Human Spindly is required for kinetochore localization of cytoplasmic dynein, which is essential for poleward movement of chromosomes and for kinetochore protein streaming. In addition, Spindly controls the activity and kinetochore abundance of the RZZ complex, which contributes to microtubule attachment and mitotic checkpoint activity.
Abstract
Spindly recruits a fraction of cytoplasmic dynein to kinetochores for poleward movement of chromosomes and control of mitotic checkpoint signaling. Here we show that human Spindly is a cell cycle–regulated mitotic phosphoprotein that interacts with the Rod/ZW10/Zwilch (RZZ) complex. The kinetochore levels of Spindly are regulated by microtubule attachment and biorientation induced tension. Deletion mutants lacking the N-terminal half of the protein (NΔ253), or the conserved Spindly box (ΔSB), strongly localized to kinetochores and failed to respond to attachment or tension. In addition, these mutants prevented the removal of the RZZ complex and that of MAD2 from bioriented chromosomes and caused cells to arrest at metaphase, showing that RZZ-Spindly has to be removed from kinetochores to terminate mitotic checkpoint signaling. Depletion of Spindly by RNAi, however, caused cells to arrest in prometaphase because of a delay in microtubule attachment. Surprisingly, this defect was alleviated by codepletion of ZW10. Thus, Spindly is not only required for kinetochore localization of dynein but is a functional component of a mechanism that couples dynein-dependent poleward movement of chromosomes to their efficient attachment to microtubules.
INTRODUCTION
For equational segregation, chromosomes have to attach to the mitotic spindle in a bipolar manner. This is achieved by the formation of stable “end-on” attachments of microtubules from one spindle pole to the kinetochore of one sister chromatid. Successful bipolar attachment, also called biorientation, is required for chromosome congression to the metaphase plate and results in stretching forces within and between the kinetochores (Waters et al., 1996; Maresca and Salmon, 2009; Uchida et al., 2009). Together with microtubule attachment these tension-induced changes are required to terminate the spindle assembly checkpoint (SAC), which otherwise inhibits sister chromatid separation and exit from mitosis (reviewed in Musacchio and Salmon (2007)).
Cytoplasmic dynein, a large microtubule minus end-directed motor, is one of many motor proteins essential for mitosis in animal cells (Walczak and Heald, 2008; Gatlin and Bloom, 2010). Dynein, localized to the spindle poles and the cellular cortex, is involved in bipolar spindle formation (Vaisberg et al., 1993; Gaglio et al., 1996; Heald et al., 1996; Merdes et al., 1996) and orientation (Li et al., 1993; Carminati and Stearns, 1997; Busson et al., 1998; Faulkner et al., 2000; O'Connell and Wang, 2000), respectively. During early mitosis, a fraction of dynein also localizes to the outer fibrillar corona of kinetochores (Pfarr et al., 1990; Steuer et al., 1990; Wordeman et al., 1991; Vorozhko et al., 2008), where it engages tangentially with microtubules (Rieder and Alexander, 1990). These initial interactions allow dynein to move chromosomes toward the microtubule dense environment of the spindle pole (Sharp et al., 2000; Yang et al., 2007), which is believed to facilitate the generation of stable end-on attachments mediated by the kinetochore NDC80 complex (Santaguida and Musacchio, 2009).
After the formation of stable end-on attachments, dynein disconnects from the kinetochore and translocates to the spindle pole (Pfarr et al., 1990; Steuer et al., 1990; King et al., 2000; Whyte et al., 2008). After this poorly understood transition from a kinetochore resident to a moving protein, dynein remains associated with many proteins of the outer kinetochore and thus, effectively removes them from kinetochores in a process called kinetochore stripping or streaming (Hoffman et al., 2001; Howell et al., 2001; Wojcik et al., 2001; Basto et al., 2004; Whyte et al., 2008). Because kinetochore stripping also affects mitotic checkpoint proteins, such as MAD2 (Howell et al., 2001), it may be an effective mechanism that controls checkpoint signaling strength in response to microtubule attachment (Karess, 2005).
To achieve its diverse functions, the catalytic dynein heavy chains (DHCs) are associated with several noncatalytic subunits that either directly interact with substrates or bind to other protein complexes, such as dynactin, the Rod/ZW10/Zwilch (RZZ) complex and other factors (for a recent review see Kardon and Vale, 2009). The precise function of these subunits and their interacting proteins is only poorly understood. Some of them directly interact with cargoes, such as the light intermediate chain 1 (DLIC1), which is required for MAD2 removal from kinetochores of bioriented chromosomes (Sivaram et al., 2009). Others are required for dynein localization to kinetochores, which, among other factors, is influenced by kinetochore-associated NUDE/NDE, which binds to one of the dynein light chains (Stehman et al., 2007) or by the interaction between ZW10 and dynein IC 1 (DIC1; Starr et al., 1998; Kops et al., 2005; Whyte et al., 2008).
Many of the factors that are required for dynein localization to, or function at, the kinetochores, including dynactin (Echeverri et al., 1996), LIS-1 (which directly interacts with DHC; Tai et al., 2002), and the RZZ complex (Basto et al., 2004) are also subject to kinetochore streaming, whereas other dynein-interacting proteins, such as NDE and NDEL (Stehman et al., 2007), remain on the kinetochores in anaphase. Whether these different dynein complexes carry out different functions or whether there is only one dynein motor that is controlled by these interactions is an important but still unresolved issue.
Dynactin is a large multisubunit complex that is known to regulate the processivity of dynein but might also function as a dynein cargo receptor (for review see Schroer, 2004). The p150glued component of dynactin directly interacts with DIC1, but whether dynactin and dynein are recruited by the same or different proteins has not been clearly deciphered. Strong overexpression of p50/dynamitin disrupts the dynactin complex, depletes dynein from kinetochores but also interferes with nonkinetochore dynein functions and causes multiple mitotic defects (Echeverri et al., 1996; Karki and Holzbaur, 1999; Howell et al., 2001; Melkonian et al., 2007; Whyte et al., 2008). At lower expression levels, dynamitin does not interfere with dynein localization but prevents anaphase, implying that checkpoint inactivation requires dynein to interact with dynactin, a process that may also be controlled by protein phosphorylation (Whyte et al., 2008). Many of these phenotypes were also seen by using dominant negative dynein heavy chain mutants, which, in addition, revealed roles for dynein in the formation of stable microtubule attachments and in kinetochore orientation (Varma et al., 2008).
The RZZ complex is required for dynein recruitment to kinetochores as well as for SAC activity (Williams et al., 1992; Scaerou et al., 2001; Karess, 2005; Yang et al., 2007). How the RZZ complex contributes to mitotic checkpoint signaling is not entirely clear, but it might serve as an activation platform for MAD2 (Karess, 2005). ZW10 directly interacts with DIC1 (Whyte et al., 2008) and p50/dynamitin (Starr et al., 1998) and is subject to kinetochore streaming, but whether ZW10 removal from kinetochores is essential and how it is controlled is still not well understood. ZW10, which is recruited to kinetochores via ZWINT-1 (Starr et al., 2000) and other mechanisms (Famulski et al., 2008), is a highly dynamic protein, whose dwelling time at kinetochores decreases upon attachment of microtubules (Famulski et al., 2008). In addition, ZW10 localization to kinetochores is also controlled by an Aurora B kinase–regulated tension sensitive pathway (Famulski and Chan, 2007), which might be an important means to resist dynein-dependent removal of the RZZ complex to maintain checkpoint signaling until tension is sensed.
In flies, worms and in human cells, the RZZ complex also interacts with Spindly, a coiled-coil domain containing protein, which is required for dynein localization to kinetochores (Griffis et al., 2007; Gassmann et al., 2008; Yamamoto et al., 2008; Chan et al., 2009). Whether Spindly acts as an RZZ specific adaptor required for kinetochore streaming or whether Spindly controls additional dynein functions on kinetochores is controversial as evidenced by different effects on SAC activity in different species.
Although fly cells depleted for Spindly arrested at metaphase with high levels of kinetochore-bound Mad2 (Griffis et al., 2007), worm embryos progressed to anaphase, because SPDL-1 depletion abrogated Mad2 and, thus, mitotic checkpoint function (Yamamoto et al., 2008). In addition, SPDL-1 appears to have a novel function in chromosome attachment, because its loss caused a chromosome congression defect in nematodes (Gassmann et al., 2008). This effect of Spindly depletion on chromosome congression was also noted in fly cells, although it was less prominent (Griffis et al., 2007). In human cells, Spindly knockdown also impaired chromosome congression and spindle positioning but was not required for MAD2 removal from kinetochores (Chan et al., 2009).
Thus, Spindly is required for dynein functions at kinetochores but how Spindly is controlled, bound to kinetochores and how it exerts its functions, is still largely unknown. Here, we show that human Spindly/CCDC99 is not merely a kinetochore tether for dynein but that Spindly is involved in the coordination of microtubule attachment and mitotic checkpoint signaling.
MATERIALS AND METHODS
Reagents
Chemicals, enzymes, and oligonucleotides were purchased from Sigma (Vienna, Austria), Promega (Mannheim, Germany), and MWG Biotech (Ebersberg, Germany), respectively, unless stated otherwise. Antibodies used in this study are listed in the Supplemental Material and Methods.
