Hsp90 Chaperone Complex
To confirm the interaction between channel subunits and chaperone proteins identified by mass spectrometry, Hsp40, Hsc70, and Hsp90 were coimmunoprecipitated with fSUR1 or HA-Kir6.2 from INS-1 cells transduced with the fSUR1 or the HA-Kir6.2 virus, respectively, as described under Materials and Methods. Results revealed that all three chaperones interact with fSUR1 (Figure 1A, left). Similarly, all three chaperones were detected in the HA-Kir6.2 immunoprecipitate (Figure 1A, right). Note that overexpression of individual channel subunits in INS-1 cells did not significantly alter the abundance of the various heat shock proteins examined (input).
Figure 1.
Interactions between KATP channel subunits and Hsp90 complexes. (A) Coimmunoprecipitation of Hsp90 complexes with SUR1 subunits was examined in uninfected (UnI) and wild-type FLAG-SUR1 (fSUR1) or HA-Kir6.2 adenovirus infected INS-1 cells by immunoprecipitation (IP) using anti-FLAG or anti-HA antibody-coated beads, respectively. Membranes were immunoblotted (IB) individually with the indicated antibody (anti-Hsp90, anti-Hsc70, and anti-Hsp40) for endogenous protein detection either before IP (bottom, input) or after IP (top). Input fSUR1 or HA-Kir6.2 is also shown on the very bottom. (B) Similar coIP experiments were performed in COSm6 cells transfected with fSUR1 or HA-Kir6.2 alone.
Because INS-1 cells express endogenous KATP channel proteins albeit at a lower level than the exogenously expressed fSUR1 or HA-Kir6.2 (Lin et al., 2006), it is possible that association with fSUR1 or HA-Kir6.2 was mediated by endogenous channel subunits interacting with the epitope-tagged exogenous channel subunits. We therefore expressed fSUR1 or HA-Kir6.2 individually in COS cells that do not have endogenous SUR1 or Kir6.2 and performed similar coimmunoprecipitation experiments. As shown in Figure 1B, Hsp90, Hsc70, and Hsp40 still coimmunoprecipitated with fSUR1, but they were not detected in the Kir6.2 immunoprecipitate. Although the lack of signal for these chaperone proteins in the Kir6.2 immunoprecipitate could be caused by factors such as the cell type used or the expression level of Kir6.2, the simplest interpretation of the results is that these chaperone proteins might interact preferentially with SUR1 subunit in the KATP channel complex.
Inhibition of Hsp90 Reduces KATP Channel Surface Expression
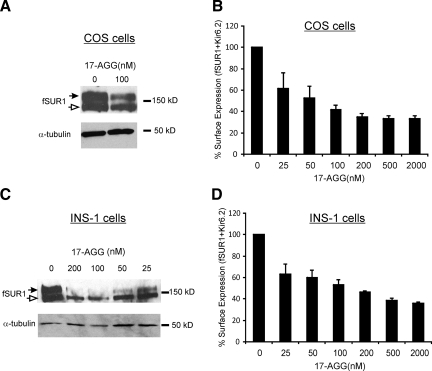
To determine the role of Hsp90 complexes in channel biogenesis, we examined the effects of 17-AGG, a geldanamycin derivative that inhibits the ATPase activity and client binding properties of Hsp90 (Kamal et al., 2003), on KATP channel biogenesis efficiency and surface expression in COS cells. SUR1 has two N-linked glycosylation sites. As the SUR1/Kir6.2 complex traffics through the Golgi network, SUR1 becomes complex glycosylated to give rise to the mature form that migrates slower on SDS-PAGE (referred to as the upper band; Figure 2) than the immature ER core-glycosylated form (the lower band). Western blots of lysates from cells expressing both fSUR1 and Kir6.2 showed that 17-AGG (100 nM for 12 h) significantly decreased the abundance of the mature complex-glycosylated SUR1 band (upper band), suggesting reduced processing efficiency of the channel protein (Figure 2A, left). In agreement, chemiluminescence assays showed that surface expression of KATP channels in COS cells treated with a range of 17-AGG concentrations (25 nM–2 μM) was reduced in a dose-dependent manner, with the effect being apparent at 25 nM and saturating at ∼200 nM (Figure 2B). Similar results were obtained in INS-1 cells infected with KATP channel subunits viruses, with the effect on surface expression being apparent at 25 nM and saturating at ∼500 nM (Figure 2, C and D). In addition to reducing the abundance of the upper band, 17-AGG also reduced the signal of the lower fSUR1 band (Figure 2, A and C), suggesting that inhibition of Hsp90 function might accelerate degradation of SUR1 in the ER as has been reported for CFTR (Loo et al., 1998).
Figure 2.
