The meiosis-specific mug28+ gene of Schizosaccharomyces pombe encodes a putative RNA-binding protein. mug28Δ cells generated spores with low viability, due to the aberrant FSM formation. Meu14-GFP in mug28Δ cells showed that the FSM formed extra membranes with buds. We conclude that Mug28 is essential for the proper maturation of the FSM and the spore wall.
Abstract
The meiosis-specific mug28+ gene of Schizosaccharomyces pombe encodes a putative RNA-binding protein with three RNA recognition motifs (RRMs). Live observations of meiotic cells that express Mug28 tagged with green fluorescent protein (GFP) revealed that Mug28 is localized in the cytoplasm, and accumulates around the nucleus from metaphase I to anaphase II. Disruption of mug28+ generated spores with low viability, due to the aberrant formation of the forespore membrane (FSM). Visualization of the FSM in living cells expressing GFP-tagged Psy1, an FSM protein, indicated that mug28Δ cells harbored abnormal FSMs that contained buds, and had a delayed disappearance of Meu14, a leading edge protein. Electron microscopic observation revealed that FSM formation was abnormal in mug28Δ cells, showing bifurcated spore walls that were thicker than the nonbifurcated spore walls of the wild type. Analysis of Mug28 mutants revealed that RRM3, in particular phenylalanin-466, is of primary importance for the proper localization of Mug28, spore viability, and FSM formation. Together, we conclude that Mug28 is essential for the proper maturation of the FSM and the spore wall.
INTRODUCTION
Meiosis, a specialized cell cycle consisting of one round of DNA synthesis followed by two successive meiotic divisions, is an important process in haploid gamete formation, which is essential for the transmission of genetic information to the next generation in sexually reproducing species. The fission yeast Schizosaccharomyces pombe is a useful model organism for studying the regulatory mechanisms during meiosis, such as recombination, chromosome behavior, and sporulation, because it synchronously enters meiosis and can easily be manipulated by a variety of genetic and cell biological techniques. The cell cycle switch from mitosis to meiosis is accompanied by a remarkable change in gene expression profiles (Mata et al., 2002), and meiosis-specific transcripts are selectively removed by Mmi1 if they are expressed during vegetative growth (Harigaya and Yamamoto, 2007). Meiosis in the fission yeast is initiated when haploid cells of opposite mating types (h+/h−) experience nitrogen starvation and conjugate (Xue-Franzén et al., 2006). After karyogamy (nuclear fusion), premeiotic DNA synthesis occurs. The nucleus then develops a horse-tail shape and moves back and forth about the axis of the cell. This behavior permits homologous pairing and recombination to take place in the cells (Asakawa et al., 2007). After the successive nuclear divisions of meiosis I (MI) and meiosis II, capsular plasma membranes form around each haploid nucleus in the cytoplasm of the mother cell. Then, spore morphogenesis occurs to generate four mature ascospores.
The most important event of sporulation is the assembly of double-layered intracellular membranes, termed forespore membranes (FSMs), which develop into the plasma membrane of spores (Shimoda, 2004). In meiosis II, FSMs are assembled by the fusion of membrane vesicles. After the two sequential meiotic divisions, FSMs expand and encapsulate the haploid nuclei, producing the membrane-surrounded spore precursors. Psy1, a plasma membrane protein that resembles the mammalian target membrane-soluble N-ethylmaleimide-sensitive factor attachment protein receptor protein syntaxin-1A, plays an essential role in FSM formation (Nakamura et al., 2001). Psy1-GFP labels four cup-shaped membranes near the spindle pole body (SPB) at metaphase II, each of which surrounds one of the nuclei during anaphase II. Then, the expanding FSM encloses and completely engulfs each haploid nucleus in a synchronous manner (Nakamura et al., 2008). Leading edge proteins are involved in the initiation, development, and closure of FSMs during the expansion of FSMs at meiosis II. Meu14, one of the meiosis-specific coiled-coil proteins (Ohtaka et al., 2007), localizes at the leading edge of the FSM and regulates spore morphogenesis, in particular the determination of FSM closure during meiosis II (Okuzaki et al., 2003). Meu14-GFP first appears inside the nuclear region at prophase II, after which it accumulates beside the two SPBs at metaphase II, forming two ring-shaped structures that surround the nucleus at early anaphase II. Meu14-GFP disappears at post-anaphase II. The basic process of sporulation is also conserved in the budding yeast Saccharomyces cerevisiae (Neiman, 2005).
Large-scale cDNA sequencing projects in mammals have revealed that eukaryotic cells contain numerous polyadenylated noncoding RNAs (ncRNAs) that are expected to play physiological roles, especially in the regulation of gene expression (Okazaki et al., 2002; Mattick, 2004; Carninci et al., 2005; Tomilin, 2008). These ncRNAs are also abundant in fission yeast, particularly during meiosis (Watanabe et al., 2001; Kakihara et al., 2003). By comparing the genome-wide distribution of peaks for double-strand DNA breaks (DSBs) to that of the prl class of polyadenylated ncRNA (Watanabe et al., 2002), it was found that peaks for DSBs almost colocalize with prl loci, which are most often situated within intergenic regions (Wahls et al., 2008); this suggests that meiotic recombination hotspots of fission yeast are directed to the loci that express polyadenylated ncRNAs. Other ncRNAs may play important roles in the progression of meiosis in fission yeast, but their functions and their putative RNA-binding protein (RBP) association partners remain elusive.
A polyadenylated ncRNA named meiRNA, a cofactor of an RNA-binding protein called Mei2, is expressed only during meiosis and is essential for MI (Watanabe and Yamamoto, 1994; Yamamoto, 1996). Mei2 forms a dot structure in meiotic prophase nuclei, and meiRNA is required for this localization pattern of Mei2 (Yamashita et al., 1998). Spo5, another meiosis-specific RNA-binding protein of S. pombe, also forms a nuclear focus at meiotic prophase I and regulates meiotic progression and homologous recombination (Kasama et al., 2006). Although the Spo5 focus colocalizes with the Mei2 dot in the nucleus at prophase I, Spo5 does not interact with meiRNA. Meu5/Crp79, yet another meiosis-specific RNA-binding protein of S. pombe (Watanabe et al., 2001), also was isolated as a novel mRNA export factor that binds to poly(A)+ RNA and regulates its export from the nucleus to the cytoplasm (Thakurta et al., 2002).
Although many polyadenylated ncRNAs have been isolated from S. pombe, a detailed understanding of their functions and their putative association partners (RBPs) remain elusive, partly due to the lack of information available on meiosis-specific RBPs. Except for Mei2, it remains unknown whether any polyadenylated ncRNA specifically associates and functions with Spo5 and Meu5/Crp79, the meiosis-specific RBPs described above. Indeed, we need a more detailed analysis of other uncharacterized meiosis-specific RBPs of S. pombe. As an initial step to understanding the putative physiological roles of these meiotic RBPs, we searched for meiosis-specific proteins that harbor putative RNA recognition motifs (RRMs). We report here the functional analysis of a novel meiosis-specific gene, mug28+, which encodes Mug28, a protein containing three RRMs. We show that Mug28 is required for spore maturation, particularly for the proper formation of FSMs.
MATERIALS AND METHODS
Yeast Strains, Media, and Molecular Biology
The S. pombe strains and plasmids used in this study are listed in Supplemental Table S1. The media used were yeast extract-peptone-dextrose or yeast extract plus histidine (75 μg/ml) complete media, Edinburgh minimal medium 2 (EMM2) synthetic medium, and malt extract, EMM2-nitrogen (EMM2-N), or SPA sporulation media. The induction of synchronous meiosis was assessed as described previously (Shimada et al., 2002).
Construction of Strains Harboring Integrated mug28+-tagged Genes
To construct the mug28+-green fluorescent protein (GFP) strain, we performed PCR as described previously (Saito et al., 2004) and obtained a DNA fragment carrying the open reading frame and 3′ downstream region of the mug28+ gene. For this purpose, we synthesized the following two oligonucleotides and used them as primers (Supplemental Table S1): mug28-5′Fw-AscI-SalI and mug28–3′Rv-NotI. The PCR product of the 3′ downstream region of the mug28+ gene was inserted into the pRGT41 vector via the XhoI-SacI sites. The PCR product of the mug28+ open reading frame, the 1.8-kb HindIII fragment containing the ura4+ cassette (Grimm et al., 1988), and the pRGT41-mug28+-3′ untranslated region vector were inserted into the pBluescriptII KS(+) vector via the EcoRI-NotI, HindIII, and NotI-SacI sites, respectively. This plasmid construct was then digested with BamHI, and the resulting construct was introduced into the TP4-1D wild-type strain (AS6). The Ura+ transformants were then screened by PCR to identify the mug28+::mug28+-gfp-ura4+ strain.