Plasmid Construction, Protein Expression, and Antibody Generation and Purification
Human Spindly and ZW10 were amplified from IRAUp969B0653D (ImaGenes, Berlin, Germany) and LIFESEQ2639836 (Open Biosystems, Thermo Fisher Scientific, Vienna, Austria), respectively, and cloned into expression vectors using GATEWAY technology (Invitrogen, Karlsruhe, Germany). pCherry-α-tubulin was constructed by subcloning α-tubulin from enhanced green fluorescent protein (pEGFP)-tubulin (Clontech, Heidelberg, Germany) into pCherry-C1. The red fluorescent protein (RFP) expression vector pCS2-mRFP was constructed by subcloning PCR-amplified mRFP (mCherry and mRFP were kindly provided by Roger Tsien, University of California, San Diego, La Jolla, CA) into the mammalian expression vector pCS2. FLAG-tagged constructs were created using pΔT-3x-FLAG-DEST (unpublished data). The expression vector for CenpB-YFP was created by PCR amplification of human CenpB (aa 1-174) from IMAGE clone 6470289 (ImaGenes), subcloning into pDONR 207 (Invitrogen) and shuttling into an yellow fluorescent protein (YFP) expression vector. Deletion and site-specific mutations were introduced by overlap extension PCR, subcloned and verified by DNA sequencing.
Cell Culture Methods, Transfection, and RNA Interference
HeLa, U2OS and human embryonic kidney HEK293 cells were grown (Wolf et al., 2006) and synchronized as described (Geley et al., 2001). Mitotic synchronization was achieved by 250 nM nocodazole treatment for 16 h, monopolar spindles induced by 100 μM monastrol for 12 h, and Aurora B kinase activity was inhibited by 2 μM ZM447439 (Tocris, Biozol, Eching, Germany) for 2 h. U2OS-H2B-GFP/mCherry-α-tubulin and U2OS cells expressing YFP-ZW10 or CENP-B-YFP were generated by transfecting cells with linearized plasmids using Lipofectamine 2000 (Invitrogen) and selecting for stable integrants using 1 mg/ml G418. For overexpression experiments subconfluent HeLa or U2OS cells were transfected with 2 μg of FLAG-Spindly expression constructs together with 0.4 μg pCS2-mRFP using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For RNA interference (RNAi) treatment, cells were transfected at 30–50% confluency using Lipofectamine 2000 with 50 nM siRNAs for 48 h: Spindly: 5′-GAAAGGGUCUCAAACUGAATT-3′, MAD2 (Martin-Lluesma et al., 2002), ZW10 (Yang et al., 2007), hKID (Wolf et al., 2006), NDC80 (Martin-Lluesma et al., 2002), NUF2 (Liu et al., 2007), DHC1 (Harborth et al., 2001; Draviam et al., 2006), SGO1 (Wolf et al., 2006), CENP-E (Tanudji et al., 2004); and control (Luciferase): 5′ GCUAUGAAACGAUAUGGGCTT, and Dharmacon small interfering RNA (siRNA) pools for ZWINT-1, CDCA5, and KNTC1/Rod.
PhosTag Gel Electrophoresis, Immunoblotting, and In Vitro Kinase Assay
PhosTag gel electrophoresis was performed using FLAG-tagged Spindly immunopurified from nocodazole-arrested mitotic and untreated asynchronous cells by using 10 μM PhosTag (NARD Institute, Hyogo, Japan)/10 μM MnCl2 in a 6% polyacrylamide gel (Kinoshita et al., 2006). For immunoprecipitation, subconfluent HEK293A cells were transfected in six-well plates with 4 μg FLAG-Spindly wild-type or S515A mutant plasmid. After 48 h, cells were harvested and lysed in immunoprecipitation (IP) buffer (150 mM NaCl, 20 mM NaF, 50 mM Tris, pH 7.4, 0.1% NP40, 1 mM EGTA, and Roche Complete protease inhibitors, Roche Applied Science, Vienna, Austria). Four hundred micrograms of total proteins were used for immunoprecipitation with 4.9 μg FLAG M2 (Sigma F3165) coupled to Affi-Prep protein A beads (Bio-Rad Laboratories, Vienna, Austria), 30 min at 4°C. The beads were extensively washed with IP buffer and split in two. One-half was eluted with SDS sample buffer and checked for IP efficiency by immunoblotting. The second half was washed once more with kinase buffer and used as a substrate in an in vitro kinase assay. Active recombinant CDK2/cyclin A was purified by mixing the cleared lysates of induced BL21[DE3] transformed with pGEX-CDK2-GST-Civ1 (kindly provided by Jane Endicott, Oxford, United Kingdom) and pET21-cyclinA3 (kindly provided by Tim Hunt, C.R.U.K, South Mimms, United Kingdom), and sequential GSH-Sepharose (GE Healthcare, Munich, Germany) and metal affinity chromatography (Qiagen, Hilden, Germany). Kinase reactions were carried out at 30°C for 10 min using recombinant Cdk2/CycA3, [γ-32P]ATP, and magnesium acetate, after which proteins were resolved by SDS-PAGE, Coomassie gel–stained, dried, and subjected to autoradiography.
Immunoprecipitation and Mass Spectrometry
Mitotically arrested cells were collected by shake-off, washed in PBS, and resuspended in 20 mM HEPES-KOH, pH 7.5, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, and 1 mM EGTA, with protease (Complete, Roche) and phosphatase inhibitors (PhosSTOP, Roche), lysed by four freeze-thaw cycles, sonicated, and centrifuged at 10,000 × g for 20 min at 4°C. For IP, 1–10 μg of primary mAb or control antibodies, covalently coupled to Affi-Prep protein A beads, was added to 1–10 mg protein lysate, at 4°C for 1 h, washed, and analyzed by SDS-PAGE followed by immunoblotting or colloidal Coomassie Brilliant Blue (cCBB) staining (modified after Neuhoff et al., 1988). For identification of co-immunoprecipitated proteins, cCBB-stained protein bands were excised from gels and in-gel digested with trypsin as described (Hellman, 2000). The in-gel digests were concentrated, desalted using microZipTipC18 (Millipore, Schwalbach, Germany), and peptides were directly eluted onto the MALDI target in 0.1% TFA/60% ACN containing 50% saturated α-cyano-4-hydroxycinnamic acid (Fluka, Buchs, Switzerland). Mass spectra were acquired on a calibrated MALDI-TOF/TOF Ultraflex instrument (Bruker Daltonics, Bremen, Germany), using a 337-nm nitrogen laser at 50 Hz and 30–50% of maximal intensity, a peptide mass tandem mass spectrometry (MS/MS) tolerance of 100 ppm and 0.6 Da, respectively. Data were processed using Flex Analysis 2.4 and Bio-Tools 3.0 and peptide mass fingerprints and MS/MS spectra of selected precursor ions interpreted using MASCOT (Matrix Science, Boston, MA) searches against the Swiss-Prot protein database (http://www.expasy.ch/sprot/). The μLC-ESI-MS/MS protocol is described in the Supplemental Materials and Methods.
Microscopy and Image Processing
Live cell microscopy was performed on an Axiovert 200M microscope (Carl Zeiss, Jena, Germany) as described (Wolf et al., 2006). Imaging of CENP-B-YFP cells in mitosis was performed using a Cascade II camera (512 × 512_EMCCD; Photometrics, Hannover, Germany) neutral density filters and imaging every 30 s (1 μm z-stacks, 10 planes, 1-μm distance) for several hours. Confocal microscopy was performed on a Zeiss LSM510 microscope as described (Wolf et al., 2006) or on a Leica TCS-SP5 DMI6000 microscope (Leica, Mannheim, Germany) equipped with a 405-nm diode, an argon ion, a 561-nm DPSS and a 633-nm HeNe laser. Images were acquired using a HCX PL APO lambda blue 63×, NA 1.4 oil UV objective at a pixel resolution of 67 nm. Images were analyzed using Metamorph 7.0 (Molecular Devices, Downingtown, PA), converted to 8-bit TIFF format and further processed using Adobe Photoshop CS2 and Illustrator CS2 (Adobe Systems, San Jose, CA). Image stacks were deconvoluted using Huygens deconvolution software (SVI, Hilversum, Netherlands). For quantification of kinetochore fluorescence signals, pixel intensities were measured in 10-pixel diameter circular regions of interest (ROIs). Mean background signals were determined from signals measured in 2-pixel-wide rings outside these ROIs and subtracted from the mean intensities of the ROIs to obtain background corrected values, which were normalized to CREST fluorescence intensities. Interkinetochore distances were measured from the outer edges of paired CREST-stained kinetochore signals. For fluorescence recovery after photobleaching (FRAP) measurements, U2OS cells expressing YFP-ZW10 were plated onto 35-mm glass-bottom dishes and monitored on a Leica SP5 microscope at 37°C using a 512 × 100-pixel area scanned at 400 Hz at 13× zoom using a 63×, NA 1.4 oil objective and a pinhole of 2 airy units. After identification of kinetochore pairs, one was photobleached with two laser pulses (60% power of the argon laser), and subsequent fluorescence recovery was recorded for 35 s at 0.57-s intervals (with 3% laser power). Five images were collected before and 60 after bleaching. For calculation of fluorescence recovery, background corrected fluorescence signals were double-normalized to the averaged prebleach levels and to the first image after the bleaching pulse. The mean values of double normalized recoveries were then used for curve fitting to determine the half-life of recovery as well as recovery efficiency as described in http://www.embl.org/cmci/downloads/FRAPmanual.htm.