Hsp90 inhibitor 17-AGG decreases surface expression of KATP channel in COS and INS-1 cells. COS cells cotransfected with fSUR1 and Kir6.2 subunits were treated with 100 nM 17-AGG overnight. (A) Western blot shows reduced complex- (upper band, solid arrow) and core-glycosylated (lower band, empty arrow) fSUR1 in 17-AGG–treated cells compared with control, indicating reduced channel biogenesis. (B) KATP channels in the plasma membrane assessed by chemiluminescence assays show significantly reduced surface expression levels in cells treated with various concentrations of 17-AGG compared with control cells. The effect of 17-AGG was evident at 25 nM. (C) Effects of 17-AGG on KATP channel maturation and surface expression were further examined in INS-1 cells infected with fSUR1 and Kir6.2 adenoviruses. As in COS cells, Western blots show that 17-AGG causes a dose-dependent reduction in the amount of complex-glycosylated fSUR1. (D) Reduction in surface expression of KATP channels was quantified by chemiluminescence assays. Each data point in the bar graphs represents the mean ± SEM of three experiments. All 17-AGG–treated cells showed statistically significant difference in surface expression compared with untreated cells (p = 0.02 for 25 nM and p <0.02 for all other groups).
Because the studies described so far were conducted on KATP channels heterologously expressed in COSm6 cells or INS-1 cells in which protein overexpression might be a concern, we examined the effect of 17-AGG on functional expression of endogenous KATP channels in INS-1 cells. Channel function was first assessed by 86Rb+ efflux assays that monitor activity in response to metabolic inhibition in intact cells (Shyng et al., 1998). Compared with control, cells treated with 100 nM 17-AGG for 12 h showed substantially reduced 86Rb+ efflux over a 40-min period, indicating reduced channel activity (Figure 3A). Next, inside-out patch voltage-clamp recordings were used to directly measure KATP current density in nucleotide-free solutions. In INS-1 cells treated with 50 or 100 nM 17-AGG, the averaged KATP current was 38.81 ± 6.07 and 37.30 ± 8.44 pA, respectively, both significantly lower than that observed in control cells (78.71 ± 12.95 pA; Figure 3, B and C). Because 17-AGG did not affect current amplitude or Mg-ADP response of KATP channels when applied acutely to isolated membrane patches at concentrations used in the above-mentioned experiments (data not shown; also see Discussion), we concluded that the reduced KATP channel activities were due to effects of 17-AGG on the number of channels expressed at the surface rather than on channel gating directly. Thus, inhibition of Hsp90 function by 17-AGG caused a decrease in endogenous KATP channels present at the INS-1 cells membrane. These results support a role of Hsp90 in the biogenesis and surface expression efficiency of native KATP channels.
Overexpression of Hsp90 Increases Surface Expression of KATP Channels in COSm6 Cells
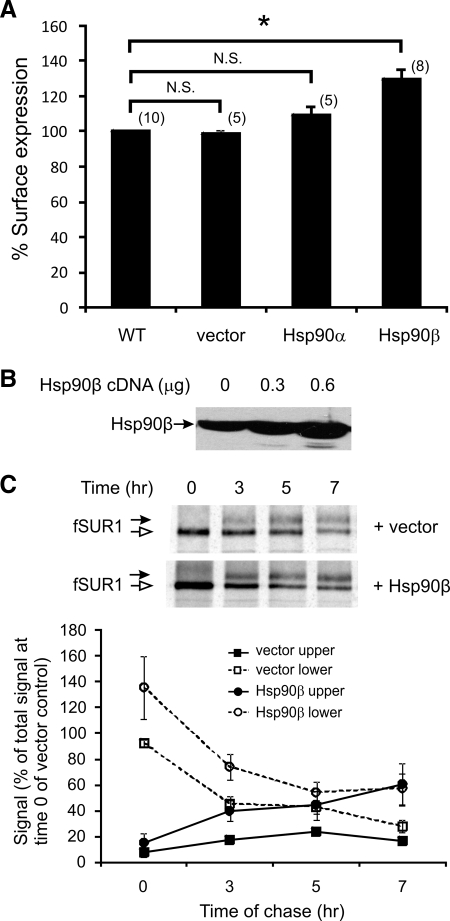
Given that the Hsp90 inhibitor reduces KATP channel processing and surface expression, we next examined whether overexpression of Hsp90 might facilitate KATP channel maturation and increase surface channel expression. There are two major cytosolic Hsp90 isoforms: the α isoform that is highly inducible and the β isoform that is thought to be more constitutive (Sreedhar et al., 2004). For our experiments, Hsp90α or β cDNA or empty vector were transfected along with WT fSUR1 and Kir6.2 into COSm6 cells, and the resulting KATP channel surface expression was analyzed by chemiluminescence assays. As shown in Figure 4A, overexpression of Hsp90β cDNA increased surface expression of KATP channels by 29.64 ± 6.29%. Overexpression of Hsp90α also improved surface expression slightly (by 9.46 ± 4.82%), but this effect did not reach statistical significance. We therefore focused on Hsp90β in the experiments described below. To determine whether the increased surface expression of KATP channels in cells overexpressing Hsp90β is a consequence of increased channel biogenesis, we performed metabolic pulse-chase labeling experiments to monitor the processing and maturation of newly synthesized SUR1 protein. Overexpression of Hsp90β was first confirmed by Western blots (Figure 4B). In metabolic pulse-chase experiments, the amount of SUR1 protein labeled at the end of the 1-h pulse period was higher in cells overexpressing Hsp90β than in control cells, indicating Hsp90β overexpression led to increased biogenesis of SUR1. Signals of both the lower and upper bands from seven experiments were quantified and are shown in Figure 4C. The signal of the mature complex-glycosylated SUR1 upper band in Hsp90β-cotransfected cells is consistently higher than that in control cells at all times during the chase period. These results provide evidence that Hsp90β function contributes to the biogenesis efficiency, thereby surface expression levels of KATP channels.