Fluorescence Microscopy
To detect Mug28-GFP in living meiotic cells, mid-log phase mug28+-GFP cells (AS108) were cultured in EMM2-N to induce meiosis as described previously (Saito et al., 2005). The living cells were then stained with 3.0 μg/ml Hoechst33342 for 5 min and spotted on a coverslip. Fluorescence images of these cells were observed using a fluorescence microscope (BX51; Olympus, Tokyo, Japan) with a Cool SNAP charge-coupled-device camera (Roper Scientific, San Diego, CA). Fluorescence images were acquired using Photoshop 7.0 (Adobe Systems, Mountain View, CA).
For time-lapse observations, YN68, YOD50, SOP091w, AS117, AS160, and AS161 cells, which expressed GFP-Psy1 and Meu14-GFP, were cultured in 10 ml of EMM2 with supplements adenine (75 mg/ml), histidine (75 mg/ml), leucine (250 mg/ml), and uracil (75 mg/ml) until they reached the mid-log phase at 28°C. They were then induced to enter meiosis by incubation in EMM2-N at 28°C. After 10 h of nitrogen starvation, live cells were placed on a glass-bottomed dish coated with 0.2% concanavalin A. Images were acquired with an IX71 fluorescence microscope (Olympus) every 3 min (with a 3-s exposure time), and three optical sections at 200-nm intervals were acquired for each time point. The projected images obtained were analyzed with MetaMorph software (Molecular Devices, Sunnyvale, CA).
RESULTS
Mug28 Is a Meiosis-specific Protein That Harbors RNA Recognition Motifs
To identify a novel meiosis-specific RBP that may associate with the meiosis-specific mRNA-like ncRNAs of S. pombe that we reported previously (Watanabe et al., 2001, 2002), we searched the S. pombe genome database (http://www.genedb.org/genedb/pombe/index.jsp) for uncharacterized genes that both harbor RNA recognition motifs and show enhanced expression during meiosis. We screened 278 genes in the genome database AmiGO (http://www.genedb.org/amigo-cgi/browse.cgi?speciesdb = GeneDB_Spombe) using “RNA binding” as a query word. In addition, we selected genes whose expression is enhanced at least 20-fold in the meiotic phase over that in the G1-arrested phase, based on the cDNA microarray data. This selection yielded the following 10 candidate genes: SPAC1610.02c (64), SPAC1610.03c (=meu5+/crp79+) (293), SPAC343.07 (=mrb2+/mug28+) (110), SPAC4G9.05 (=mpf1+) (93), SPBC28E12.02 (286), SPBC29A10.02 (=mrb1+/spo5+/mug12+) (214), SPBC609.01 (30), SPCC1682.08c (=mcp2+) (20.7), SPCC320.07c (= mde7+) (112), SPCC74.09 (415), with the numbers in parentheses indicating the ratio of the maximum expression to the expression in the G1-arrested phase. We initiated the functional analysis of SPAC343.07 by naming it mrb2+ after meiotic RNA-binding protein 2; however, we hereafter refer to it as mug28+ (Martín-Castellanos et al., 2005), which is its official name. We selected the mug28+ gene for further analysis because it resembled SPAC1610.03c (=meu5+/crp79+), which we previously isolated as one of the meu (meiotic expression upregulated) genes (Watanabe et al., 2001) and later identified as a regulator of RNA transport from the nucleus to the cytoplasm (Thakurta et al., 2002).
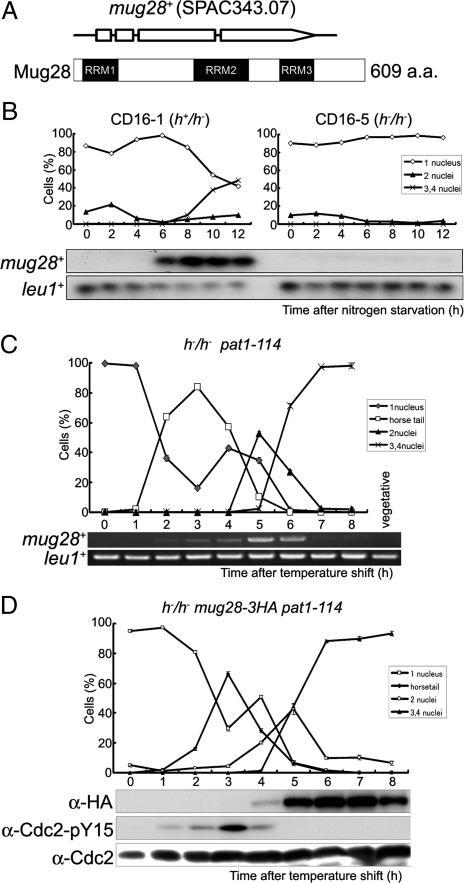
Mug28 (mug28+ protein) consists of 609 amino acids, harbors three RNA recognition motifs, and has no homologous genes in other species (Figure 1A). To examine whether the transcription of mug28+ is meiosis specific, we performed Northern blot and reverse transcribed (RT)-PCR analyses. For Northern blot analysis, we used RNA obtained from CD16-1 (h+/h−) and CD16-5 (h−/h−) cells harvested at various times after the induction of meiosis by nitrogen starvation; CD16-1, but not CD16-5 (negative control), cells enter meiosis. We found that mug28+ transcripts were detected exclusively from 6 to 12 h after meiotic induction (Figure 1B). We next assessed the mRNA level of mug28+ by RT-PCR using RNA obtained from JZ670 (h−/h− pat1-114) cells, which enter meiosis in a highly synchronous manner when shifted to the restrictive temperature. We found that mug28+ transcripts are observed exclusively from 2 to 6 h after meiotic induction, peaking 5 h after the temperature shift (Figure 1C), timing that is consistent with that revealed by Northern blot analysis. These results indicate that the transcription of the mug28+ gene is meiosis specific.
Figure 1.
Mug28 is a meiosis-specific protein with three RRMs. (A) Schematic depictions of the Mug28 protein (609 amino acids) and the mug28+ gene, which consists of four exons (represented by three open boxes and an open arrow). The predicted RRMs were identified by SMART (http://smart.embl-heidelberg.de/) and are indicated by black boxes. (B) Northern blot analysis of mug28+ expression during meiosis. Total RNA was extracted from diploid h+/h− (CD16-1) and h−/h− (CD16-5) cells at the indicated times after the cells were subjected to nitrogen starvation, which induces CD16-1, but not CD16-5, cells to enter meiosis. The cells were collected at 2 h intervals and the total RNAs were blotted and probed with the mug28+ open reading frame (ORF) and leu1+ ORF (loading control). The graphs indicate the meiotic profiles of the cells used for RNA extraction. (C) RT-PCR analysis of mug28+ transcripts during meiosis. The h−/h− pat1-114 strain (JZ670) was induced to enter meiosis synchronously, and the total RNA was harvested, used for RT-PCR, and the PCR products were resolved by 1% agarose gel electrophoresis. (D) Meiotic expression of Mug28-3HA. The h−/h− pat1-114 mug28+-3HA strain (AS122) was induced to enter meiosis synchronously by a temperature shift and the cells were collected at 1 h intervals for protein extraction, blotting, and probing with the anti-HA antibody. Phosphorylation of Cdc2-Y15 was also analyzed using an anti-Cdc2-Y15 antibody to help identify the meiotic stage at each time point. The Cdc2 levels were also examined, as a loading control. At each time point in B–D, the frequency of cells with one, two, three or four nuclei was determined by counting at least 200 Hoechst33342-stained cells under a microscope (top).
To accurately examine the expression of Mug28 during meiosis, we constructed a mug28+-3HA strain, which expresses Mug28 tagged with three copies of the HA epitope at its C-terminal end. To attain synchronous meiosis, we used the pat1-114 temperature-sensitive strain. We first replaced the mug28+ gene of the pat1-114 strain with the mug28+-3HA fusion gene. Then, h−/h− pat1-114 mug28+-3HA diploid cells were induced to enter synchronized meiosis by temperature shift, and their lysates were subjected to Western blot analysis using the anti-hemagglutinin (HA) antibody. This Western blot analysis detected the Mug28-3HA protein of the expected size, which was expressed only during meiosis, 4–8 h after the temperature shift, and peaking at 6–7 h (Figure 1D). As a control to monitor the timing of meiosis, we tested for tyrosine 15 phosphorylation of Cdc2, which appears around the premeiotic S phase.