RESULTS
Spindly Is a Mitotic Phosphoprotein That Interacts with the RZZ Complex
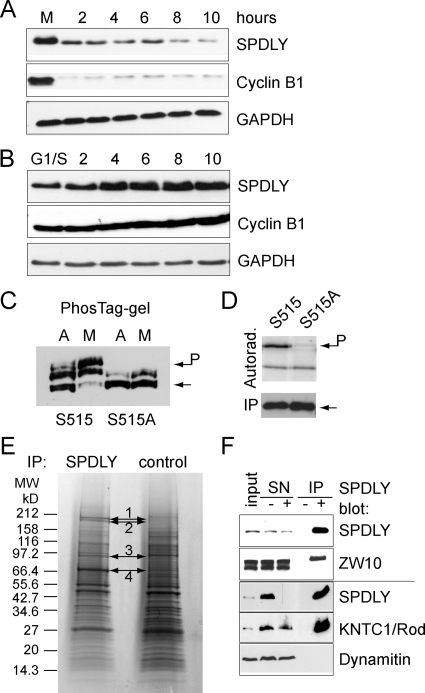
To investigate the function and regulation of human Spindly, we first analyzed its expression pattern during the cell cycle. When cell extracts prepared from synchronized cultures of HeLa and U2OS cells were probed by immunoblotting, we found that Spindly protein levels oscillated during the cell cycle and peaked in mitosis (Figure 1, A and B), mirroring its mRNA expression profile (Whitfield et al., 2002). The electrophoretic mobility of Spindly was slightly slower in mitosis than in interphase, indicative of mitotic phosphorylation, which was verified by the identification of serine 515 phosphorylation by mass spectrometry of FLAG-tagged Spindly immunopurified from mitotically arrested HEK293 cells (Supplemental Figure 1A). PhosTag gel-electrophoresis (Kinoshita et al., 2006) of FLAG-tagged-Spindly and the S515A mutant confirmed that serine 515 was a major phospho-acceptor site in mitosis (Figure 1C). Because S515 lies within a CDK consensus motif, we purified FLAG-Spindly from interphase HEK293 cells and incubated it with radiolabeled ATP and recombinant cyclinA/CDK2. As can be seen in Figure 1D, active CDK2 phosphorylated the wild-type but not the S515A mutant. Thus, Spindly is a mitotic phosphoprotein that can be phosphorylated by CDKs at S515.
Figure 1.
Spindly associates with the RZZ complex. (A and B) Spindly is cell cycle regulated. Total cell extracts prepared from nocodazole block/release U2OS (A) and double thymidine block/release HeLa cells (B) at the indicated time points (h) were analyzed by immunoblotting for Spindly, Cyclin B1, and GAPDH. (C) Serine 515 is a major mitotic phosphorylation site. HEK293A cells were transfected with FLAG-tagged versions of wild-type and S515A mutant Spindly and treated with nocodazole for 12 h (mitotic, M) or left untreated (asynchronous, A). FLAG-Spindly immunoprecipitates were separated on 6% PAGE/10 μM PhosTag gels and immunoblotted using an anti FLAG antibody. (D) Immunoprecipitated FLAG-tagged wild-type or S515A Spindly was incubated with recombinant cyclinA/CDK2 and radiolabeled proteins detected by autoradiography. (E) Spindly was immunoprecipitated from 10 mg mitotic cell lysate. Proteins were visualized by cCBB staining and analyzed by mass spectrometry. (F) Spindly immunoprecipitates were probed for ZW10 (top two panels) or Rod and dynamitin (bottom panels).
To identify Spindly-interacting proteins, we immunoaffinity-purified endogenous Spindly from nocodazole-arrested mitotic HeLa cells and analyzed the precipitated proteins by SDS-PAGE and mass spectrometry. As can be seen in Figure 1E, several protein bands were enriched in the Spindly immunoprecipitate, which were identified, in at least two independent experiments (Supplemental Table 1), as the members of the RZZ complex (Karess, 2005): KNTC1/Rod (bands 1 and 2), ZW10 (band 3), and ZWILCH (band 4). To confirm the association of Spindly with the RZZ complex, we performed coimmunoprecipitation (coIP) as well as cofractionation experiments using cell extracts obtained from mitotically arrested and asynchronous HeLa cells. CoIP confirmed the interaction of Spindly with ZW10 and KNTC1/Rod in mitosis under low-stringency conditions (low salt, no detergent), but did not reveal an interaction between Spindly and dynamitin (Figure 1F), dynein heavy chain 1 (DHC1), DIC1 or p150glued (data not shown). Gel filtration using ACA34 and Superose 6 columns revealed that Spindly partly cofractionated with RZZ components in a large (>600 kDa) protein complex in mitosis (Supplemental Figure 1B), but not in interphase (not shown). In summary, these experiments demonstrate that Spindly is associated with, or is part of, the RZZ complex.
The interaction of Spindly with the RZZ complex was further supported by colocalization of ZW10 and Spindly on kinetochores of HeLa cells (Supplemental Figure 1C). Kinetochore localization was first detectable after nuclear envelope breakdown, strongest during prometaphase, and lowest during ana- and telophase (Supplemental Figure 1D). In addition, Spindly stained the mitotic spindle and its poles during prometa- and metaphase (Supplemental Figure 1, E and F). In cells depleted for ZWINT-1 or ZW10, Spindly did not localize to kinetochores (Supplemental Figure 1, E and F). In contrast to ZWINT RNAi treatment, depletion of ZW10 also reduced the total protein level of Spindly (and that of KNTC1/Rod; Supplemental Figure 1G), a finding that further supported the hypothesis that Spindly is part of the RZZ complex, whose disruption caused destabilization of its subunits.
Knockdown of Spindly depleted DHC1 and DIC as well as the dynactin component p150glued from kinetochores (Supplemental Figure 2, A and B, and data not shown), but had no depleting effect on more proximal kinetochore components, including ZWINT-1 and ZW10 (Supplemental Figure 1, E and F, and data not shown), indicating that Spindly might act in a linear RZZ-Spindly-dynein pathway, consistent with previous publications (Griffis et al., 2007; Gassmann et al., 2008; Chan et al., 2009). To clarify the role of Spindly in this pathway, we first determined how Spindly binds to kinetochores and then probed its function by RNAi experiments and overexpression of dominant negative mutants.
Spindly Localization to Kinetochores Is Controlled by Attachment and Tension
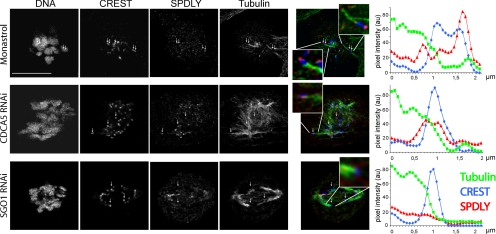
Because the kinetochore levels of Spindly declined during the process of chromosome alignment, we investigated whether they were regulated by microtubule attachment, tension, or both. To this end, we used three different approaches to induce monopolarly attached chromosomes to compare Spindly binding to the attached versus the unattached kinetochore. First, we treated HeLa cells for 12 h with 100 μM monastrol to arrest them in mitosis with monopolar spindles. Chromosomes attach to monastrol-induced asters either in a monopolar manner, displaying one unattached kinetochore, or in a syntelic manner, in which both kinetochores are attached. As can be seen in Figure 2, monopolarly attached chromosomes displayed asymmetric levels of Spindly, which were found to be higher on the unattached kinetochore. Second, we knocked down CDCA5/sororin in HeLa cells to prevent the establishment of sister chromatid cohesion during S-phase to obtain mitotic cells with unpaired sister chromatids (Schmitz et al., 2007). When CDCA5 RNAi-treated cells were analyzed by immunofluorescence, Spindly levels on kinetochores were low, but still detectable. In a third set of experiments, we depleted SGO1 by RNAi, which induces loss of mitotic sister chromatid cohesion (Salic et al., 2004; McGuinness et al., 2005), to induce precocious sister chromatid separation in response to tension after biorientation. In contrast to the above two experiments, Spindly could not be detected on individual kinetochores in SGO1 knockdown cells (compare red lines, which correspond to Spindly levels, in the intensity plots shown Figure 2). We concluded from these experiments that, in contrast to SGO1-depleted cells, the kinetochores in the monastrol and CDCA5-RNAi experiments were not under sufficient tension to completely deplete them from Spindly.
Figure 2.
Spindly levels on kinetochores are reduced upon microtubule attachment. HeLa cells were treated with 100 μM monastrol for 12 h or transfected with 50 nM CDCA5 or SGO1 siRNA for 24 h before fixation and immunostaining for DNA, kinetochores (blue), Spindly (red), and tubulin (green). Confocal image stacks taken on a Leica SP5 microscope were deconvoluted and fluorescence signals measured along microtubules attached to a kinetochore. Signals of tubulin (green), CREST (blue), and Spindly (red) of kinetochores interacting with microtubules were measured by line scans and are displayed as line plots of mean intensities of three microtubule–kinetochore junctions each. Size bar, 10 μm.