Figure 4.
Overexpression of Hsp90 increases surface expression of KATP channels by improving biogenesis efficiency of the channel. (A) COS cells were cotransfected with fSUR1 and Kir6.2 subunits and either 0.6 μg Hsp90 (α or β) or empty vector as control. Surface expression of KATP channels was analyzed by chemiluminescence assays 48–72 h after transfection. In cells cotransfected with Hsp90β, channel surface expression levels were significantly higher (29.65 ± 6.29%) than cells cotransfected with either empty vector or Hsp90α. *p < 0.0003; N.S., not significant. (B) Western blot of Hsp90 from COS cells transfected with 0, 0.3, or 0.6 μg of Hsp90β cDNA (per 35-mm culture dish). (C) Metabolic pulse-chase experiments were performed in COS cells transfected with KATP channel subunits and Hsp90β or empty vector to assess channel maturation. Cells were pulse-labeled with [35S]methionine for 1 h and chased for the indicated times, and fSUR1 was immunoprecipitated using anti-FLAG antibodies. An autoradiograph shows the changes in the signal of fSUR1 mature upper band and the core-glycosylated immature lower band over the 7-h chase period. The signal of the upper and lower band was quantified using a PhosphorImager and is expressed as percentage of signal of total signal observed at time 0 for the empty vector-transfected control sample (n = 7 for each data point). The Hsp90β-transfected samples showed significantly higher upper band signal at 3 and 7 h (p = 0.02 and 0.03, respectively); the signal was also higher for the Hsp90β sample at 5 h, although the difference did not reach statistical significance (p = 0.07).
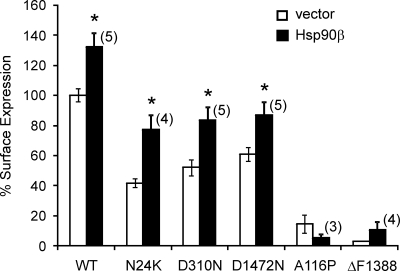
To test whether overexpression of Hsp90β can alleviate the folding/surface expression defects of mutant KATP channels, we examined the effect of Hsp90β on surface expression of several mutant channels characterized previously as being processing/trafficking-deficient (Cartier et al., 2001; Yan et al., 2004, 2007). The five mutants examined harbor mutation N24K, A116P, D310N, ΔF1388, or D1472N in the SUR1 subunit. Of these, the N24K, D310N, and D1472N mutants exhibited improved surface expression levels in cells cotransfected with Hsp90β cDNA relative to cells cotransfected with a control empty vector. However, the A116P and ΔF1388 mutants did not show improved surface expression (Figure 5). Interestingly, the N24K, D310N and D1472N mutants have relatively milder processing/trafficking defects in that they do express at the cell surface to some extent even under control conditions, in contrast to A116P and ΔF1388 that show virtually no surface expression. These results suggest that although up-regulation of Hsp90 can rescue the processing and surface expression of some mutants, effects are dependent on the nature of the mutation and severity of processing defect.
Figure 5.
Hsp90 improves surface expression of select misprocessed mutant KATP channels. The effect of Hsp90β abundance on surface expression of KATP channels harboring SUR1 mutations known to disrupt normal processing and trafficking of the channel was assessed by chemiluminescence assays in COS cells cotransfected with various KATP channel subunits (0.3 μg of WT or mutant fSUR1, 0.2 μg of Kir6.2 per 35-mm dish of cells) and either 0.6 μg of Hsp90β or empty vector as control. Although Hsp90β improved surface expression of the N24K, D310N, and D1472N mutant (p = 0.01, 0.01, 0.05, and 0.03 for WT, N24K, D310N, and D1472N, respectively), it did not significantly increase surface expression of the A116P or ΔF1388 mutants.