Mug28 Localizes to the Cytoplasm during Meiosis I and to the FSM during Meiosis II
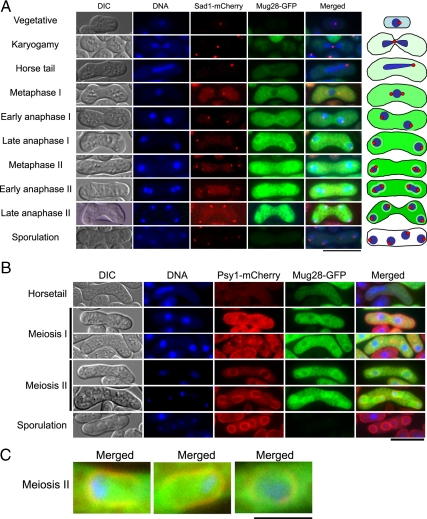
To examine the subcellular localization of the Mug28 protein during meiosis, we prepared an S. pombe (h90) strain expressing both Mug28-GFP and Sad1-mCherry, a SPB marker. We induced it to enter zygotic meiosis, and observed the GFP and mCherry signals in living cells, without fixation, and determined the timing of meiotic progression with a fluorescence microscope by tracking the nucleus with Hoechst33342 staining, and the SPB by monitoring the mCherry signal. As shown in Figure 2A, no Mug28-GFP signal was detected when cells were in the vegetative growth phase and undergoing karyogamy. In contrast, after meiotic progression, Mug28-GFP faintly appeared in the cytoplasm at the horsetail phase. At metaphase I, Mug28-GFP signal was strongly detected uniformly in the cytoplasm but not in the nucleus. Then, the signal gradually accumulated around the nucleus until the cell reached anaphase I. Thereafter, Mug28-GFP signals became apparent not only around the nucleus but also in the nucleus. After late anaphase II and during sporulation, the Mug28-GFP signal disappeared. This subcellular localization of Mug28-GFP corresponds with the results of the time course Western blot analysis (Figure 1D), showing that expression of Mug28-3HA declines after peaking 6–7 h after the meiotic induction.
Figure 2.
Subcellular localization of Mug28-GFP in living S. pombe cells. (A) Fluorescence signals from Mug28-GFP and Sad1-mCherry were observed at multiple stages of meiosis after meiotic induction by nitrogen starvation in AS127 (h90 mug28+-GFP sad1+-mCherry) cells. Differential interference contrast (DIC) images are shown in the leftmost panels. Fluorescence microscopy images showing DNA (blue), Sad1-mCherry (red), Mug28-GFP (green), and the corresponding merged images are shown. The schematically depicted images are shown in the rightmost panel. Bar, 10 μm. (B) Fluorescence signals from Mug28-GFP and Psy1-mCherry were observed at multiple stages of meiosis after meiotic induction by nitrogen starvation in AS181 (h90 mug28+-GFP psy1+-mCherry) cells. DIC images are shown in the leftmost panels. Fluorescence microscopy images showing DNA (blue), Psy1-mCherry (red), Mug28-GFP (green), and the corresponding merged images are shown. The schematically depicted images are shown in the rightmost panel. Bar, 10 μm. (C) Enlarged images of Mug28-GFP colocalized with Psy1-mCherry during meiosis II. Bar, 2.5 μm.
We also examined the detailed subcellular localization of Mug28 using an S. pombe (h90) strain that expresses both Mug28-GFP and Psy1-mCheery, an FSM marker. As shown in Figure 2, B and C, Mug28-GFP was faintly visible in the cytoplasm at the horsetail phase as described above. At metaphase I, Mug28-GFP signal was uniformly detected in the cytoplasm, but not in the nucleus. Then, the signal gradually accumulated around the nucleus until the cell reached anaphase I. After that, the Mug28-GFP signal became visible not only in the area around the nucleus, which displayed colocalization with Psy1-mCherry, but also in the nucleus at anaphase II. After late anaphase II and during sporulation, the Mug28-GFP signal disappeared. This result suggests that Mug28 protein plays a role in formation of the FSM during meiosis II.
mug28Δ Cells Generate Abnormal Spores with a Snowman-like Morphology
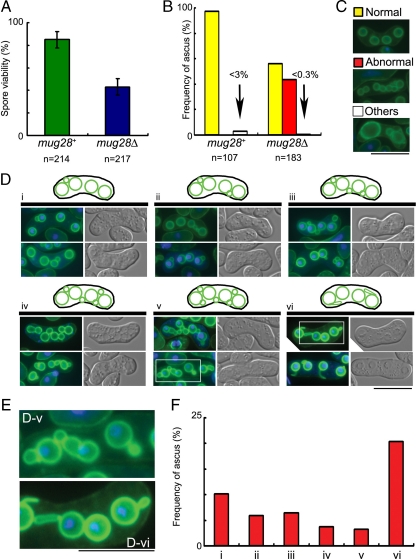
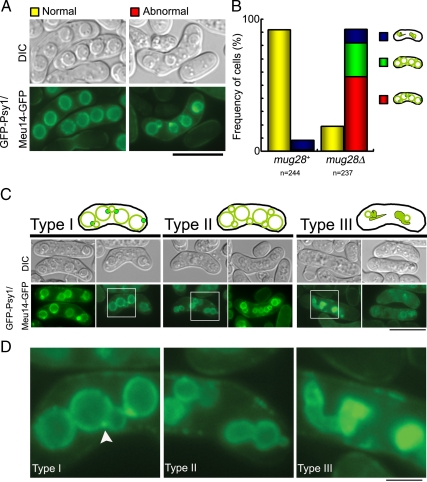
To assess the physiological role of mug28+, a mug28+ disrupted strain (mug28Δ) was constructed by one-step gene replacement. Diploid cells in which one of the mug28+ genes had been replaced by ura4+ were sporulated and germinated. The generated spores were viable, indicating that the mug28+ gene is not essential for vegetative growth. However, many of the diploid h90 mug28Δ cells failed to form proper spores 18 h after meiotic induction, and the spore viability of the mug28Δ mutant was only half that of the mug28+ cells (Figure 3A). We confirmed that the ectopic expression of Mug28-GFP suppressed this abnormal sporulation phenotype of mug28Δ cells (Supplemental Figure S1). To investigate the cause of abnormal spore formation in mug28Δ cells, the morphologies of FSMs in these mutant cells were analyzed by fluorescence microscopy using an FSM marker, GFP-Psy1 (Nakamura et al., 2001). We found that about half of the mug28Δ spores showed the abnormal snowman-like morphology, whereas most of the mug28+ spores were normal (Figure 3B). After incubation on sporulation plates for 18 h, many of the wild-type cells displayed GFP-Psy1 fluorescence as circles of uniform brightness, representing FSMs with a haploid nucleus already enclosed. In contrast, many mug28Δ cells displayed buds of various sizes, or spores with large buds forming a shape resembling a snowman (Figure 3C). When we classified these abnormal morphologies into six types (i∼vi), we noticed that many mug28Δ cells displayed more than five GFP-Psy1 circles, which is due to the budded shape of the FSMs. Interestingly, some spores had FSMs extending from the FSM-encapsulated nucleus (Figure 3, D-v and vi). Enlarged images of such spores are surrounded by squares and presented in Figure 3D-v (bottom row) and Figure 3D-vi (top row) (Figure 3E). Frequencies of each abnormal spore type are presented as bar graphs in Figure 3F. These results indicate that Mug28 is required for normal spore formation.
Figure 3.
The mug28Δ cells display reduced spore viability and abnormal morphology of FSM formation. (A) Spore viability of mug28+ (AS6) and mug28Δ (AS99) strains. Spore viability was measured by random spore analysis. Bar graphs show the averages of three independent experiments with SEs (error bars). At least 200 spores (n = 214 and 217 for mug28+ and mug28Δ, respectively) were measured. (B) Quantification of the classified spores in the ascus of the mug28+ or mug28Δ strain, summarized as a graph. Yellow, red, and white bars represent the frequency of asci with normal spores carrying four FSMs (normal), more than four FSMs (abnormal), and less than four FSMs (others), respectively, as visualized by GFP-Psy1. The red bar represents the frequency of asci containing FSMs with an abnormal morphology. After the YN68 (GFP-psy1+) and AS117 (GFP-psy1+ mug28Δ) cells were induced to enter meiosis by nitrogen starvation, they were stained with Hoechst33342 (blue) and GFP-Psy1 (green) and were observed by fluorescence microscopy. (C) Typical images of the FSMs in the ascus, representing the yellow, red, and white bars in B, as visualized by GFP-Psy1. When GFP-Psy1 localized to four circular FSMs, the FSMs were classified as normal (yellow bar). When GFP-Psy1 localized to more than four circular FSMs, the FSMs were classified as abnormal (red bar). When GFP-Psy1 localized to fewer than four circular FSMs, the FSMs were classified as others (white bar). Bar, 10 μm. (D) Typical images (bottom) and illustrations (top) of the FSMs of mug28Δ cells that were classified as abnormal in C are further categorized into six groups (i–vi). Left, merged images of GFP-Psy1 (green) and DNA (blue). Right, DIC images. Bar, 10 μm. (E) Enlarged images of the regions indicated by white squares in D-v and D-vi. Bar, 10 μm. (F) The histogram shows the percentage of each abnormal type of FSMs, as categorized in D.