To more directly assay the effect of attachment and tension on kinetochore Spindly levels, we treated cells with 20 μM MG132 for 4 h, to enrich for metaphase cells, before addition of 1 μM nocodazole or taxol for 2 h in the presence or absence of 2 μM ZM447439, an inhibitor of Aurora kinase B (Ditchfield et al., 2003). As can be seen in Supplemental Figure 3, disruption of microtubules by nocodazole caused strong localization of Spindly to kinetochores that was not affected by the activity of Aurora kinase B. In contrast, kinetochore binding of Spindly in the presence of taxol was weaker than in nocodazole-treated cells but sensitive to the kinase inhibitor, consistent with previous reports (Chan et al., 2009). Thus, the levels of Spindly are mainly regulated by microtubule attachment, but until tension starts, Aurora kinase B keeps some Spindly at the kinetochore.
Spindly Knockdown Causes Multiple Spindle Defects and Induces a MAD2-dependent Prometaphase Arrest
To analyze the function of Spindly in cell cycle progression, we performed siRNA transfection experiments in HeLa and U2OS cells expressing histone H2B-GFP. Spindly knockdown caused cells to accumulate in prometaphase (81.3% of mitotic cells), whereas only 15.4% were in metaphase (see below Figure 4C, Movies 1 and 2, and Supplemental Figure 4, A and B). When MAD2 was codepleted with Spindly, cells exited mitosis, demonstrating that Spindly knockdown caused a MAD2-dependent mitotic arrest (Supplemental Figure 4C).
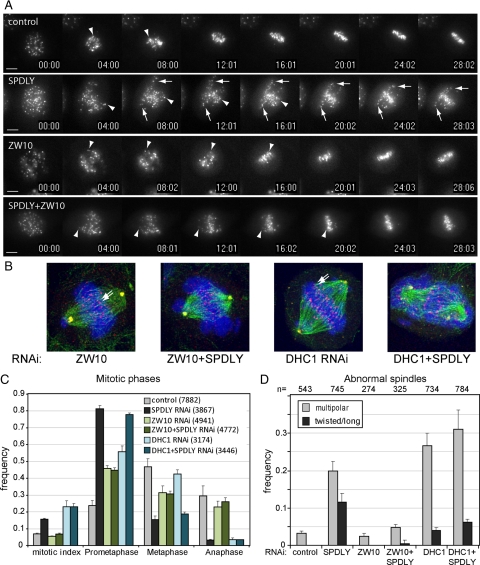
Figure 4.
Spindly RNAi effects depend on ZW10. (A) CenpB-YFP–expressing cells were either transfected with 50 nM control or Spindly, ZW10-specific siRNAs or a combination of both. Thirty-six hours after transfection mitotic cells were monitored using live cell fluorescence imaging, taking images every 30 s. Arrows, slowly congressing; arrowheads, fast congressing chromosomes. Scale bar, 10 μm.(B) HeLa cells transfected with siRNAs for Spindly, ZW10, or DHC, alone or in combination were fixed 48 h after transfection, stained, and imaged as in A. Shown are maximum intensity projections (MIPs) of representative ZW10, ZW10+Spindly, DHC, and DHC+Spindly siRNA-transfected arrested cells stained for tubulin (green), kinetochores (red), pericentrin (yellow), and DNA (blue). (C) Mitotic index and phases were determined by visual inspection, and classification of images of fixed cells stained as in (B) taken on an a Axiovert200M widefield microscope with a 40× Plan Neofluar0.75 NA objective in three independent experiments with total cell numbers indicated in parentheses. (D) The same images were analyzed for spindle morphology according to the defined main phenotypes (see text), and the frequency of multipolar and long or twisted spindles shown in relation to the total number of mitotic cells was analyzed.
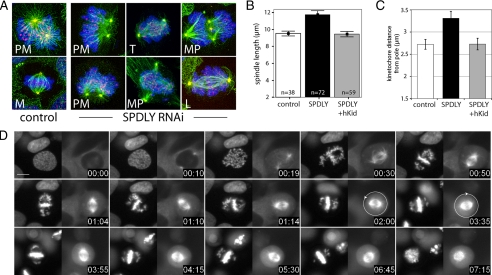
To determine the cause of this mitotic arrest, we first looked at spindle formation and morphology in Spindly RNAi-treated cells. As shown in Figure 3A, mitotic spindles of fixed HeLa Spindly knockdown cells exhibited various degrees of abnormal morphology. The mildest and most frequent form (81% in Spindly RNAi and 24% in controls; see Figure 4C) displayed nonaligned chromosomes similar to those seen in prometaphase of control cells. The second category encompassed multipolar spindles (MP) that were more frequent in Spindly RNAi cells (20%) than in controls (3%; see Figure 4D), whereas the strongest phenotype comprised spindles that were either very long (L), bent, or twisted (T) with their spindle poles (pericentrin signals) often displaced off the spindle axis (long and/or twisted spindles, 11.6% in Spindly knockdown, 0% in controls; see Figure 4D).
Figure 3.
Spindly RNAi affects spindle morphology. (A) HeLa cells transfected with Spindly siRNA for 36 h stained for DNA (blue), kinetochores (red), tubulin (green), and pericentrin (yellow). Images show maximum intensity projections (MIPs) of deconvoluted confocal z-stacks (step size, 120 nm). PM, prometaphase; M, metaphase; T, twisted spindle; MP, multipolar spindle; L, elongated spindle. (B) Spindle length in control, Spindly, and Spindly+hKid siRNA-transfected cells (48 h) was calculated as the distance between the pericentrin stained poles in z-stacks (mean, ± 95% confidence interval). (C) HeLa cells transfected with control, Spindly, and Spindly+hKid siRNA for 36 h before addition of 100 μM monastrol for 12 h. Cells were stained and imaged as in A, and average distances of kinetochores to the spindle pole in a total of 60 cells in three independent experiments per treatment were measured (mean ± SE). (D) U2OS H2B-GFP/Cherry-tubulin cells were transfected with Spindly siRNA for 36 h, and images were taken every 5 min over 10 h. Size bar, 10 μm. Time = h:min. Arrows indicate direction of spindle rotation.
On average, metaphase spindles were found to be longer (11.7 ± 1.95 μm) in Spindly RNAi-treated than in control cells (9.5 ± 0.88 μm, Figure 3B). To determine whether this elongation was due to cortical forces pulling at the spindle poles or pushing forces within the spindle, we performed double RNAi experiments to knockdown Spindly together with the chromokinesin hKid (KIF22), a kinesin that pushes spindle poles apart (Tokai-Nishizumi et al., 2005). As can be seen in Figure 3B, the length of the mitotic spindles was restored in double RNAi cells (9.4 ± 1.24 μm), suggesting that Spindly-dependent kinetochore dynein counteracts the polar ejection force of chromokinesins (Levesque and Compton, 2001). To test this antagonism between plus- and minus-end–directed motors in an independent setting, we treated HeLa cells again with monastrol to induce monopolar spindles in which chromosomes surround the spindle poles at a distance determined by the balance between these motor proteins. As can be seen in Figure 3C, knockdown of Spindly increased the average distance of kinetochores to the poles, whereas codepletion of hKid restored it to levels found in control cells. Thus, loss of Spindly function causes loss of a spindle contracting force, probably due to a loss of kinetochore dynein activity.
In the elongated spindles of Spindly knockdown cells, the interkinetochore distances of aligned chromosomes were found to be shorter (mean = 1.1 ± 0.2 μm, n = 28) than those of congressed chromosomes in control cells (1.3 ± 0.19 μm, n = 37), suggesting that stable end-on attachment of microtubules was not fully established. However, because the ultrastructure of kinetochores appeared normal in Spindly RNAi cells (Supplemental Figure 5) and because most spindles in RNAi-treated cells were of normal length, we concluded that the spindle elongation phenotype in Spindly knockdown cells was only transient and possibly was due to a combination of delayed microtubule attachment and a predominance of polar ejection forces exerted by chromokinesins.
To obtain more dynamic data about the roles of Spindly in establishing a bipolar spindle, we performed RNAi experiments in U2OS cells stably expressing histone H2B-GFP and mCherry-α-tubulin. Analysis of time-lapse recordings confirmed chromosome congression defects but also highlighted a spindle orientation defect in Spindly knockdown cells. As can be seen in Figure 3D (and in the Supplemental Movies 3 and 4), the mitotic spindle failed to orient properly and frequently started to rotate. However, a similar spindle rotation phenotype could also be observed in cells arrested in prometaphase by other means, e.g., knockdown of CENP-E and NUF2 (Movies 5 and 6; Yao et al., 2000; Liu et al., 2007), but not in cells arrested at metaphase by using the proteasome inhibitor MG132 or low doses of nocodazole (Movies 7 and 8). Thus, the spindle orientation defect seemed to be common in prometaphase-arrested cells and was not specific for the depletion of Spindly.