Hsp90 Exerts Its Effect on KATP Channel Biogenesis via the SUR1 Subunit
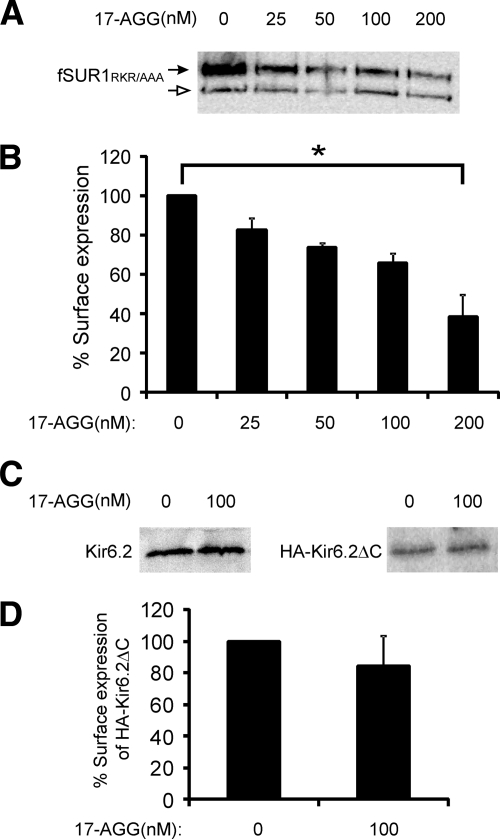
Unlike CFTR or hERG channels, the KATP channel complex is formed by two membrane proteins of very different size and topological complexity. In coimmunoprecipitation experiments conducted in COS cells expressing fSUR1 or HA-Kir6.2 alone, we detected association of the Hsp90 chaperone complex proteins with SUR1 but not Kir6.2 (Figure 1), suggesting the chaperone complex might target the SUR1 subunit to exert its effect on channel biogenesis. To address the question of whether Hsp90 promotes channel maturation and surface expression by facilitating the folding and processing of SUR1, Kir6.2 or both, we examined how manipulation of Hsp90 function affects surface expression of SUR1 and Kir6.2 individually. For these studies, we used an fSUR1 construct in which the RKR ER retention/retrieval motif has been mutated to AAA (fSUR1RKR→AAA) and a HA-Kir6.2 construct in which the RKR motif at the C terminus has been deleted (HA-Kir6.2ΔC36). Previous studies have shown that mutation of the RKR motif to AAA or deletion of this signal allows individual subunits to express at the cell surface independently of the other subunit (Zerangue et al., 1999). Also, the FLAG- and HA-epitope tags are inserted in the extracellular domain of SUR1 and Kir6.2 respectively, to permit quantification of surface expression of the individual protein by using the chemiluminescence assay described under Materials and Methods (Cartier et al., 2001; Lin et al., 2005). In COSm6 cells expressing fSUR1RKR→AAA alone, inhibition of Hsp90 function by 17-AGG led to reduced steady state mature fSUR1RKR→AAA upper band level as well as the core-glycosylated immature lower band (Figure 6A), similar to that observed for fSUR1 coexpressed with Kir6.2 (Figure 2A). Chemiluminescence assays also revealed reduced surface expression of fSUR1RKR→AAA in a 17-AGG concentration-dependent manner (Figure 6B). In contrast, the steady state protein level of Kir6.2 or HA-Kir6.2ΔC36 expressed in COSm6 cells in the absence of SUR1 was not affected by the 17-AGG treatment based on Western blots (Figure 6C), and surface expression of HA-Kir6.2 also unaffected as assessed by chemiluminescence assays (Figure 6D).
Figure 6.
Inhibition of Hsp90 function by 17-AGG reduces surface expression of fSUR1RKR→AAA but not HA-Kir6.2ΔC36 expressed in the absence of the other channel subunit. (A) COS cells were transfected with fSUR1RKR→AAA only and treated with various concentrations of 17-AGG for 12 h. Western blot shows reduced complex-glycosylated as well as core-glycosylated fSUR1RKR→AAA in 17-AGG–treated cells compared with control. (B) Chemiluminescence assays also showed reduced surface expression of fSUR1RKR→AAA in cells treated with 17-AGG compared with untreated cells (p = 0.02 for 25 nM and p <0.001 for all other groups; n = 3). (C) Similar experiments as described in A were performed in COS cells transfected with Kir6.2 or HA-Kir6.2ΔC36 (HA-Kir6.2ΔC) only. Western blots showed no difference in steady-state Kir6.2 protein expression level between control and 17-AGG–treated group. (D) Chemiluminescence assays performed using cells expressing the HA-Kir6.2ΔC36 mutant construct (as Kir6.2 does not traffic to the cell surface in the absence of SUR1) showed no significant difference between control and 17-AGG–treated group (n = 3).
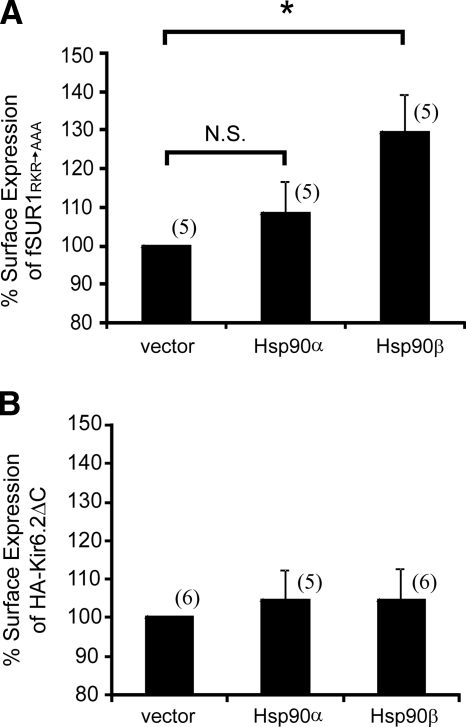
Next, either fSUR1RKR→AAA or HA-Kir6.2ΔC36 were coexpressed with Hsp90α or β in COSm6 cells and surface expression of each channel subunit measured by chemiluminescence assays by using anti-FLAG or anti-HA antibody respectively. As shown in Figure 7A, coexpression of Hsp90β resulted in 29.52 ± 9.87% increase in surface expression of fSUR1RKR→AAA, similar to that observed for WT SUR1/Kir6.2 channel complex (Figure 4A). By contrast, no significant difference in surface expression of HA-Kir6.2Δ36 was observed between control cells (vector) and cells overexpressing either Hsp90α or β (Figure 7B). These results suggest that Hsp90 improves the surface expression of KATP channels by facilitating folding and processing efficiency of SUR1 rather than Kir6.2.