Meu10-GFP Localizes to the Snowman-like FSM Structure
Because the snowman-like phenotype of mug28Δ cells, shown in Figure 3, is strikingly similar to that of meu10Δ cells (Tougan et al., 2002), we investigated whether Mug 28 regulates the protein expression or subcellular distribution of Meu10, which functions in normal spore wall assembly as a glycosylphosphatidylinositol (GPI)-anchored cell surface protein. We first examined meu10+ mRNA expression in mug28Δ cells during meiosis by microarray analysis and found no apparent difference from wild-type cells (Supplemental Figure S2iv). We next determined the subcellular localization of Meu10 by constructing a Meu10-GFP-expressing strain in the h90 mug28Δ genetic background and inducing it to undergo meiosis by nitrogen starvation. Fluorescence microscopy during meiosis II showed that Meu10-GFP signals colocalized to the snowman-like FSM visualized by mCherry-tagged Psy1 (arrowheads in Supplemental Figure S3A). However, Meu10-GFP appeared in the spore cytoplasm and near the spore wall when wild-type or mug28Δ cells entered meiosis II (right panels in Supplemental Figure S3A, i and ii). Furthermore, Meu10-GFP showed similar signal intensity in wild-type and mug28Δ cells. Thus, although the abnormal spore wall of mug28Δ cells is similar to that of Meu10 mutants (Tougan et al., 2002), our results indicate that Meu10 is not a regulatory target of Mug28.
Abnormal Behavior of Meu14-GFP in mug28Δ Cells during Sporulation
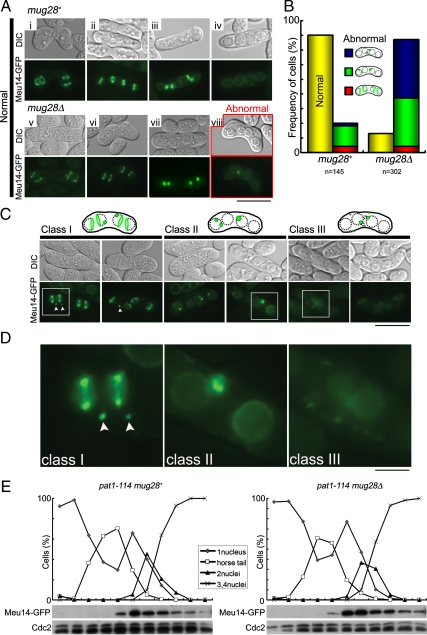
Meu14 is known to regulate the proper formation of FSMs by localizing at the leading edge of the FSM during meiosis II in fission yeast; the Meu14 protein then disappears as FSMs enclose the nucleus (Okuzaki et al., 2003). To explore the cause of abnormal FSM formation in mug28Δ cells, we monitored the subcellular localization of Meu14-GFP in mug28Δ and mug28+ cells by fluorescence microscopy (Figure 4A). We found that the disappearance of Meu14-GFP was incomplete in most (85.8% in Figure 4B) of the mug28Δ cells (panels surrounded by red squares in Figure 4A) at a time when most (80.0% in Figure 4B) of the mug28+ cells no longer carried Meu14-GFP signals (Figure 4Aiv). This also suggests that the sporulation process is delayed in mug28Δ cells, because more than five times as many mug28Δ cells as mug28+ cells (20.0% in Figure 4B) had a residual Meu14-GFP signal at the closing aperture of the FSM. The residual Meu14-GFP signal was observed even after completion of FSM formation at the end of sporulation in mug28Δ cells (Figure 4Aviii).
Figure 4.
Abnormal behavior of Meu14-GFP in mug28Δ cells. (A) Subcellular localization of Meu14-GFP as visualized by fluorescent microscopy from meiosis II to sporulation in mug28+ and mug28Δ cells. YOD50 (meu14+-GFP) and AS160 (meu14+-GFP mug28Δ) cells were induced to enter meiosis by nitrogen starvation, and then Meu14-GFP (green) and DIC images were acquired by fluorescence microscopy, until sporulation occurred. Images in i–iv and v–viii represent the subcellular localization of Meu14 from meiosis II to sporulation in mug28+ and mug28Δ cells, respectively. Except for image viii, which was acquired at sporulation, all other Meu14-GFP images are similar between mug28Δ and mug28+ cells. Bar, 10 μm. (B) The histogram shows the percentage of normal and abnormal cells expressing Meu14-GFP in mug28+ and mug28Δ strains. (C) Typical images of abnormal mug28Δ cells expressing Meu14-GFP. Images where Meu14-GFP localized to satellite dots in the vicinity of the Meu14-GFP rings were classified as class I. White arrowheads indicate the satellite dots near the Meu14-GFP rings. Images where the Meu14-GFP signal accumulated at the leading edge of FSM even after the completion of FSM formation were classified as class II. Images where Meu14-GFP localized abnormally to uncharacterized dots in the vicinity of FSM in the cytoplasm were classified as class III. Bar, 10 μm. (D) Enlarged views of the regions indicated by white squares in C. Bar, 10 μm. (E) Meiotic expression of Meu14-GFP in pat1-114 mug28+ and pat1-114 mug28Δ cells. These cells were induced to enter meiosis synchronously by a temperature shift and were collected at 1-h intervals for protein extraction, blotting, and probing with the anti-GFP and anti-Cdc2 (loading control) antibodies. At each time point, the frequency of cells with one, two, three, or four nuclei was determined by counting at least 200 Hoechst33342-stained cells under a microscope (top).
To analyze this abnormality in more detail, we categorized the cells into three classes (Figure 4C). Class I cells contain satellite dots (white arrowheads) in the vicinity of the major Meu14-GFP rings. In class II cells, Meu14-GFP signals are detected at the leading edge of the FSM, even after the completion of FSM formation. These signals usually disappeared at the end of anaphase II in mug28+ cells. Class III cells show abnormally localized dots at uncharacterized locations in the cytoplasm, adjacent to the FSM. The phenotypes marked with squares in Figure 4C are enlarged in Figure 4D. The fact that class II and class III cells are frequently produced from mug28Δ cells suggests that Mug28 is required for the degradation of Meu14 and/or the evacuation of Meu14 from the FSM at the last phase of the sporulation process. These results suggest that Meu14 aberrantly remaining at the leading edge of FSM induced the formation of bud-like extra FSM in mug28Δ cells.
Western blot analysis indicated that an ineligible amount of Meu14 remained even at the end of sporulation in mug28Δ cells (Figure 4E). This was not due to remaining meu14+ transcription, because microarray analysis showed no apparent change in the meu14+ mRNA level in mug28Δ cells (Supplemental Figure S2). These data indicate that Mug28 regulates the stability of Meu14 protein but is not involved in transcriptional regulation of the meu14+ gene.
Behavior of Both Meu14 and Psy1 Are Abnormal in mug28Δ Cells
We next examined the behavior of FSM (depicted by GFP-Psy1) and Meu14-GFP simultaneously in mug28+ and mug28Δ cells. Because the mCherry signal, which is weak and short lived, is not suitable for monitoring the behavior of Psy1 and Meu14 over a long period, such as sporulation, we observed the GFP signal alone using a mug28Δ strain (AS161) that expresses both GFP-Psy1 and Meu14-GFP. This is by taking advantage of the fact that the subcellular localizations of GFP-Psy1 and Meu14-GFP are distinct, and we compared the behavior of Psy1 and Meu14 simultaneously using the same fluorescent tag. We first confirmed that the frequency of abnormal cells (83.1%) was similar to that presented in Figure 4B (85.8%), in which the disappearance of the Meu14-GFP signal at the end of the FSM formation process was incomplete (Figure 5, A and B).
Figure 5.
Simultaneous observation of Meu14-GFP and GFP-Psy1 signals in mug28+ and mug28Δ cells at sporulation. (A) Typical images of normal and abnormal mug28+ and mug28Δ cells expressing Meu14-GFP/GFP-Psy1 at sporulation. SOP091w (meu14+-GFP GFP-psy1+) and AS161 (meu14+-GFP GFP-psy1+ mug28Δ) cells were induced to enter meiosis by nitrogen starvation, and GFP signals (green) and differential interference contrast (DIC) images were acquired. Bar, 10 μm. (B) Histogram of the normal and abnormal Meu14-GFP/GFP-Psy1 expressing cells in mug28+ and mug28Δ strains. Red, green, and blue areas indicate type I, II, and III categories of abnormal Psy1/Meu14 localization, respectively. (C) Typical fluorescence images of abnormal Meu14-GFP/GFP-Psy1 expressing cells. (type I) mug28Δ cells that display a strong signal of residual Meu14-GFP at the junction between mother and daughter FSMs are classified as type I. (type II) mug28Δ cells that display more than four snowman-like FSMs without Meu14-GFP dots are classified as type II. (type III) mug28Δ cells that display extraordinarily deformed Meu14-GFP signals are classified as type III. Bar, 10 μm. (D) Enlarged views of the regions enclosed by white squares in C. White arrowheads indicate the abnormal behavior of Meu14-GFP at the bud-neck between mother and daughter FSMs. Bar, 10 μm.
