Bim1 promotes microtubule assembly in vitro, primarily by decreasing the frequency of catastrophes. In contrast, Bik1 inhibits microtubule assembly by slowing growth and, consequently, promoting catastrophes. These proteins interact to form a complex that affects microtubule dynamics in much the same way as Bim1 alone.
Abstract
Microtubule dynamics are regulated by plus-end tracking proteins (+TIPs), which bind microtubule ends and influence their polymerization properties. In addition to binding microtubules, most +TIPs physically associate with other +TIPs, creating a complex web of interactions. To fully understand how +TIPs regulate microtubule dynamics, it is essential to know the intrinsic biochemical activities of each +TIP and how +TIP interactions affect these activities. Here, we describe the activities of Bim1 and Bik1, two +TIP proteins from budding yeast and members of the EB1 and CLIP-170 families, respectively. We find that purified Bim1 and Bik1 form homodimers that interact with each other to form a tetramer. Bim1 binds along the microtubule lattice but with highest affinity for the microtubule end; however, Bik1 requires Bim1 for localization to the microtubule lattice and end. In vitro microtubule polymerization assays show that Bim1 promotes microtubule assembly, primarily by decreasing the frequency of catastrophes. In contrast, Bik1 inhibits microtubule assembly by slowing growth and, consequently, promoting catastrophes. Interestingly, the Bim1-Bik1 complex affects microtubule dynamics in much the same way as Bim1 alone. These studies reveal new activities for EB1 and CLIP-170 family members and demonstrate how interactions between two +TIP proteins influence their activities.
INTRODUCTION
The microtubule cytoskeleton is essential for a variety of cellular processes that influence cell shape and organization, as well as chromosome segregation during mitosis. In most dividing cells, polarized microtubule arrays are arranged with their minus ends located at the microtubule organizing center, whereas their plus ends extend out in the cytoplasm. Microtubule plus ends alternate rapidly between states of polymerization and depolymerization in a process known as dynamic instability (Desai and Mitchison, 1997). This process is central to the biological function of microtubules, allowing them to probe the cell for specific targets such as kinetochores and cortical sites. A central question in biology is how the dynamics of microtubule plus ends are precisely regulated to achieve the correct configuration of microtubule arrays.
Microtubule dynamics are regulated, in large part, by a group of proteins known as plus end tracking proteins (+TIPs) because they associate with growing microtubule plus ends (Schuyler and Pellman, 2001; Lansbergen and Akhmanova, 2006; Howard and Hyman, 2007; Akhmanova and Steinmetz, 2008). A number of +TIPs families have been identified and these are evolutionarily conserved from yeast to humans. Interestingly, most +TIPs have the ability to physically associate with a number of other +TIPs, creating a complex web of interactions (Akhmanova and Hoogenraad, 2005; Akhmanova and Steinmetz, 2008). These interactions likely play important roles in integrating +TIP activities at the microtubule plus end.
A complete understanding of how +TIPs regulate microtubule dynamics will require knowledge of the intrinsic biochemical activities of each +TIP and how +TIP interactions affect these activities. +TIPs can influence microtubule turnover through a variety of methods, such as altering the rate of polymerization or depolymerization, or the frequency of transitions between assembly and disassembly. However, for most +TIPs, the mechanisms by which they exert their influence are not yet clear. This information is difficult to obtain from in vivo loss-of-function (mutation or depletion) experiments, because loss of a targeted +TIP may decrease the activity of other +TIPs that rely on it for localization or increase the activity of other +TIPs that compete with it for access to microtubule plus ends. Therefore, deciphering +TIP activities through in vitro experiments is essential to understanding their roles in controlling microtubule dynamics.
In this article, we focus on two +TIPs from the budding yeast Saccharomyces cerevisiae, a model organism that has been used extensively to elucidate the molecular mechanisms that govern chromosome segregation. Bim1 and Bik1 are members of the EB1 and CLIP-170 +TIP families, respectively. Both proteins are present on astral and spindle microtubules and play roles in spindle orientation and chromosome segregation (Berlin et al., 1990; Schwartz et al., 1997; Tirnauer et al., 1999; Hwang et al., 2003; Miller et al., 2006; Wolyniak et al., 2006; Gardner et al., 2008; Zimniak et al., 2009). Here, we describe the activities of Bim1 and Bik1 on microtubule dynamics in vitro and compare these activities to those of the human EB1 and CLIP-170 proteins. We also show that these proteins, like EB1 and CLIP-170 (Honnappa et al., 2006; Bieling et al., 2008), form a stable complex and examine the activity of this Bim1-Bik1 complex. We find that Bim1 and Bik1 possess opposing activities and that the Bim1 activity predominates in the Bim1-Bik1 complex.
MATERIALS AND METHODS
Reaction Buffers
The following reaction buffers were used: buffer A: 20 mM Tris, 500 mM KCl, 20 mM imidazole, and 5 mM β-mercaptoethanol, pH 8.5; buffer B: 20 mM Tris, 1 M KCl, 20 mM imidazole, and 5 mM β-mercaptoethanol, pH 8.5; buffer D: 20 mM Tris, 200 mM KCl, and 5 mM β-mercaptoethanol, pH 8.0; SGF buffer: 25 mM Tris, 180 mM KCl, 1 mM MgCl2, and 1 mM EGTA, pH 7.5; BRB80K: 80 mM Pipes, 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 5 mM β-mercaptoethanol, pH 6.8; PBS: 4.3 mM Na2HPO4, 1.47 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.4; and PEM: 100 mM Pipes, 1 mM EGTA, and 1 mM MgCl2, pH 6.8.