Figure 7.
Overexpression of Hsp90 increases surface expression of fSUR1RKR→AAA but not HA-Kir6.2ΔC36. (A) COS cells were transfected with fSUR1RKR→AAA and 0.6 μg of either Hsp90α, Hsp90β, or empty vector (as control). Surface expression of fSUR1RKR→AAA was analyzed by chemiluminescence assays by using anti-FLAG antibody. In cells overexpressing Hsp90β, surface expression levels of fSUR1RKR→AAA were significantly higher (29.52 ± 9.87%) than cells cotransfected with either empty vector or Hsp90α. *p = 0.02; N.S., not significant. (B) Same as A except that surface expression of HA-Kir6.2ΔC36 was examined using anti-HA antibody. No difference was observed between the control and Hsp90α- or Hsp90β-cotransfected cells.
DISCUSSION
Pancreatic β-cell KATP channels have been extensively studied in the past two decades. However, as the key signal transduction protein complex linking glucose metabolism to insulin secretion remarkably little is known about the molecular mechanisms involved in the channel's biogenesis process. In this work, we have begun to address these issues by identifying molecular chaperones that associate with the channel subunits in β-cells, by using the insulinoma cell line INS-1 as an experimental platform. We provide evidence that the Hsp90 chaperone complex participates in the biogenesis of KATP channels. First, KATP channels are associated with Hsp90, Hsc70, and Hsp40, components of the Hsp90 chaperone complex. Second, inhibition of Hsp90 function by 17-AGG markedly reduced surface expression of KATP channels, whereas overexpression of Hsp90β significantly increased surface expression. In addition, we show that overexpression of Hsp90β improved surface expression of select misprocessing/mistrafficking SUR1 mutants. Finally, we present evidence that Hsp90 facilitates channel biogenesis by increasing the folding/processing efficiency of SUR1.
Comparison with Other Hsp90 Client Proteins
Hsp90 is an abundant cytosolic chaperone (Pearl and Prodromou, 2006). Unlike Hsc/p70, which acts by holding newly synthesized proteins in a folding competent state, Hsp90 is required for the folding of a subset of client proteins that are thought to have difficulties in reaching a native conformation (Nathan et al., 1997; Young et al., 2001). Even with proteins that share structural similarity or complexity, there can be a high degree of specificity. We now identify the KATP channel as another Hsp90 client. KATP channels are unique in that they are heterooligomers of SUR1 and Kir6.2—two membrane proteins with very different topological complexity (Inagaki et al., 1995; Conti et al., 2001). In understanding the biogenesis of such heteromeric protein complex, one has to consider the folding and processing of both channel subunits. Our study using SUR1 and Kir6.2 mutants that are able to express at the cell surface in the absence of the other assembly partner subunit shows that the two subunits exhibit differential dependence on Hsp90 for surface expression. Although surface levels of SUR1 are reduced by 17-AGG and augmented by overexpression of Hsp90β, those of Kir6.2 are not affected. These findings are consistent with the coimmunoprecipitation results obtained in COS cells in which the Hsp90 complex proteins were only detectable in the SUR1 but not Kir6.2 immunoprecipitates. To our knowledge, this is the first example in which differential dependences of individual components of a membrane protein complex on Hsp90 has been documented.
Identification of SUR1 as an Hsp90 client protein also underlines the question of how substrate specificity for this molecular chaperone is determined (Pearl and Prodromou, 2000). Although Hsp90 is important for the folding and maturation of CFTR and SUR1, it does not seem critical for the processing and maturation of two other ABC transporters, P-glycoprotein and MRP (Loo et al., 1998). In terms of sequence homology, P-glycoprotein belongs to the ABCB subfamily, whereas CFTR, SUR1, and MRP are in the ABCC subfamily (Vasiliou et al., 2009). With respect to membrane topology, CFTR and P-glycoprotein both contain a tandem repeat of six transmembrane helices, each set followed by an ATP-binding domain. SUR1 and MRP, in contrast, have an additional N-terminal transmembrane domain of five transmembrane helices that precedes the ABC core structure seen in the CFTR (Tusnady et al., 2006). Thus, prediction of Hsp90 substrates cannot be made simply based on similarity in sequence or topology. Future structure-function analyses of these closely related proteins may reveal features recognized by the Hsp90 chaperone complex.