To further analyze this abnormality, we classified the abnormal cells into three types, and representative images of each type are shown in Figure 5C. Enlarged images show that type I mug28Δ cells emit a strong signal of residual Meu14-GFP at the junction between mother and daughter FSMs, which strongly suggests that budding of extra FSM emerges from the closing FSM aperture due to residual Meu14. Type II mug28Δ cells harbor FSM (or GFP-Psy1) with four or more snowman-like abnormal spores structures, and without Meu14-GFP dots, which suggests that Meu14 is no longer required for the maintenance of the budded spore morphology. Type III mug28Δ cells harbor an extraordinarily abnormal spore morphology, which is probably not due to Mug28 abolishment, because even a small percentage of mug28+ spores exhibit this abnormality (Figure 5C). Enlarged views of the squared portion of Figure 5C are presented in Figure 5D. These results suggest that aberrantly retained Meu14 at the leading edge of FSM induced the formation of additional FSM in mug28Δ cells.
We also examined the effects of overexpression of Meu14 protein, which was directed from the nmt promoter during meiosis. However, we found none of the abnormal phenotypes observed in mug28Δ cells (Supplemental Figure S3B). Thus, although the aberrant amount of remaining Meu14 is an indication of abnormal spores of mug28Δ cells, it alone does not explain its abnormality (see Discussion). We also examined whether the meu14 mutation was genetically epistatic to the mug28 mutation by constructing a mug28Δ meu14Δ double mutant strain. However, we found no genetic interactions between these mutations either with respect to the abnormal spores of mug28Δ cells or the abnormal nuclear fragmentation of meu14Δ cells (Supplemental Figure S4).
Time-Lapse Analysis of Dynamic FSM Formation
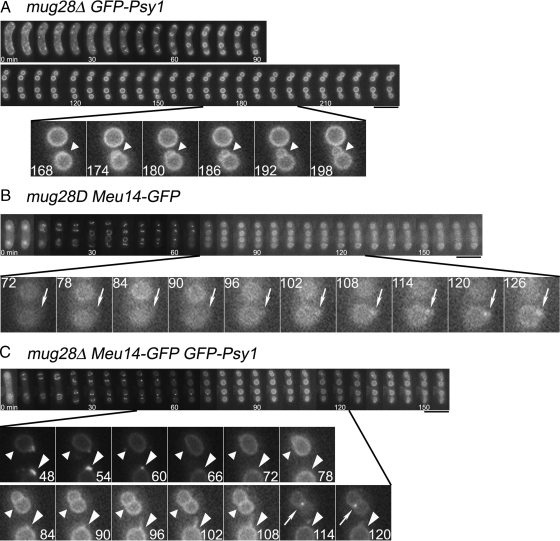
To investigate the live movement of FSM formation and the movement of leading edge proteins of mug28Δ cells in detail, we performed time-lapse observations of GFP-Psy1 and/or Meu14-GFP signals, either separately or simultaneously, in both mug28+ and mug28Δ cells. When we performed a time-lapse analysis of GFP-Psy1, we found that new FSM initiated budding from already enclosed FSM only in mug28Δ cells (Figure 6A and Supplemental Movie 1) but not in mug28+ cells, which resulted in the formation of a snowman-like morphology of the spore after the FSM was completely enclosed (Supplemental Figure S5A and Supplemental Movie 2). When we similarly observed Meu14-GFP, we continually detected the residual Meu14-GFP signal at the leading edge of the FSM only in mug28Δ cells (Figure 6B and Supplemental Movie 3), even when all Meu14-GFP signals disappeared in mug28+ cells (Supplemental Figure S5B and Supplemental Movie 4). Simultaneous time-lapse analysis of both GFP-Psy1 and Meu14-GFP signals confirmed the above-mentioned observations, which are tightly related to the residual Meu14-GFP signals at the leading edge of the FSM in mug28Δ cells (Figure 6C and Supplemental Movie 5), but not in mug28+ cells (Supplemental Figure S5C and Supplemental Movie 6). Together, these results indicate that Meu14 is not properly degraded in mug28Δ cells.
Figure 6.
Time-lapse images of FSM formation and/or leading edge protein in mug28Δ cells at 28°C. These images show a subset of GFP images that was captured every 6 min. Numbers indicated at the bottom of each photograph denote the time, in minutes, from the onset of FSM formation. (A) GFP-Psy1 signals in mug28Δ (AS117) cells revealed the process of budding (denoted by arrowheads), which resulted in the formation of a snowman-like FSM. (B) Meu14-GFP signals in mug28Δ (AS160) cells (denoted by arrows) were unusually retained at the enclosed leading edge of the FSM. (C) Simultaneous observation of Meu14-GFP (arrows) and GFP-Psy1 (arrowheads) in mug28Δ (AS161) cells unveiled the process of new FSM formation (budding) that initiated from the already enclosed mother FSM, generating the snowman-like morphology of FSM. Meu14-GFP signals (arrows) were detected at the junction of mother and daughter FSMs. The GFP-Psy1 signal in the bottom spore (large arrowheads) indicates the fiberlike elongation of the FSM from the already enclosed FSM. Bars, 5 μm.
Ectopic expression of Meu14DD-GFP (Meu14DD-R148AL151A or -R161AL164A), in which Arg and Leu in the two putative destruction boxes (RxxL) are replaced by Ala, caused no apparent abnormalities in FSM formation as visualized by Psy1-GFP (Supplemental Figure S6). Thus, the ubiquitin-proteasome pathway is apparently not involved in the dysregulation of timely degradation of Meu14 at the end of meiosis.
Observation of Aberrant Spore Morphology by Electron Microscopy
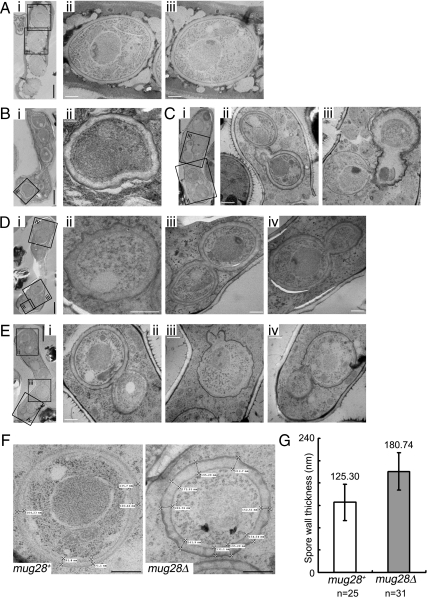
To understand the detailed spore structures of mug28Δ cells, we examined their morphologies by thin-section electron microscopy (TEM). We treated sporulating mug28+ and mug28Δ cells with rapid freezing to minimize the artifacts that can arise during the prefixation process (Kasama et al., 2006). The substituted cells were then fixed and observed by electron microscopy. As shown in Figure 7A, TEM observation in mug28+ cells revealed four normal spores that possess nuclei, mitochondria, rough endoplasmic reticulum, and Golgi apparatuses. In contrast, many of the single mug28Δ spores possess a spore wall that is thicker than that of mug28+ spores (Figure 7, Bii and Dii). We also found that some of the budded mug28Δ spores retained a canal-like passage of cytoplasm between the mother and daughter spores (Figure 7, Cii, Div, and Eiv), whereas in snowman-like spores, which may represent mature spores, the cytoplasm of the mother and daughter spores was separated by a common spore wall (Figure 7, Diii and Eii). Notably, many of the budded mug28Δ spores displayed ruffled and seemingly fragile spore walls (Figure 7, Ciii, Dii, and Eiii and iv). In contrast, the snowman-like spores typically seemed to have a solid spore wall (Figure 7, Bii, Diii, and Eii). For quantitative analysis, we selected four typical TEM images of both mug28+ and mug28Δ spores and measured the thickness of their spore walls (Figure 7F and Supplemental Figure S7). Indeed, the spore walls of the mug28Δ strain were ∼1.5 times thicker than those of the mug28+ strain (Figure 7G). These results indicate that Mug 28 is required for proper spore wall formation.
Figure 7.
Thin-section EM of mug28+ (AS6) and mug28Δ (AS99) cells at sporulation. (A) i, typical EM image of the mug28+ strain. ii and iii, enlarged views of the indicated regions, showing normal spores. (B) i, typical electron microscopy image of mug28Δ cell. ii, enlarged view of the indicated region shows an abnormal spore with a thick spore wall. (C) i, typical EM image of a mug28Δ cell. ii and iii, enlarged views of the indicated regions showing abnormally shaped snowman-like spores. (D) i, typical EM image of a mug28Δ cell carrying six spores. ii–iv, enlarged views of the indicated regions showing abnormally shaped snowman-like spores. (E) i, typical EM image of a mug28Δ cell. ii–iv, enlarged views of the indicated regions show abnormally shaped snowman-like spores. White bar, 0.5 μm. Black bar, 2 μm. (F) Wall thickness of mug28+ and mug28Δ spores. Bar, 0.5 μm. (G) Histogram of the average spore wall thickness of mug28+ and mug28Δ cells. Spore wall thickness was measured at 25 and 31 positions for mug28+ and mug28Δ spores, respectively. The errors bars show the SD.