Spindly Is Required for the Poleward Movement of Chromosomes and for Their Rapid Attachment
The time-lapse recordings as well as the end-point analysis of mitotic Spindly RNAi cells revealed that, although the majority of chromosomes were found at the metaphase plate, some chromosomes were either strongly delayed or failed to congress at all. To be able to monitor chromosome congression more precisely, we generated U2OS cells whose kinetochores were labeled by CENP-B-YFP. As can be seen in Figure 4A (and Supplemental Movies 9–12), in control prometaphase cells, chromosomes rapidly moved toward the spindle equator (exemplary congressing chromosomes are marked by arrowheads). In Spindly knockdown cells, however, chromosomes located at the periphery of the cell (Figure 4A arrows point to noncongressing chromosomes) moved toward the spindle equator only slowly, if at all, and, thus, failed to align at the metaphase plate, whereas more centrally localized chromosomes aligned normally. Thus, loss of Spindly function did not prevent chromosome attachment, biorientation and congression per se but impaired the “capture” of peripheral chromosomes and their rapid integration into the mitotic spindle.
This failure is consistent with Spindly's role in recruiting dynein to the kinetochores, but previous studies on the function of the dynein-recruitment factor ZW10 or dynein itself (Zhu et al., 2005, 2007; Varma et al., 2008; Sivaram et al., 2009) have not reported a similarly strong chromosome congression defect as could be seen in Spindly knockdown cells. This discrepancy prompted us to perform “epistatic” RNAi experiments in U2OS-CENP-B-YFP and HeLa-H2B-GFP cells to examine the function of Spindly in the proposed RZZ-Spindly-dynein pathway. As can be seen in Figure 4A, in comparison to control U2OS-CENP-B-YFP cells, chromosome congression and metaphase plate formation were much more efficient in ZW10 RNAi-treated than in Spindly knockdown cells. Importantly, codepletion of ZW10 rescued the chromosome congression defect in Spindly RNAi cells.
Similar results were obtained in HeLa H2B-GFP cells as shown in Supplemental Figure 6, which shows selected time frames of time-lapse recordings of siRNA-transfected cells. As already mentioned above, chromosome congression was severely affected in Spindly knockdown cells but to a much lesser degree in either ZWINT-1 or ZW10 RNAi cells. Again, codepletion of ZW10 in Spindly RNAi cells ameliorated the chromosome congression defect. This difference in chromosome congression was also evident when cells were fixed 48 h after transfection and stained for tubulin, kinetochores, and DNA (Figures 3A and 4B). As can be seen in Figure 4C, the mitotic index was increased in Spindly- but not in ZW10-RNAi–treated cells, consistent with the known dependency of checkpoint signaling on ZW10 (Chan et al., 2000). The majority of mitotic Spindly RNAi cells was found to be in prometaphase (81.3%), whereas in ZW10 knockdown cells only 45.8% were in prometaphase but 31.4% in metaphase.
When the dynein motor complex was targeted by RNAi against its catalytic DHC1 subunit, the mitotic arrest phenotypes were more complex, consistent with the many functions of dynein in mitosis. Metaphase plate formation was, however, more efficient (42.4%) than in Spindly single (15.4%) or DHC+Spindly double RNAi cells (18.8%; Figure 4, B and C; Supplemental Figure 6), but whether DHC1 depletion was complete in these experiments was difficult to assess. Similar to the knockdown of Spindly, DHC RNAi caused cells to arrest in mitosis with a high frequency of cells with multipolar spindles (26.6%). In contrast, spindle abnormalities were less frequent in ZW10 RNAi cells (Figure 4D). The high frequency of elongated spindles in Spindly knockdown cells was, however, not seen in ZW10- or DHC-RNAi–treated cells.
Although chromosomes congressed more rapidly in either ZW10 or DHC1 than in Spindly-RNAi cells, the metaphase plates appeared more irregular than in control cells and contained erroneously aligned chromosomes, such as syntelically attached chromosomes (arrows in Figure 4B). These data indicate that Spindly not only is a kinetochore recruitment factor for dynein but that it may have an additional, RZZ-dependent function in regulating chromosome attachment important for the formation of a correct metaphase plate.
Spindly Is Required for ZW10 Turnover But Not for MAD2 Removal from Kinetochores of Bioriented Chromosomes
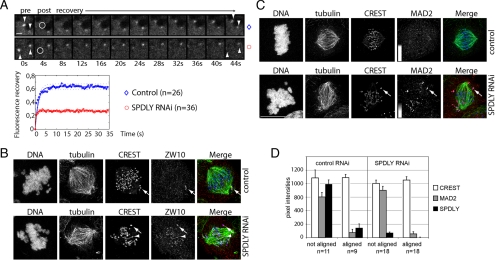
To further investigate how Spindly might regulate the RZZ complex, we first examined whether Spindly controlled ZW10 levels at kinetochores by establishing and analyzing a U2OS cell line expressing YFP-ZW10. Like the endogenous protein, the tagged version of ZW10 strongly localized to kinetochores during prometaphase and gradually declined during chromosome congression to accumulate at the spindle poles. As reported previously for MAD2 (Howell et al., 2004), intense fibrillar signals could occasionally be seen moving toward the spindle pole (Supplemental Figure 7; Movie 13), which could be due to a sudden removal of larger amounts of YFP-ZW10 from kinetochores, e.g., in response to rapid biorientation.
Next, we performed photobleaching experiments on one of the paired YFP-ZW10 kinetochore signals (Figure 5A, arrowheads), which revealed that YFP-ZW10 recovery was reduced from 62% in controls to 27% in Spindly knockdown cells, indicating that kinetochores in Spindly-depleted cells contained a higher fraction of stably bound ZW10. Consistent with these dynamic measurements, we found higher ZW10 signals on kinetochores of congressed chromosomes in Spindly knockdown than in control cells (arrowhead, Figure 5B) as determined by immunofluorescence staining.
Figure 5.
Spindly depletion affects ZW10 but not MAD2. (A) Live cell confocal imaging of YFP-ZW10 expressing U2OS cell. Arrowheads point to paired YFP-ZW10 signals at kinetochores. The circle indicates the bleaching area. Scale bar, 1 μm. Fluorescence intensities were recorded on 26 kinetochores in control and 36 in Spindly RNAi cells. The mean values of double normalization pixel intensities were plotted as a function of time. (B) HeLa cells were transfected with control and Spindly siRNAs for 48 h and stained for DNA (Hoechst), kinetochores (CREST, blue), microtubules (tubulin, green), and ZW10 (red) and imaged on a Leica SP5 microscope. Arrow, a kinetochore of a noncongressed; arrowhead, a kinetochore of a congressed chromosome. (C) HeLa cells were transfected as in B and stained for DNA, microtubules, kinetochores, and MAD2. Top panel, a control metaphase cell. MAD2 signals were contrast enhanced as indicated by the gray scale bar. Bottom panel, arrows point to an unattached kinetochore with crescent MAD2 staining. Scale bar, 10 μm. (D) Control and Spindly RNAi HeLa cells stained for MAD2, Spindly, kinetochores, and DNA were imaged on a Zeiss LSM510 microscope. Background-corrected kinetochore signals of aligned and not aligned chromosomes in control and Spindly RNAi cells (mean ± SE).
Because dynein has been shown to control MAD2 levels, we stained control and Spindly knockdown cells for MAD2. In both treatments, MAD2-staining was most intense on unattached kinetochores (Figure 5C, arrow), weaker on attached and low on kinetochores of congressed, bioriented chromosomes (Figure 5C, arrowhead, and quantification of MAD2 signals on kinetochores of aligned vs. nonaligned chromosomes in D). Unlike in control cells, however, MAD2 did not localize to spindle poles in Spindly RNAi cells, demonstrating that dynein-dependent transport was effectively blocked. Thus, consistent with previous reports (Chan et al., 2009), knockdown of human Spindly did not affect MAD2 levels on kinetochores of bioriented chromosomes, unlike what has been observed in Drosophila cells depleted of Spindly, in which kinetochores of aligned chromosomes stained brightly for Mad2 (Griffis et al., 2007). In summary, these loss-of-function RNAi experiments suggest that human Spindly controls the steady state kinetochore levels of ZW10 but not those of MAD2, confirming the hypothesis that Spindly might function as a specific adaptor between the RZZ and dynein motor complexes.
The Kinetochore Localization of Spindly Requires Multiple Domains But Is Independent of S515 Phosphorylation
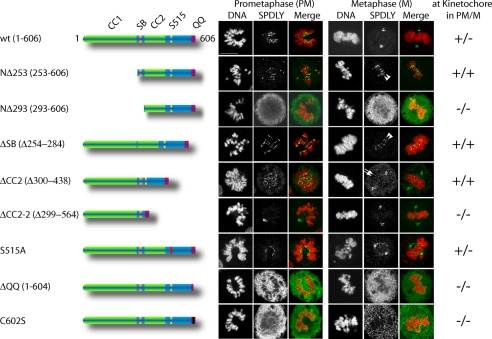
Spindly contains two putative coiled-coil domains, separated by the conserved Spindly box (Griffis et al., 2007), and a feature-less C-terminal third of the protein. To define domains important for kinetochore localization and protein function, we constructed N- and C-terminal deletion mutants and expressed them as N-terminally FLAG-tagged proteins in HeLa cells. As shown in Figure 6, a mutant lacking the N-terminal 253-amino acid residues (including the first coiled-coil domain, NΔ253) still localized to kinetochores, whereas a slightly larger deletion mutant (NΔ293) did not, indicating a critical role of the Spindly box (residues 254-284) for kinetochore localization. Surprisingly, however, deletion of the Spindly box alone (ΔSB) had no effect on kinetochore localization. In contrast to the wild-type protein, however, NΔ253 and ΔSB failed to translocate to the spindle poles and remained on kinetochores of congressed chromosomes at metaphase (Figure 6, arrowheads). Thus, the N-terminal coiled-coil domain and the Spindly box contribute to kinetochore localization and are required for Spindly displacement from kinetochores in response to microtubule attachment.