Many cochaperone proteins interact with Hsp90 to regulate client protein folding and maturation. For example, Aha-1 is a cochaperone that stimulates the ATPase activity of Hsp90 and has been shown to play a role in CFTR folding. Knocking down Aha-1 by RNA interference rescued the trafficking defect caused by ΔF508 (Wang et al., 2006). Another cochaperone, FK506-binding protein of 38-kDa (FKBP8; also called FKBP38), has been reported as a cochaperone of both CFTR and hERG potassium channels and knockdown of FKBP8 caused a reduction of processing and trafficking for both hERG and CFTR (Wang et al., 2006; Walker et al., 2007). Curiously, we did not observe Aha-1 or FKBP8 associated with wild-type KATP channels in our proteomics experiments, and further studies are needed to determine whether these cochaperones are involved in the folding/processing of wild-type and mutant KATP channels in INS-1 cells.
Role of Hsp90 in Modulating Surface Expression of Misprocessing/Mistrafficking KATP Channel Mutants
Studies of mutations identified in patients with congenital insulin secretion disease have shown that many SUR1 and Kir6.2 mutations affect channel function by preventing normal processing and trafficking of the channel (Ashcroft, 2005; Lin et al., 2006; Yan et al., 2007). In the present study, we have examined five such SUR1 mutations identified in congenital hyperinsulinism. Interestingly, only mutants that are considered to have mild processing/trafficking defects showed improved surface expression upon overexpression of Hsp90. Because Hsp90 is thought to act on substrates at a late stage of folding, it is possible that the A116P and ΔF1388-SUR1 mutations render folding difficulties at an early stage that cannot be overcome by upregulation of Hsp90 function. Examination of more disease mutations that disrupt channel processing and trafficking to different degrees will further test this idea. Given our results, we speculate that the Hsp90 chaperone complex may play a role in modulating disease phenotype by modulating the expression of mutant channels at the cell surface. We have shown previously that many disease mutations disrupt both channel gating and biogenesis/surface expression (Lin et al., 2006; Yan et al., 2007; Pratt et al., 2009). In this case, the expression level of a mutant will modulate the severity of β-cell dysfunction by modulating the total channel functional output (Lin et al., 2006). It is conceivable that changes in Hsp90 expression under different physiological or pathological conditions could affect manifestation of certain disease mutations. This could potentially help explain why disease symptoms can vary among patients carrying the same mutations (Ashcroft, 2005).
Effects of the Hsp90β Inhibitor 17-AGG on KATP Channels
In recent years, Hsp90 inhibitors such as 17-AGG have shown great promise in treating cancer by targeting oncogenic kinases for degradation (Solit and Rosen, 2006). Our study suggests that the use of these drugs may reduce the number of KATP channels to alter glucose-stimulated insulin secretion response in pancreatic β-cells. It is also worth noting that although 17-AGG has little effect on KATP channel gating at concentrations below 100 nM (those used in Figure 3), it does inhibit channel response to the stimulatory effect of Mg-nucleotides when applied to the cytoplasmic face of inside-out patches at concentrations above 200 nM (Pratt and Shyng, unpublished). Because KATP channel stimulation by Mg-nucleotides is mediated by the NBDs of SUR1, 17-AGG may disrupt Mg-nucleotide binding or hydrolysis at NBDs to alter channel gating. Thus, 17-AGG at high concentrations probably targets ATPases other than Hsp90 (Chene, 2002). In this respect, functional data obtained with the use of 17-AGG need to be viewed with biochemical data and interpreted with caution. Recently, it has been reported that 17-AGG partially rescues the processing defect of a disease-causing aquaporin-2 mutation, although the precise mechanism of rescue is unknown (Yang et al., 2009). We, too, tested whether 17-AGG has a salutary effect on the five SUR1 trafficking mutants but saw no effects.
In summary, we have demonstrated in this study that Hsp90 plays a role in the biogenesis of SUR1/Kir6.2 KATP channels, in particular the maturation of the SUR1 subunit. Alterations in Hsp90 expression or function affect the number of KATP channels in the plasma membrane. Up-regulation of Hsp90 alleviates the processing/trafficking defects in some but not all disease-associated SUR1 mutations. Future studies examining the effect of overexpression or knockdown of other chaperones and cochaperones on the biogenesis of wild-type and mutant channel proteins will probably shed additional light on the molecular and structural mechanisms of channel folding and processing. Besides the SUR1/Kir6.2 KATP channel subtype, several other KATP channel subtypes containing the SUR2 isoforms are expressed in cardiac, vascular, and skeletal muscle tissues where they play important roles in linking metabolic changes to muscle activities (Minami et al., 2004). It would be important to determine whether Hsp90 similarly affects biogenesis and expression of these channels by interacting with the SUR2 protein.