RRM3 Is Predominantly Required for Full Function of Mug28
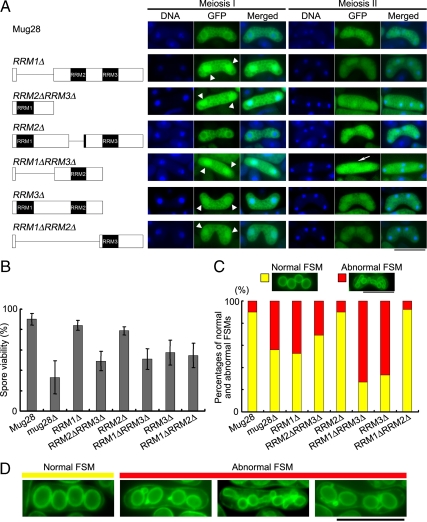
To analyze the role of each RRM domain in Mug28 function, we prepared Mug28 deletion mutants that express GFP fused to truncated Mug28 proteins (Figure 8A, left). We induced these mutant cells to enter meiosis by nitrogen starvation and observed the GFP signals of living cells without fixation, tracking the nucleus by staining with Hoechst33342. Apart from the Mug28-RRM2Δ-GFP mutant, the proper perinuclear localization of Mug28-GFP was disturbed at meiosis I in all mutants, and the Mug28-GFP signal was also observed in the nucleus (arrowheads in middle panels of Figure 8A). This result indicates that RRM2 does not contribute to the proper subcellular localization of Mug28. In contrast, at meiosis II, Mug28-GFP localization was almost normal in all mutants except for the Mug28-RRM1ΔRRM3Δ mutant, in which the nuclear accumulation of GFP signal was abolished (rightmost panel of Figure 8A). This result suggests that the RRM1 and RRM3 domains are required for the proper subcellular localization of Mug28.
Figure 8.
Functional analysis of the RRM deletion mutants of Mug28. (A) Schematic representations of the truncated forms of Mug28 that are expressed as C-terminal GFP fusion proteins are shown in the left panels. Fluorescence images of the Mug28-GFP deletion mutants (green) and of DNA (blue), and merged images, observed at meiosis I and meiosis II, are shown in the middle and right panels, respectively. Arrowheads in the middle panel indicate the abnormal localization of Mug28-GFP in the nucleus during meiosis I. An arrow in the right panel indicates the aberrant uniform localization of Mug28-RRM1ΔRRM3Δ-GFP in the cytoplasm during meiosis II. Bar, 10 μm. (B) Bar graphs of the spore viability of the deletion mutants, as measured by random spore analysis. Bar graphs show the averages of three independent experiments with SEs. At least 200 spores were measured for each strain. (C) Histogram of normal and abnormal FSM formation in the indicated deletion mutants (bottom) and their typical corresponding phenotypes (top). Bar, 10 μm. (D) Typical images of normal and abnormal FSM formation in the deletion mutants of Mug28, in which the YFP-Psy1 signals (green) were observed by fluorescence microscopy. Bar, 10 μm.
The spore viabilities of the Mug28-RRM1Δ and Mug28-RRM2Δ mutants were almost normal, whereas the Mug28-RRM3Δ mutant displayed a spore viability that was reduced to the level found in the Mug28-RRM1ΔRRM2Δ double mutant (Figure 8B). Mug28-RRM1ΔRRM3Δ and Mug28-RRM2ΔRRM3Δ double mutants exhibited a further reduction in spore viability, whereas Mug28Δ, which corresponds to the triple mutant (RRM1ΔRRM2ΔRRM3Δ), showed the most remarkable reduction of spore viability. These results indicate that the RRM3 domain is most important for viable spore formation, and that the RRM1 and RRM2 domains play subsidiary roles in spore viability.
To further assess the requirements of each RRM domain for proper FSM formation, we prepared mutants expressing YFP-Psy1 and the truncated form of Mug28-3HA, and we observed the YFP signals in living cells after meiotic induction. We used YFP-Psy1 instead of GFP-Psy1 here to exclude the possibility that the above-mentioned observations were due to artifacts linked to the fluorescent signals of GFP. Indeed, we could confirm that YFP-Psy1 behaved similarly to GFP-Psy1 during fluorescence microscopy (our unpublished data). To visualize the YFP-Psy1 signals alone and to examine the influence of the GFP tag on the phenotype, we replaced the GFP tag fused to the deletion mutants with the 3HA tag. We first confirmed that Mug28-RRM3Δ-3HA cells had a reduced spore viability at a similar level of the Mug28-RRM3Δ-GFP mutant (Supplemental Figure S8). This indicates that GFP-tagged and HA-tagged deletion mutants show similar results. We also confirmed that abnormal YFP-Psy1 localization occurred at a similar frequency (bar graph of mug28Δ in Figure 8C) as abnormal GFP-Psy1 localization and that YFP-Psy1 and GFP-Psy1 labeled similar abnormal morphologies (panels indicated as abnormal FSM of mug28Δ cells in Figure 8D) (Figure 3, B and D). As for the other deletion mutants, nearly 90% of Mug28-RRM2Δ (AS199) cells displayed normal YFP-Psy1 localization, whereas only 33% of Mug28-RRM3Δ (AS200) and 27% of Mug28-RRM1ΔRRM3Δ (AS202) cells exhibited normal YFP-Psy1 localization. Percentages of normal cell populations in Mug28-RRM1Δ (AS198), Mug28-RRM2ΔRRM3Δ (AS201), and Mug28-RRM1ΔRRM2Δ (AS203) cells ranged between 53 and 92%. Notably, these percentages are similar to those of spore viability of these mutants. Together, we conclude that the RRM3 domain is pivotal for the full activity of Mug28, whereas the RRM1 and RRM2 domains have supplementary roles.
The F466A Mutation in the RRM3 Domain Causes Abnormal Phenotypes
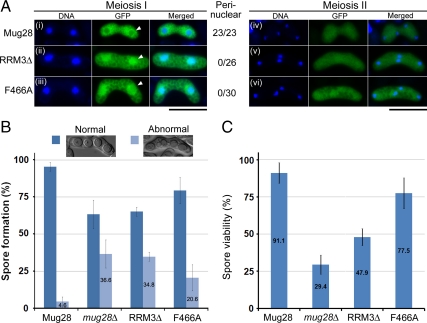
To examine whether the abnormal phenotype of Mug28-RRM3Δ cells was due to defective Mug28 RNA binding ability rather than due to conformational changes, we replaced Phe-466 (F466) with alanine in the conserved RNA-binding motif RRM3 to generate mutant cells that expressed GFP-Mug28F466A (Figure 9A). Then, we compared the GFP localization of Mug28F466A-GFP (NP252) with that of Mug28-GFP (AS164), Mug28RRM3Δ-GFP (AS176), and in mug28Δ (AS99) cells after meiotic induction by nitrogen starvation. We found that the perinuclear localization of Mug28F466A-GFP and Mug28RRM3Δ-GFP was disrupted to a similar degree during meiosis I (arrowheads in the middle panels of Figure 9A, ii and iii). In contrast, all Mug28-GFP signals were observed in the peri-nucleus (arrowhead in middle panels of Figure 9Ai). This result indicates that F466, a putative RNA binding site, is required for proper subcellular localization of Mug28-GFP. In contrast, Mug28F466A-GFP localized normally during meiosis II, as did Mug28-GFP and Mug28RRM3Δ-GFP (rightmost panel of Figure 9A).
Figure 9.
Functional analysis of the Mug28F466A mutant. (A) Subcellular localization of GFP-tagged Mug28 protein (green) during meiosis I and meiosis II in mug28Δ (AS99) cells: Mug28-GFP (AS164; i and iv), Mug28RRM3Δ-GFP (AS176; ii and v), and Mug28F466A-GFP (NP252; iii and vi). Merged images with DNA (blue) are shown in the rightmost panels. Arrowheads indicate normal (i) or abnormal localization of Mug28RRM3Δ-GFP (ii) and Mug28F466A-GFP (iii) to the perinucleus during meiosis I. Numbers represent the proportion of cells displaying perinuclear GFP signals per examined cell. Bar, 10 μm. (B) Frequency of normal and abnormal spore formation in WT (Mug28) and mutants (mug28Δ, RRM3Δ, and F466A). Bar graphs show the averages of three independent experiments with SEs. At least 200 spores were measured for each strain. (C) Spore viability of WT (Mug28) and mutants (mug28Δ, RRM3Δ, and F466A), measured by random spore analysis. Bar graphs show the averages of three independent experiments with SEs. At least 200 spores were measured for each strain.
We also examined spore formation and found that the frequency of abnormal spores from F466A mutant cells (20.6%) was greater than that from WT cells (4.6%) but slightly less than that from mug28Δ (36.6%) and RRM3Δ (34.8%) cells (Figure 9B). Spore viability of F466A mutant cells (77.5%) was also intermediate between WT cells (91.1%) and mug28Δ (29.4%)/RRM3Δ (47.9%) cells (Figure 9C). These results indicate that the F466 residue in RRM3 of Mug28 F466 is required for full function of Mug28.