Figure 6.
Identification of Spindly domains required for kinetochore binding. HeLa cells were transfected with expression plasmids for FLAG-epitope–tagged wild-type and Spindly mutants for 24 h, methanol-fixed, and stained for DNA (Hoechst, red) and for overexpressed Spindly using anti-FLAG-antibody (SPDLY, green). Images were taken on a Leica SP5 confocal microscope using a 63× HCX PL APO lambda blue, 1.4 NA objective. Size bar, 10 μm.
Deletion of the second coiled-coil domain (ΔCC2, 299-438) did not interfere with the localization of Spindly but, in contrast to the wild-type, slightly higher levels were retained on kinetochores of aligned chromosomes (arrows in Figure 6), suggesting that streaming was delayed. A larger deletion in the C-terminal part of the protein (ΔCC2-2, 299-564) impaired kinetochore localization but not association with the mitotic spindle and its poles. This suggested the presence of a critical kinetochore binding domain between amino acid residues 440 and 564, encompassing the mitotic phosphorylation site. However, no effect on kinetochore localization could be observed by analyzing the nonphosphorylatable (S515A, Figure 6) and phosphomimetic (S515E, data not shown) Spindly mutants.
The C-terminal region was found to be less tolerant to modifications than the N-terminal domain, because deletion of the two C-terminal glutamines (ΔQQ), addition of a single Asp residue (not shown), or mutation of the cysteine residue at position 602 into a serine, abrogated kinetochore binding (Figure 6). Thus, the C-terminal region is required but not sufficient for the localization of Spindly to kinetochores. In summary, the localization of Spindly to the kinetochore seems to depend on multiple domains, which suggests that Spindly might interact with more than one protein, whose identities remain to be discovered.
Dominant Negative Spindly Prevents the Removal of ZW10 and Blocks Cells at Metaphase
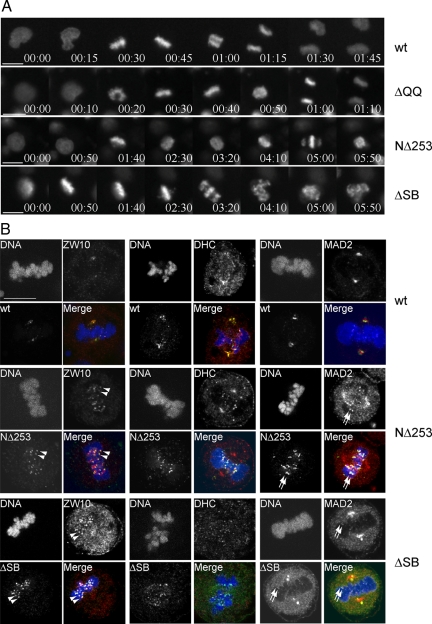
The above described RNAi experiments confirmed the role of Spindly in recruiting dynein to kinetochores which is important for chromosome movement and congression but failed to confirm dynein's function in mitotic checkpoint silencing (Howell et al., 2001). Because our domain mapping experiments revealed that Spindly NΔ253 and ΔSB remained kinetochore-bound even after microtubule attachment and biorientation (see Figure 6), we hypothesized that these mutants might act as dominant negative inhibitors of kinetochore protein streaming.
To investigate Spindly's role in mitotic checkpoint silencing, we performed live cell imaging experiments in HeLa-H2B-GFP cells transfected with Spindly expression plasmids together with an RFP-expression tracer plasmid. When RFP-positive cells were followed by live cell imaging, 51% (n = 31) and 27% (n = 26) of RFP-positive cells transfected with NΔ253 and ΔSB expression plasmids, respectively, became arrested in mitosis. In contrast, only 1 of 29 RFP-positive cells transfected with wild type and none of 16 ΔQQ Spindly-transfected cells arrested in mitosis. As can be seen in Figure 7A (Movies 14–17), NΔ253- and ΔSB-transfected cells arrested in mitosis with aligned chromosomes. Although chromosome congression was also impaired in NΔ253-transfected cells, all cells that were arrested by ΔSB expression showed normal chromosome congression to the metaphase plate but then arrested at metaphase for prolonged periods of time before their chromosomes became scattered (Figure 7A; Movie 17). Thus, in contrast to Spindly depletion, overexpression of the Spindly box deletion mutant did not interfere with chromosome congression but delayed the onset of anaphase.
Figure 7.
Removal of Spindly is required for silencing of the mitotic checkpoint. (A) HeLa H2B-GFP–expressing cells were transfected with expression plasmids for FLAG-epitope–tagged wild-type, ΔQQ, NΔ253, and ΔSB Spindly along with an RFP expression vector for 36 h. Red fluorescent cells were monitored for progression through mitosis by live cell imaging. Selected image frames of Movies 14–17 are shown. Time = h:min. Scale bar, 10 μm. (B) HeLa cells transfected for 24 h with FLAG-epitope–tagged wild-type, NΔ253 and ΔSB Spindly were methanol-fixed and stained using anti-FLAG antibody (Spindly, green) antibodies specific for ZW10, DHC1, or MAD2 (red), and Hoechst dye (DNA, blue). Arrowheads, kinetochores of aligned chromosomes positive for Spindly mutants and ZW10; arrows, colocalization of Spindly mutants and MAD2. Scale bar, 10 μm.
When transfected cells were stained for FLAG-tagged Spindly proteins and either ZW10, MAD2, or DHC1 (Figure 7B), it became evident that ZW10 (arrowheads), and to a lesser extent dynein and MAD2 (arrows), were still readily detectable on kinetochores of congressed chromosomes in cells expressing ΔN253 or ΔSB but not in cells expressing the wild-type protein or the non-kinetochore-binding mutant ΔQQ. The interkinetochore distance of congressed chromosomes in cells expressing either ΔN253, ΔSB, wild-type Spindly, or ΔQQ were all similar (1.36 ± 0.15 μm, n = 47; 1.4 ± 0.13 μm, n = 17; 1.33 ± 0.18 μm, n = 29; 1.32 ± 0.14 μm, n = 16) and wider than those of nonaligned chromosomes (0.91 ± 0.1 μm; n = 18). We concluded from this analysis that expression of ΔN253 and ΔSB blocked SAC inactivation in response to microtubule attachment and biorientation, which confirmed the essential role of dynein-dependent kinetochore streaming for SAC inactivation but also highlighted an important function of Spindly in the regulation of the RZZ complex.
DISCUSSION
Spindly proteins are conserved in metazoans but, similar to the members of the RZZ complex (Karess, 2005), have not been identified in yeasts and higher plants. Spindly and the RZZ complex are linked to functions of cytoplasmic dynein and thus, may have evolved along with the specialized functions of dynein at kinetochores in animal organisms with an open mitosis. Here, we provide evidence that human Spindly is not simply required for recruiting dynein to kinetochores but that it serves as an adaptor between the RZZ and the dynein complex, which is important for chromosome alignment and mitotic checkpoint signaling.
Spindly Associates with the RZZ Complex
By using immunoaffinity purification and mass spectrometry, we showed that human Spindly interacted with the RZZ complex. This interaction was unstable in the presence of high salt (>140 mM) and low amounts of detergent (0.1% NP40). A similarly weak interaction between the RZZ complex and Spindly was reported for worm SPDL-1 (Gassmann et al., 2008), suggesting that Spindly proteins only weakly interact with the RZZ complex at least when purified from mitotically arrested nocodazole-treated cells.
The sensitivity to salt suggests that Spindly might use ionic interactions for binding to the RZZ complex and that these interactions could be regulated by protein phosphorylation. Although we found Spindly to be phosphorylated on serine 515, a CDK substrate in vitro and major phosphorylation site in mitosis, this modification did not control the localization of Spindly to kinetochores but additional possible functions of this phosphorylation, e.g., in regulating ZW10 function, remain to be investigated.
The high sensitivity of the Spindly–RZZ interaction to detergent hints to a contribution of hydrophobic residues in these contacts. Our domain mapping approach revealed that the C-terminal amino acid residues of Spindly, which are conserved among vertebrates, are essential for kinetochore localization. The C-terminal CPQQ sequence is remotely similar to the CAAX farnesylation consensus motif and the deleterious effect of the cysteine 602 mutation to serine on Spindly localization points to an essential function of the sulfhydryl group, raising the possibility that Spindly might be a potential substrate for farnesyltransferases, as suggested recently (Hougland et al., 2009). Farnesylation is not only involved in targeting proteins to cellular membranes but also in protein–protein interactions and has been shown to be essential for the kinetochore localization of CENP-E and CENP-F (Ashar et al., 2000). Whether Spindly is indeed farnesylated remains to be determined, but protein lipidation could explain the high detergent sensitivity of the protein–protein interactions between Spindly and the RZZ complex. In summary, the weak interaction of Spindly with the RZZ complex suggests that Spindly is not a core component of the RZZ complex but may function as a regulator of the RZZ complex, e.g., by serving as an adaptor between RZZ and dynein.