ACKNOWLEDGMENTS
The INS-1E cell line clone 832/13 was kindly provided by Dr. Christopher B. Newgard. Rat Kir6.2 was from Dr. Carol A. Vandenberg (University of California, Santa Barbara, CA). Hsp90α and β cDNAs were generously provided by Dr. Solomon Snyder (Johns Hopkins University, Baltimore, MD). This work was supported by National Institutes of Health (NIH) grant DK-57699 (to S.-L. S.), the March of Dimes Research Grant Foundation grant 1-2001-707 (to S.-L. S.), and a Collins Medical Trust Foundation grant (to F.F.Y.). This work was supported by the Proteomics Shared Resource that is generously funded by the Oregon Opportunity, and NIH center grants 5P30CA-069533 and 5P30EY-010572.
Abbreviations used:
- 17-AGG
17-allylamino-17-demethoxygeldanamycin
- CFTR
cystic fibrosis transmembrane conductance regulator
- KATP
ATP-sensitive potassium
- Kir6.2
inwardly rectifying potassium channel 6.2
- KV
voltage-gated potassium channel
- NBD
nucleotide binding domain
- SUR1
sulfonylurea receptor 1
- TMD
transmembrane domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-02-0116) on April 28, 2010.
REFERENCES
- Aguilar-Bryan L., Bryan J. Molecular biology of adenosine triphosphate-sensitive potassium channels. Endocr. Rev. 1999;20:101–135. doi: 10.1210/edrv.20.2.0361. [DOI] [PubMed] [Google Scholar]
- Ashcroft F. M. ATP-sensitive potassium channelopathies: focus on insulin secretion. J. Clin. Invest. 2005;115:2047–2058. doi: 10.1172/JCI25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Cartier E. A., Conti L. R., Vandenberg C. A., Shyng S. L. Defective trafficking and function of KATP channels caused by a sulfonylurea receptor 1 mutation associated with persistent hyperinsulinemic hypoglycemia of infancy. Proc. Natl. Acad. Sci. USA. 2001;98:2882–2887. doi: 10.1073/pnas.051499698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chene P. ATPases as drug targets: learning from their structure. Nat. Rev. 2002;1:665–673. doi: 10.1038/nrd894. [DOI] [PubMed] [Google Scholar]
- Conti L. R., Radeke C. M., Shyng S. L., Vandenberg C. A. Transmembrane topology of the sulfonylurea receptor SUR1. J. Biol. Chem. 2001;276:41270–41278. doi: 10.1074/jbc.M106555200. [DOI] [PubMed] [Google Scholar]
- Ficker E., Dennis A. T., Wang L., Brown A. M. Role of the cytosolic chaperones Hsp70 and Hsp90 in maturation of the cardiac potassium channel HERG. Circulation Res. 2003;92:e87–e100. doi: 10.1161/01.RES.0000079028.31393.15. [DOI] [PubMed] [Google Scholar]
- Flanagan S. E., Clauin S., Bellanne-Chantelot C., de Lonlay P., Harries L. W., Gloyn A. L., Ellard S. Update of mutations in the genes encoding the pancreatic beta-cell K(ATP) channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism. Hum. Mutat. 2009;30:170–180. doi: 10.1002/humu.20838. [DOI] [PubMed] [Google Scholar]
- Higgins C. F. The ABC of channel regulation. Cell. 1995;82:693–696. doi: 10.1016/0092-8674(95)90465-4. [DOI] [PubMed] [Google Scholar]
- Hohmeier H. E., BeltrandelRio H., Clark S. A., Henkel-Rieger R., Normington K., Newgard C. B. Regulation of insulin secretion from novel engineered insulinoma cell lines. Diabetes. 1997;46:968–977. doi: 10.2337/diab.46.6.968. [DOI] [PubMed] [Google Scholar]
- Inagaki N., Gonoi T., Clement J.P.t., Namba N., Inazawa J., Gonzalez G., Aguilar-Bryan L., Seino S., Bryan J. Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- Kamal A., Thao L., Sensintaffar J., Zhang L., Boehm M. F., Fritz L. C., Burrows F. J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Lin C. W., Lin Y. W., Yan F. F., Casey J., Kochhar M., Pratt E. B., Shyng S. L. Kir6.2 mutations associated with neonatal diabetes reduce expression of ATP-sensitive K+ channels: implications in disease mechanism and sulfonylurea therapy. Diabetes. 2006;55:1738–1746. doi: 10.2337/db05-1571. [DOI] [PubMed] [Google Scholar]
- Lin C. W., Yan F., Shimamura S., Barg S., Shyng S. L. Membrane phosphoinositides control insulin secretion through their effects on ATP-sensitive K+ channel activity. Diabetes. 2005;54:2852–2858. doi: 10.2337/diabetes.54.10.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. W., Bushman J. D., Yan F. F., Haidar S., MacMullen C., Ganguly A., Stanley C. A., Shyng S. L. Destabilization of ATP-sensitive potassium channel activity by novel KCNJ11 mutations identified in congenital hyperinsulinism. J. Biol. Chem. 2008;283:9146–9156. doi: 10.1074/jbc.M708798200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo M. A., Jensen T. J., Cui L., Hou Y., Chang X. B., Riordan J. R. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami K., Miki T., Kadowaki T., Seino S. Roles of ATP-sensitive K+ channels as metabolic sensors: studies of Kir6.x null mice. Diabetes. 2004;53(suppl 3):S176–S180. doi: 10.2337/diabetes.53.suppl_3.s176. [DOI] [PubMed] [Google Scholar]