DISCUSSION
Mug28 Is Required for the Proper Formation of FSM in Sporulation
In meiosis, the spatiotemporal coordination of meiotic nuclear divisions and FSM formation is essential for accurate distribution of the genome into four haploid spores. Here, we showed that expression of meiosis-specific Mug28, which has three RRM domains, is detected in the cytoplasm from metaphase I to anaphase II, and accumulates particularly at the periphery of the nucleus during meiosis I (Figures 1 and 2). Disruption of the mug28+ gene caused abnormal FSM formation, which may be one of the causes of the reduced spore viability (Figure 3A) and the formation of abnormal spores with the snowman-like morphology, as revealed by an FSM marker, GFP-Psy1 (Figure 3, B–F). Visualization of the FSM in living cells by GFP-Psy1 indicated that mug28Δ cells harbored abnormal FSMs with buds, and displayed the retarded disappearance of Meu14 (Figures 4–6).
The abnormal FSM phenotype of mug28Δ cells, displaying the snowman-like morphology, resembles that of sst4/vps27Δ cells (Onishi et al., 2007), in which the expression of the FSM is inefficient and bubble-like structures emerge from the leading edge of the FSM. However, whereas sst4/vps27Δ cells generating FSMs with bubbles produce oval-shaped spores with fuzzy rims, mug28Δ cells generating FSMs with buds produce spores with an abnormal morphology, due to their abnormal FSM formation. Micorarray analysis showed no alteration in the mRNA level of sst4 in mug28Δ cells (Supplemental Figure S2vii). Moreover, we found no genetic interaction between the genes mug28+ and sst4+ (Supplemental Figure S9). Thus, Mug28 and Sst4 seem to function independently during meiosis. Sst4/Vps27 is a downstream factor of the phosphatidylinositol 3-kinase pathway and functions in FSM formation. Vps27p, the Sst4/Vps27 homologue of the budding yeast S. cerevisiae, is a component of the endosomal sorting complex required for transport during protein sorting at the multi vesicular body (Bowers and Stevens, 2005). Because most of the genes required for vacuolar biogenesis and protein transport are conserved between S. pombe and S. cerevisiae (Takegawa et al., 2003), it is possible that Sst4/Vps27 is also a component of endosomal sorting complexes in S. pombe.
Spo20, a phosphatidylinositol/phosphatidylcholine transfer protein of S. pombe, is also notable because some of the abnormal FSM morphologies resembled those of mug28Δ cells in spo20-KC104 mutant cells (Nakase et al., 2001). However, DNA micorarray analysis showed no change in the mRNA level of spo20 in mug28Δ cells (Supplemental Figure S2vii). The result suggests that not only the phosphatidylinositol 3-kinase pathway but also the meiosis-specific RNA-binding protein Mug28 is independently involved in the proper formation of the FSM via regulation of the sorting factors of FSMs. Because the expression of an uncharacterized gene termed meu6+ (Watanabe et al., 2001) was higher in mug28Δ cells at the sporulation stage (Supplemental Figure S2), its significance as a downstream target of Mug28 will be analyzed in a future study.
Electron microscopy revealed that mug28Δ cells displayed not only abnormal snowman-like FSMs but also thicker spore walls than those of mug28+ cells (Figure 7). After formation of FSMs, spore wall assembly begins in the lumen between the two membranes derived from the FSM, where there is no preexisting wall structure to serve as a template. Because bgs2+ and chs1+, which encode a β-glucan synthase and a chitin synthase, respectively, are required for formation of viable spores, β-glucan and chitin must be necessary components of spore walls (Arellano et al., 2000; Liu et al., 2000; Martin et al., 2000). Thus, although the mRNA levels of these genes were not regulated by Mug28 (Supplemental Figure S2vii), Mug28 could still be involved in spore wall assembly.
Mug28 May Contribute to the Proper Disappearance of the Leading Edge Protein Meu14 in Sporulation
In sporulation, leading edge proteins are involved in the initiation and development of the FSM. In fission yeast, a leading edge protein, Meu14, functions in the expansion and closure of four FSMs in a synchronous manner; closure of the FSM must be coordinated with the completion of meiosis II, to ensure proper cell division (Okuzaki et al., 2003). In mug28Δ cells, abnormal behavior of Meu14 was observed during sporulation, compared with mug28+ cells (Figure 4). Although most Meu14 formed a ring-shape during early anaphase II in the mug28+ strain, both Meu14 dots and Meu14 rings were detected in mug28Δ cells (Figures 4 and 5). Moreover, Meu14 could not properly disappear in mug28Δ cells, even after the guidance of leading FSM formation, and was thus aberrantly retained at the edge of the FSM (Figures 5 and 6). This abnormal behavior of Meu14 in mug28Δ cells suggests that Mug28 might somehow regulate the proper disappearance of Meu14 from the leading edge of the FSM during sporulation. The aberrant disappearance of Meu14 is considered to cause the formation of additional FSMs in mug28Δ cells. Notably, Cam2 (type I myosin light chain calmodulin), Myo1 (type I myosin heavy chain), F-actin, and Meu14 colocalize at the leading edge of FSM (Itadani et al., 2006); this suggests that the actin-mediated endocytosis machinery may regulate the closure of the leading edge. Thus, Mug28 also may control the actin-mediated endocytosis machinery by controlling the proper disappearance of Meu14 at sporulation.
Interestingly, this abnormal timing of Meu14 disappearance in mug28Δ cells resembles that of Ssp1p in the ama1Δ strain of S. cerevisiae (Diamond et al., 2009). Three leading edge proteins have been identified in budding yeast, namely, Don1p, Ssp1p, and Ady3p (Moreno-Borchart et al., 2001). Among these three proteins, Ssp1p is essential for sporulation in S. cerevisiae (Nickas and Neiman, 2002). Ssp1p is required for localization of both Ady3p and Don1p to the leading edge of the prospore membrane (PSM), an analogue of FSM in budding yeast, whereas PSM growth is abnormal and spore formation is blocked in ssp1Δ cells. Notably, PSM closure is defective in the absence of ama1, a meiosis-specific activator of the anaphase promoting complex/cyclosome (Diamond et al., 2009). Moreover, the leading edge protein Ssp1p is stabilized in ama1Δ mutants, suggesting that an essential function of Ama1p is to remove Ssp1p from the PSM by degradation. Because ectopic expression of Meu14DD-GFP caused no detectable abnormality in FSM formation (Supplemental Figure S6), the ubiquitin-proteasome pathway may not be involved in the untimely degradation of Meu14.
We also showed here that Mug28 localizes to the cytoplasm, particularly to the area around the nucleus, between metaphase I and anaphase II, and also to the nucleus during meiosis II (Figure 2). Considering that mug28Δ cells have both abnormal FSM formation and perinuclear localization of Mug28-GFP during meiosis, it is possible that the FSM and the nuclear membrane are coordinately regulated by Mug28 in the perinuclear region, which controls the disappearance of the leading edge protein Meu14 at sporulation. Nonetheless, because overexpression of Meu14 (Supplemental Figure S3B) and a putatively stabilized Meu14DD (Supplemental Figure S6) in wild-type cells did not reverse the morphological defects of mug28Δ cells, and because these morphological defects are also observed in mug28Δ meu14Δ double mutant cells (Supplemental Figure S4), we cannot exclude the possibility that the effects of Mug28 on Meu14 and on FSM morphology are independent.
Role of RRM Motifs for Full Function of Mug28
Analysis of Mug28 mutants indicated that the RRM3 domain that harbors F466 is pivotal for the full functioning of Mug28, which is essential for proper maturation of the FSM and the spore wall (Figure 8). Indeed, abnormal Mug28 localization, spore formation, and spore viability were observed in F466A mutant cells (Figure 9). Homology between the RRMs of Mug28 and Meu5/Crp79, another meiosis-specific RNA-binding protein (Watanabe et al., 2001), may be noteworthy. Meu5/Crp79 also was isolated as a novel mRNA export factor from the synthetic lethal screen of a mutation in rae1+, an essential gene required for mRNA export, during vegetative growth in S. pombe; indeed, Meu5/Crp79 binds to and exports mRNA from the nucleus to the cytoplasm (Thakurta et al., 2002). Although the RRM1 and RRM3 domains of Meu5/Crp79 also have an essential role in poly(A)+ RNA binding (Thakurta et al., 2002), it remains unclear whether Mug28 also can bind to and export any RNA during meiosis, because there is no clear homology between Mug28 and Meu5/Crp79 beyond that seen in the RRMs.
A similar common association partner may exist between Mug28 and Meu5/Crp79. The present study indicates that meiosis-specific proteins harboring RRMs are involved not only in meiotic progression but also in the proper formation or maturation of FSMs and spore walls. It is expected that Mug28 binds to some target RNAs, interaction partners, or both via its RRM domains and thus functions in the proper development and formation of the FSM during meiosis. Identifying the target RNAs and interaction partners of Mug28 will provide important information about the novel function of RBPs in the proper development and formation of FSMs during meiosis. We plan to address these issues in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. C. Shimoda, M. Yamamoto, T. Nakamura, W. Z. Cande, A. Ohtaka, and R. Tsien and the National BioResource Project (http://yeast.lab.nig.ac.jp/nig/) (Osaka City University) for S. pombe strains and plasmids. We are also indebted to Dr. P. Hughes for critically reading the manuscript. This work was supported in part by grants-in-aid for Scientific Research on Priority Areas “Applied Genomics,” Scientific Research (S), Exploratory Research, and the Science and Technology Incubation Program in Advanced Regions from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H. N.).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-12-0997) on April 21, 2010.