Spindly Is Essential for Dynein-dependent and-independent Functions at the Kinetochore
The knockdown of Spindly in human cells caused a MAD2-dependent delay in progression through mitosis, which was characterized by a chromosome congression defect as well as spindle abnormalities. When chromosome movements were analyzed in Spindly knockdown cells, we observed that mainly peripheral chromosomes were affected, whereas the chromosomes more close to the center of the cell rapidly bioriented. The failure of peripheral chromosomes to become rapidly integrated into the mitotic spindle can be explained by their reduced chance to engage with microtubules. This chance might further be reduced by the unopposed polar ejection forces of chromokinesins in cells lacking kinetochore dynein. This selective effect on peripheral chromosomes emphasizes the importance of dynein-mediated poleward chromosome movement in establishing stable microtubule binding to kinetochores. Once established, however, microtubule attachment is normal (as determined by electron microscopy), stable (as determined by the resistance to cold treatment; unpublished data and Chan et al., 2009), and strong enough to transform pulling forces into interkinetochore tension in Spindly RNAi cells.
Surprisingly, however, the strength of this phenotype could not be explained by the loss of dynein function alone, because when the upstream components of the proposed ZWINT-RZZ-Spindly-dynein pathway, ZWINT1 and ZW10, were knocked down, chromosome congression was not severely impaired. More impressively even, the chromosome congression defect of Spindly RNAi cells could be rescued by codepletion of ZW10. This suggests a direct role of Spindly in the early steps of microtubule attachment, e.g., by regulating an RZZ function in microtubule attachment, as proposed for nematode Spindly by Gassmann et al. (2008).
In addition to the chromosome congression defect, malformed spindles were another hallmark of Spindly knockdown cells. Some of the spindle defects, however, may be nonspecific and may be due to a mechanical damage induced by the rotation of the mitotic spindle. The multipolar spindles, for example, were most likely caused by disintegration of the spindle poles during mitosis, because Spindly RNAi cells did not exhibit supernumerary centrosomes in interphase cells (data not shown). In many cells the spindle poles were dislocated off the main axis of the spindle, as if they were displaced by pulling forces, e.g., by increased cortical dynein activity as suggested by Chan et al. (2009). In contrast, the elongated spindles that also displayed signs of reduced interkinetochore tension might be specific to a loss of Spindly function in microtubule attachment as discussed below.
On kinetochores, the function of dynein seems to depend on the mode of microtubule attachment. During early mitosis, when kinetochores are laterally attached, dynein is a resident kinetochore protein, able to move chromosomes along microtubules toward the spindle poles. On stable attachment, a switch occurs that allows dynein to leave the kinetochore to transport proteins, such as ZW10 and MAD2, from the kinetochore to the spindle poles, as has been shown in a variety of experimental systems (Howell et al., 2001; Wojcik et al., 2001; Basto et al., 2004; Griffis et al., 2007; Mische et al., 2008; Varma et al., 2008; Sivaram et al., 2009).
The dynamics of Spindly localization during progression through mitosis is very similar to that of ZW10 but different from MAD2. All three proteins show high levels on unattached kinetochores and are absent on bioriented chromosomes. In contrast to MAD2, whose kinetochores localization only responds to microtubule attachment (Waters et al., 1998), the localization of ZW10 and Spindly is sensitive to tension as well (Famulski and Chan, 2007; Chan et al., 2009; this study). Because Spindly controls the turnover rate of ZW10, this suggests that Spindly might specifically connect the RZZ complex to the dynein motor. MAD2, on the other hand, might be removed via DLIC1 (Sivaram et al., 2009), which would allow it to be controlled independently of ZW10. Consistent with the role of Spindly as a ZW10-specific adaptor, depletion of Spindly maintained residual levels of ZW10 (but not MAD2) on bioriented chromosomes. How this differential release of MAD2 and ZW10 is regulated is currently unknown but could occur at the level of kinetochore-binding strength, interaction with the dynein motor, or regulation of dynein motility. Our findings argue that the switch of dynein from a resident to a mobile protein complex might be controlled by the binding affinity of its anchoring proteins, i.e., the RZZ-Spindly complex, to the kinetochores and weakening of this binding, e.g., in response to microtubule attachment in a Spindly-dependent manner, might release dynein from kinetochores.
Spindly Couples Attachment to Checkpoint Silencing
Spindly binding to kinetochores requires the RZZ complex, but how these interactions take place is currently unknown. By using a domain-mapping approach, we could show that Spindly binds to its kinetochore partner proteins in a complex manner. Spindly localization to the kinetochores requires an intact C-terminus, whereas the coiled-coil domains are not essential for localization. The conserved Spindly box, which is only essential for kinetochore binding when deleted together with the N-terminal coiled-coil domain, seems to have an important function in the regulation of ZW10 at kinetochores (see below). Similarly, by deleting 126 additional residues downstream of the second coiled-coil domain, kinetochore localization was lost but not its ability to localize to the spindle poles. Thus, Spindly binding to kinetochores involves contacts made by the N-terminal domain containing a coiled-coil and the Spindly box, as well as by two domains in the C-terminal half of the protein.
Surprisingly, expression of NΔ253 and ΔSB blocked progression through mitosis by inhibiting the onset of anaphase. Both mutants colocalized with ZW10 and partly with dynein and MAD2 at kinetochores of aligned chromosomes, suggesting that they acted as dominant negative proteins on dynein-dependent kinetochore stripping in response to microtubule attachment and biorientation. Thus, in contrast to the RNAi-induced loss of function experiments, the presence of the dominant negative Spindly mutants also maintained MAD2 at kinetochores. Human Spindly might, therefore, be involved in the regulation of the binding affinity of kinetochore proteins to the MAD1/MAD2 complex.
Although Spindly mutants lacking N-terminal domains remained kinetochore bound, a mutant lacking the C-terminal half of the protein (ΔCC2-2) failed to localize to kinetochores but localized to the mitotic spindle and the spindle poles, suggesting that it was still able to associate with dynein. Thus, the C-terminal part seems to interact with kinetochore proteins (most likely a member of the RZZ complex), whereas the N-terminal domain, in addition to its contribution to kinetochore binding, might also interact with a component of the dynein motor, but confirmation of this interaction is still lacking.
Because overexpression of full-length Spindly did not block the release of ZW10 from kinetochores, these results suggest that Spindly might exist in different conformations, one of which is required for RZZ removal from kinetochores. Although speculative, the function of the Spindly box could be to control this transition, e.g., in response to microtubule attachment, via interactions with other kinetochore proteins. This transition, in turn, might control the function of the RZZ complex in regulating microtubule attachment and in mitotic checkpoint signaling.
In contrast to the role of Spindly in dynein-dependent processes, such as poleward chromosome movement and kinetochore stripping, its function in microtubule attachment remains obscure. In Spindly-depleted cells, peripheral chromosomes have a severe delay in becoming bioriented. Because ZW10 depletion has a milder chromosome congression phenotype than Spindly RNAi in worms (Gassmann et al., 2008) and human cells, this suggests that the RZZ complex can delay the formation of microtubule attachments. The removal of the RZZ complex is, however, not required for microtubule attachment because blocking kinetochore streaming by expression of dominant negative Spindly ΔSB did not interfere with attachment or chromosome congression. However, by blocking RZZ removal from kinetochores by expression of NΔ253 or ΔSB, SAC inactivation after successful biorientation was effectively blocked. Thus, these Spindly mutants dissected the function of the RZZ complex by locking it in an inactive state with respect to its role in microtubule attachment but kept it active with respect to checkpoint signaling.
Spindly is required for dynein recruitment to the kinetochores, but evidence for a physical interaction with dynein or dynactin complexes is still lacking. It remains, therefore, a possibility that Spindly exerts its function on dynein indirectly, e.g., by controlling the conformation and function of the RZZ complex. Spindly could function as a relay that controls RZZ functions in response to the microtubule-binding state of kinetochores. In response to dynein-mediated lateral attachments, Spindly could inactivate the RZZ complex to promote stable NDC80-dependent microtubule attachments. After the formation of these stable attachments, Spindly might then loosen the binding of the RZZ complex to kinetochores to allow it to be removed by dynein to dampen the signaling strength of the mitotic checkpoint. This process could favor the establishment of initial monopolar attachments over syntelic or merotelic ones to prevent chromosome segregation errors and aneuploidy. In summary, the main function of human Spindly is to control the kinetochore levels and function of the RZZ complex, which is important for chromosome congression and mitotic checkpoint signaling.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Sigl, K. Gutleben, H.-L. Ebner, and M. Offterdinger for help; A. Helmberg for reading the manuscript, and W. Sachsenmaier and R. Kofler for support. This work was supported by Austrian Science Fund (FWF) Grant SFB021 “Cell proliferation and cell death in tumors” to S.G. and G.B., FWF Grant P16400, FWF-DK-W11 “MCBO,” EU-Grant LSHS-CT-2004-503438 “TRANSFOG” to S.G., and FWF-Grant P19486-B12 to M.H. M.B. was supported by a fellowship of the Österreichische Krebshilfe/Krebshilfe Tirol. This work was further supported by additional grants of the Tiroler Zukunftssiftung and Tiroler Wissenschaftsfonds (TWF) and by means from the assets of Eva v. Lachmüller, Brixen, Italy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0356) on April 28, 2010.