- Nathan D. F., Vos M. H., Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl. Acad. Sci. USA. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neagoe I., Schwappach B. Pas de deux in groups of four–the biogenesis of KATP channels. J. Mol. Cell. Cardiol. 2005;38:887–894. doi: 10.1016/j.yjmcc.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Nichols C. G. KATP channels as molecular sensors of cellular metabolism. Nature. 2006;440:470–476. doi: 10.1038/nature04711. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Prodromou C. Structure and in vivo function of Hsp90. Curr. Opin. Struct. Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- Pearl L. H., Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Powers E. T., Morimoto R. I., Dillin A., Kelly J. W., Balch W. E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Pratt E. B., Yan F. F., Gay J. W., Stanley C. A., Shyng S. L. Sulfonylurea receptor 1 mutations that cause opposite insulin secretion defects with chemical chaperone exposure. J. Biol. Chem. 2009;284:7951–7959. doi: 10.1074/jbc.M807012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe S. M., Miller S., Sorscher E. J. Cystic fibrosis. N. Engl. J. Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- Shyng S. L., Ferrigni T., Shepard J. B., Nestorowicz A., Glaser B., Permutt M. A., Nichols C. G. Functional analyses of novel mutations in the sulfonylurea receptor 1 associated with persistent hyperinsulinemic hypoglycemia of infancy. Diabetes. 1998;47:1145–1151. doi: 10.2337/diabetes.47.7.1145. [DOI] [PubMed] [Google Scholar]
- Solit D. B., Rosen N. Hsp 90, a novel target for cancer therapy. Curr. Top. Med. Chem. 2006;6:1205–1214. doi: 10.2174/156802606777812068. [DOI] [PubMed] [Google Scholar]
- Sreedhar A. S., Kalmar E., Csermely P., Shen Y. F. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562:11–15. doi: 10.1016/s0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- Taschenberger G., Mougey A., Shen S., Lester L. B., LaFranchi S., Shyng S. L. Identification of a familial hyperinsulinism-causing mutation in the sulfonylurea receptor 1 that prevents normal trafficking and function of KATP channels. J. Biol. Chem. 2002;277:17139–17146. doi: 10.1074/jbc.M200363200. [DOI] [PubMed] [Google Scholar]
- Tucker S. J., Gribble F. M., Zhao C., Trapp S., Ashcroft F. M. Truncation of Kir6.2 produces ATP-sensitive K+ channels in the absence of the sulphonylurea receptor. Nature. 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- Tusnady G. E., Sarkadi B., Simon I., Varadi A. Membrane topology of human ABC proteins. FEBS Lett. 2006;580:1017–1022. doi: 10.1016/j.febslet.2005.11.040. [DOI] [PubMed] [Google Scholar]
- Vasiliou V., Vasiliou K., Nebert D. W. Human ATP-binding cassette (ABC) transporter family. Hum. Genomics. 2009;3:281–290. doi: 10.1186/1479-7364-3-3-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker V. E., Atanasiu R., Lam H., Shrier A. Co-chaperone FKBP38 promotes HERG trafficking. J. Biol. Chem. 2007;282:23509–23516. doi: 10.1074/jbc.M701006200. [DOI] [PubMed] [Google Scholar]
- Wang X., et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Welch W. J. Role of quality control pathways in human diseases involving protein misfolding. Semin. Cell Dev. Biol. 2004;15:31–38. doi: 10.1016/j.semcdb.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Yan F., Lin C. W., Weisiger E., Cartier E. A., Taschenberger G., Shyng S. L. Sulfonylureas correct trafficking defects of ATP-sensitive potassium channels caused by mutations in the sulfonylurea receptor. J. Biol. Chem. 2004;279:11096–11105. doi: 10.1074/jbc.M312810200. [DOI] [PubMed] [Google Scholar]
- Yan F. F., Lin C. W., Cartier E. A., Shyng S. L. Role of ubiquitin-proteasome degradation pathway in biogenesis efficiency of {beta}-cell ATP-sensitive potassium channels. Am J. Physiol. Cell Physiol. 2005;289:C1351–C1359. doi: 10.1152/ajpcell.00240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F. F., Lin Y. W., MacMullen C., Ganguly A., Stanley C. A., Shyng S. L. Congenital hyperinsulinism associated ABCC8 mutations that cause defective trafficking of ATP-sensitive K+ channels: identification and rescue. Diabetes. 2007;56:2339–2348. doi: 10.2337/db07-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Zhao D., Verkman A. S. Hsp90 inhibitor partially corrects nephrogenic diabetes insipidus in a conditional knock-in mouse model of aquaporin-2 mutation. FASEB J. 2009;23:503–512. doi: 10.1096/fj.08-118422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. C., Moarefi I., Hartl F. U. Hsp 90, a specialized but essential protein-folding tool. J. Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerangue N., Schwappach B., Jan Y. N., Jan L. Y. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]