REFERENCES
- Arellano M., Cartagena-Lirola H., Nasser Hajibagheri M. A., Duran A., Henar Valdivieso M. Proper ascospore maturation requires the chs1+ chitin synthase gene in Schizosaccharomyces pombe. Mol. Microbiol. 2000;35:79–89. doi: 10.1046/j.1365-2958.2000.01678.x. [DOI] [PubMed] [Google Scholar]
- Asakawa H., Haraguchi T., Hiraoka Y. Reconstruction of the kinetochore: a prelude to meiosis. Cell Div. 2007;2:17. doi: 10.1186/1747-1028-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers K., Stevens T. H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Diamond A., Park J., Inoue I., Tachikawa H., Neiman A. M. The APC targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2009;20:134–145. doi: 10.1091/mbc.E08-06-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Kohli J., Murray J., Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable maker. Mol. Gen. Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Harigaya Y., Yamamoto M. Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res. 2007;15:523–537. doi: 10.1007/s10577-007-1151-0. [DOI] [PubMed] [Google Scholar]
- Itadani A., Nakamura T., Shimoda C. Localization of type I myosin and F-actin to the leading edge region of the FSM in Shizosaccharomyces pombe. Cell Struct. Funct. 2006;31:181–195. doi: 10.1247/csf.06027. [DOI] [PubMed] [Google Scholar]
- Kakihara Y., Nabeshima K., Hirata A., Nojima H. Overlapping omt1+ and omt2+ genes are required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells. 2003;8:547–558. doi: 10.1046/j.1365-2443.2003.00654.x. [DOI] [PubMed] [Google Scholar]
- Kasama T., Shigehisa A., Hirata A., Saito T., Tougan T., Okuzaki D., Nojima H. Spo5/Mug12, a putative meiosis-specific RNA-binding protein, is essential for meiotic progression and forms Mei2 dot-like nuclear foci. Eukaryot. Cell. 2006;5:1301–1313. doi: 10.1128/EC.00099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Tang X., Wang H., Balasubramanian M. Bgs2p, a 1,3-beta-glucan synthase subunit, is essential for maturation of ascospore wall in Schizosaccharomyces pombe. FEBS Lett. 2000;478:105–108. doi: 10.1016/s0014-5793(00)01828-7. [DOI] [PubMed] [Google Scholar]
- Martín-Castellanos C., et al. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr. Biol. 2005;15:2056–2062. doi: 10.1016/j.cub.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Lyne R., Burns G., Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- Martin V., Ribas J. C., Carnero E., Duran A., Sanchez Y. Bgs2+, a sporulation-specific glucan synthase homologue is required for proper ascospore wall maturation in fission yeast. Mol. Microbiol. 2000;38:308–321. doi: 10.1046/j.1365-2958.2000.02118.x. [DOI] [PubMed] [Google Scholar]
- Mattick J. S. RNA regulation: a new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Moreno-Borchart A. C., Strasser K., Finkbeiner M. G., Shevchenko A., Shevchenko A., Knop M. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 2001;20:6946–6957. doi: 10.1093/emboj/20.24.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Nakamura-Kubo M., Hirata A., Shimoda C. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+-encoding Syntaxin-like protein. Mol. Biol. Cell. 2001;12:3955–3972. doi: 10.1091/mbc.12.12.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Asakawa H., Nakase Y., Kashiwazaki J., Hiraoka Y., Shimoda C. Live observation of forespore membrane formation in fission yeast. Mol. Biol. Cell. 2008;8:3544–3553. doi: 10.1091/mbc.E08-04-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase Y., Nakamura T., Hirata A., Routt S. M., Skinner H. B., Bankaitis V. A., Shimoda C. The Shizosaccharomyces pombe spo20+ gene encoding a homologue of Saccharomyces cerevisiae Sec14 plays an important role in FSM formation. Mol. Biol. Cell. 2001;12:901–917. doi: 10.1091/mbc.12.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;4:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas M. E., Neiman A. M. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics. 2002;160:1439–1450. doi: 10.1093/genetics/160.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaka A., Saito T., Okuzaki D., Nojima H. Meiosis specific coiled-coil proteins in Shizosaccharomyces pombe. Cell Div. 2007;18:2–14. doi: 10.1186/1747-1028-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Okuzaki D., Satake W., Hirata A., Nojima H. Fission yeast meu14+ is required for proper nuclear division and accurate FSM formation during meiosis II. J. Cell Sci. 2003;116:2721–2735. doi: 10.1242/jcs.00496. [DOI] [PubMed] [Google Scholar]
- Onishi M., Iida M., Koga T., Yamada S., Hirata A., Iwaki T., Takegawa K., Fukui Y., Tachikawa H. Schizosaccharomyces pombe Sst4p, a conserved Vps27p/Hrs homolog, functions downstream of phosphatidylinositol 3-kinase Pik3p to mediate proper spore formation. Eukaryot. Cell. 2007;6:2343–2353. doi: 10.1128/EC.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Tougan T., Kasama T., Okuzaki D., Nojima H. Mcp7, a meiosis-specific coiled-coil protein of fission yeast, associates with Meu13 and is required for meiotic recombination. Nucleic Acids Res. 2004;32:3325–3339. doi: 10.1093/nar/gkh654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Tougan T., Okuzaki D., Kasama T., Nojima H. Mcp6, a meiosis-specific coiled-coil protein of Schizosaccharomyces pombe, localizes to the spindle pole body and is required for horsetail movement and recombination. J. Cell Sci. 2005;118:447–459. doi: 10.1242/jcs.01629. [DOI] [PubMed] [Google Scholar]
- Shimada M., Nabeshima K., Tougan T., Nojima H. The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. EMBO J. 2002;21:2807–2818. doi: 10.1093/emboj/21.11.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda C. FSM assembly in yeast: coordinating SPBs and membrane trafficking. J. Cell Sci. 2004;117:389–396. doi: 10.1242/jcs.00980. [DOI] [PubMed] [Google Scholar]
- Takegawa K., Iwaki T., Fujita Y., Morita T., Hosomi A., Tanaka N. Vesicle-mediated Protein Transport Pathways to the Vacuole in Schizosaccharomyces pombe. Cell Struct. Funct. 2003;28:399–417. doi: 10.1247/csf.28.399. [DOI] [PubMed] [Google Scholar]
- Thakurta A. G., Whalen W. A., Yoon J. H., LiborKozak A. B., Whiteford C., Love D. C., Hanover J. A., Dhar R. Crp79p, like Mex67p, is an auxiliary mRNA export factor in Shizosaccharomyce pombe. Mol. Biol. Cell. 2002;13:2571–2584. doi: 10.1091/mbc.E01-11-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomilin N. V. Regulation of mammalian gene expression by retroelements and non-coding tandem repeats. Bioessays. 2008;30:338–348. doi: 10.1002/bies.20741. [DOI] [PubMed] [Google Scholar]
- Tougan T., Chiba Y., Kakihara Y., Hirata A., Nojima H. Meu10 is required for spore wall maturation in Schizosaccharomyces pombe. Gene Cells. 2002;7:217–231. doi: 10.1046/j.1356-9597.2001.00511.x. [DOI] [PubMed] [Google Scholar]
- Wahls W. P., Siegel E. R., Davidson M. K. Meiotic recombination hotspots of fission yeast are directed to loci that express non-coding RNA. PloS One. 2008;3:2887–2894. doi: 10.1371/journal.pone.0002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Miyashita K., Saito T., Yoneki T., Kakihara Y., Nabeshima K., Kishi Y. K., Shimoda C., Nojima H. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 2001;29:2327–2337. doi: 10.1093/nar/29.11.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Miyashita K., Saito T., Nabeshima K., Nojima H. Abundant poly(A)-bearing RNAs that lack open reading frames in Schizosaccharomyces pombe. DNA Res. 2002;9:209–215. doi: 10.1093/dnares/9.6.209. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Yamamoto M. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- Xue-Franzén Y., Kjærulff S., Holmberg C., Wright A., Nielsen O. Genomewide identification of pheromone-targeted transcription in fission yeast. BMC Genomics. 2006;7:303. doi: 10.1186/1471-2164-7-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M. The molecular control mechanisms of meiosis in fission yeast. Trends Biochem. Sci. 1996;21:18–22. [PubMed] [Google Scholar]
- Yamashita A., Watanabe Y., Nukina N., Yamamoto M. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell. 1998;95:115–123. doi: 10.1016/s0092-8674(00)81787-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.