REFERENCES
- Ashar H. R., James L., Gray K., Carr D., Black S., Armstrong L., Bishop W. R., Kirschmeier P. Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J. Biol. Chem. 2000;275:30451–30457. doi: 10.1074/jbc.M003469200. [DOI] [PubMed] [Google Scholar]
- Basto R., Scaerou F., Mische S., Wojcik E., Lefebvre C., Gomes R., Hays T., Karess R. In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr. Biol. 2004;14:56–61. doi: 10.1016/j.cub.2003.12.025. [DOI] [PubMed] [Google Scholar]
- Busson S., Dujardin D., Moreau A., Dompierre J., De M., Jr Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 1998;8:541–544. doi: 10.1016/s0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- Carminati J. L., Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G. K., Jablonski S. A., Starr D. A., Goldberg M. L., Yen T. J. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- Chan Y. W., Fava L. L., Uldschmid A., Schmitz M. H., Gerlich D. W., Nigg E. A., Santamaria A. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 2009;185:859–874. doi: 10.1083/jcb.200812167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V. L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S. S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V. M., Shapiro I., Aldridge B., Sorger P. K. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. EMBO J. 2006;25:2814–2827. doi: 10.1038/sj.emboj.7601168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C. J., Paschal B. M., Vaughan K. T., Vallee R. B. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski J. K., Chan G. K. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr. Biol. 2007;17:2143–2149. doi: 10.1016/j.cub.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Famulski J. K., Vos L., Sun X., Chan G. Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint. J. Cell Biol. 2008;180:507–520. doi: 10.1083/jcb.200708021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner N. E., Dujardin D. L., Tai C. Y., Vaughan K. T., O'Connell C. B., Wang Y., Vallee R. B. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Gaglio T., Saredi A., Bingham J. B., Hasbani M. J., Gill S. R., Schroer T. A., Compton D. A. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J. Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22:2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatlin J. C., Bloom K. Microtubule motors in eukaryotic spindle assembly and maintenance. Semin. Cell Dev. Biol. 2010;21:248–254. doi: 10.1016/j.semcdb.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S., Kramer E., Gieffers C., Gannon J., Peters J. M., Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E. R., Stuurman N., Vale R. D. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J., Elbashir S. M., Bechert K., Tuschl T., Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hellman U. Sample preparation by SDS/PAGE and in-gel digestion. EXS. 2000;88:43–54. doi: 10.1007/978-3-0348-8458-7_3. [DOI] [PubMed] [Google Scholar]
- Hoffman D. B., Pearson C. G., Yen T. J., Howell B. J., Salmon E. D. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougland J. L., Lamphear C. L., Scott S. A., Gibbs R. A., Fierke C. A. Context-dependent substrate recognition by protein farnesyltransferase. Biochemistry. 2009;48:1691–1701. doi: 10.1021/bi801710g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., McEwen B. F., Canman J. C., Hoffman D. B., Farrar E. M., Rieder C. L., Salmon E. D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., Salmon E. D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Kardon J. R., Vale R. D. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 2005;15:386–392. doi: 10.1016/j.tcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Karki S., Holzbaur E. L. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol. 1999;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- King J. M., Hays T. S., Nicklas R. B. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J. Cell Biol. 2000;151:739–748. doi: 10.1083/jcb.151.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteom. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Kops G. J., Kim Y., Weaver B. A., Mao Y., McLeod I., Yates J. R., III, Tagaya M., Cleveland D. W. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque A. A., Compton D. A. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 2001;154:1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Y., Yeh E., Hays T., Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., et al. Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J. Biol. Chem. 2007;282:21415–21424. doi: 10.1074/jbc.M609026200. [DOI] [PubMed] [Google Scholar]
- Maresca T. J., Salmon E. D. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V. M., Nigg E. A. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- McGuinness B. E., Hirota T., Kudo N. R., Peters J. M., Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS. Biol. 2005;3 doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian K. A., Maier K. C., Godfrey J. E., Rodgers M., Schroer T. A. Mechanism of dynamitin-mediated disruption of dynactin. J. Biol. Chem. 2007;282:19355–19364. doi: 10.1074/jbc.M700003200. [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J. D., Cleveland D. W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Mische S., He Y., Ma L., Li M., Serr M., Hays T. S. Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation. Mol. Biol. Cell. 2008;19:4918–4929. doi: 10.1091/mbc.E08-05-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E. D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D., Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- O'Connell C. B., Wang Y. L. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol. Biol. Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr C. M., Coue M., Grissom P. M., Hays T. S., Porter M. E., McIntosh J. R. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature. 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Alexander S. P. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A., Waters J. C., Mitchison T. J. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaerou F., Starr D. A., Piano F., Papoulas O., Karess R. E., Goldberg M. L. The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J. Cell Sci. 2001;114:3103–3114. doi: 10.1242/jcs.114.17.3103. [DOI] [PubMed] [Google Scholar]
- Schmitz J., Watrin E., Lenart P., Mechtler K., Peters J. M. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr. Biol. 2007;17:630–636. doi: 10.1016/j.cub.2007.02.029. [DOI] [PubMed] [Google Scholar]
- Schroer T. A. Dynactin. Annu. Rev. Cell Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Sharp D. J., Rogers G. C., Scholey J. M. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- Sivaram M. V., Wadzinski T. L., Redick S. D., Manna T., Doxsey S. J. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J. 2009;28:902–914. doi: 10.1038/emboj.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A., Saffery R., Li Z., Simpson A. E., Choo K. H., Yen T. J., Goldberg M. L. HZwint-1, a novel human kinetochore component that interacts with HZW10. J. Cell Sci. 2000;113(Pt 11):1939–1950. doi: 10.1242/jcs.113.11.1939. [DOI] [PubMed] [Google Scholar]
- Starr D. A., Williams B. C., Hays T. S., Goldberg M. L. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehman S. A., Chen Y., McKenney R. J., Vallee R. B. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J. Cell Biol. 2007;178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer E. R., Wordeman L., Schroer T. A., Sheetz M. P. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature. 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Tai C. Y., Dujardin D. L., Faulkner N. E., Vallee R. B. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanudji M., Shoemaker J., L'Italien L., Russell L., Chin G., Schebye X. M. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell. 2004;15:3771–3781. doi: 10.1091/mbc.E03-07-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokai-Nishizumi N., Ohsugi M., Suzuki E., Yamamoto T. The chromokinesin Kid is required for maintenance of proper metaphase spindle size. Mol. Biol. Cell. 2005;16:5455–5463. doi: 10.1091/mbc.E05-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K. S., Takagaki K., Kumada K., Hirayama Y., Noda T., Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg E .A., Koonce M. P., McIntosh J. R. Cytoplasmic dynein plays a role in mammalian mitotic spindle formation. J. Cell Biol. 1993;123:849–858. doi: 10.1083/jcb.123.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Monzo P., Stehman S. A., Vallee R. B. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J. Cell Biol. 2008;182:1045–1054. doi: 10.1083/jcb.200710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorozhko V. V., Emanuele M. J., Kallio M. J., Stukenberg P. T., Gorbsky G. J. Multiple mechanisms of chromosome movement in vertebrate cells mediated through the Ndc80 complex and dynein/dynactin. Chromosoma. 2008;117:169–179. doi: 10.1007/s00412-007-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C. E., Heald R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Waters J. C., Chen R. H., Murray A. W., Salmon E. D. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Skibbens R. V., Salmon E. D. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J. Cell Sci. 1996;109(Pt 12):2823–2831. doi: 10.1242/jcs.109.12.2823. [DOI] [PubMed] [Google Scholar]
- Whitfield M. L., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J., et al. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J. Cell Biol. 2008;183:819–834. doi: 10.1083/jcb.200804114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. C., Karr T. L., Montgomery J. M., Goldberg M. L. The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J. Cell Biol. 1992;118:759–773. doi: 10.1083/jcb.118.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaerou F., Karess R., Hays T. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 2001;3:1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- Wolf F., Wandke C., Isenberg N., Geley S. Dose-dependent effects of stable cyclin B1 on progression through mitosis in human cells. EMBO J. 2006;25:2802–2813. doi: 10.1038/sj.emboj.7601163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L., Steuer E. R., Sheetz M. P., Mitchison T. Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes. J. Cell Biol. 1991;114:285–294. doi: 10.1083/jcb.114.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T. G., Watanabe S., Essex A., Kitagawa R. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J. Cell Biol. 2008;183:187–194. doi: 10.1083/jcb.200805185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tulu U. S., Wadsworth P., Rieder C. L. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Abrieu A., Zheng Y., Sullivan K. F., Cleveland D. W. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J. D., Bossy-Wetzel E., Fan J. B., Abraham R. T., Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.