Bim1 and Bik1 Protein Purification
Bik1 and Bim1 were purified from insect cells using a baculovirus expression system. Recombinant baculoviruses were made using pFastBacHT, where Bik1 and Bim1 were cloned, respectively, downstream of a 6xHis tag and TEV protease cleavage sequence (Invitrogen, Carlsbad, CA). SF9 insect cells (200 ml; 106 cells/ml) were infected with 2 ml of recombinant baculovirus and harvested after ∼65 h. Cells were lysed in 50 mM Tris, 300 mM KCl, 5% glycerol, 10 mM imidazole, and 1% NP40, pH 8.5, supplemented with EDTA-free complete protease inhibitor tablets (Roche, Rotkreuz, Switzerland). The extracts were cleared by centrifugation at 20,000 × g for 15 min at 4°C. Cleared extracts were incubated with NiNTA resin (Qiagen, Valencia, CA) and then washed with buffer A, followed by buffer B, buffer A, and buffer D and then eluted with buffer D plus 150 mM imidazole. The 6xHis tags were removed from the eluted proteins with AcTEV protease (Invitrogen) supplemented with EDTA-free complete protease inhibitors for 3–5 h at 16°C. The sample was dialyzed against buffer D, and the cleaved mixture again was passed over NiNTA resin to remove the AcTEV and 6xHis tag. Bik1 and Bim1 were dialyzed into SGF buffer or BRB80K. After dialysis, proteins were spun for 20 min at 20,000 × g at 4°C to remove aggregates. Protein concentrations were determined by Bradford assay and by visual comparison of purified proteins to a BSA standard on a Coomassie-stained gel. Proteins were snap-frozen in liquid nitrogen and stored at −80°C. Before use, proteins were precleared by centrifugation at 128,000 × g for 6 min at 4°C.
Bim1-GFP and Bik1-GFP Protein Purification
Bik1-GFP (green fluorescent protein) and Bim1-GFP were cloned, respectively, downstream of a glutathione S-transferase (GST) tag and TEV protease cleavage sequence in pET-GST-TEV (Moseley et al., 2004) and transformed into BL21 DE3 Escherichia coli cells (Stratagene, La Jolla, CA). One liter of cells was grown to an OD600 of ∼0.5 at 16°C and induced with 0.2 mM IPTG for 5 h. Cells were harvested, resuspended in PBS containing complete protease inhibitor tablets, and incubated for 45 min on ice with 1 mg/ml lysozyme (Sigma, St. Louis, MO). Cells were lysed by sonication, and lysates were cleared by centrifugation at 20,000 × g at 4°C. Cleared lysates were incubated with glutathione-Sepharose beads for 1 h at 4°C (GE Healthcare, Piscataway, NJ). Beads were then washed three times with PBS followed by three washes with BRB80K. GFP-fusion proteins were released from the glutathione resin by treatment with AcTEV protease in BRB80K supplemented with protease inhibitors 16 h at 4°C. Aggregate removal, protein concentration determination, protein storage, and preclearing were done as with insect cell purified proteins.
Sources of Tubulin
Porcine brain tubulin was purified as previously described (Vasquez et al., 1994) and used for microtubule-dynamics assays. Lyophilized bovine brain tubulin and rhodamine-labeled bovine brain tubulin were purchased from Cytoskeleton (Boulder, CO) and used for all other assays.
Preparation of Yeast Extracts
CUY28 (250 ml; MATα his3Δ200 leu2-3, 112 lys2-801 trp1-1 ura3-52) were grown in yeast peptone dextrose (YPD) media (United States Biological, Swampscott, MA) to log phase, washed with SGF buffer, and resuspended in SGF buffer to ∼500 μl with protease inhibitor tablets and PMSF (Alexis Biochemicals, Läufelfingen, Switzerland). Cells were rapidly frozen in liquid nitrogen, and extracts were made by grinding (Sorger et al., 1995). Extracts were stored at −80°C. Before use, yeast extracts were precleared by centrifugation at 128,000 × g for 6 min at 4°C.
Gel Filtration
Gel filtration was performed using a Superose 6 10/300 GL column (GE Healthcare) and an AKTA fast protein liquid chromatography (FPLC) machine. Yeast whole cell extract or purified proteins were run in SGF buffer at a flow rate of 0.2 ml/min. For protein binding experiments, the proteins were incubated together for 10 min at 4°C to allow complexes to form before loading. Elution was monitored by absorbance at 280 nm, and fractions were analyzed by SDS-PAGE. Western blotting used an anti-Bim1 or anti-Bik1 primary antibody (Wolyniak et al., 2006) and goat anti-rabbit HRP-conjugated secondary antibody (Bio-Rad, Hercules, CA). The column was calibrated using the following proteins (Sigma) with the indicated Stokes radii: cytochrome c (cyt c; 1.0 nm), carbonic anhydrase (2.4 nm), BSA (3.6 nm), alcohol dehydrogenase (4.6 nm), β-amylase (4.8 nm), apoferritin (6.1 nm), and thyroglobulin (8.5 nm). Blue Dextran (Sigma) identified the void volume. Stokes radii were determined as averages of the Porath and Laurent-Kilander methods (Porath, 1963; Laurent and Killander, 1964).
Sucrose Gradient Sedimentation
Sucrose gradients were performed as described in van Brugel et al. (2003) with slight modifications. Yeast whole cell extract or purified proteins were spun through 2-ml 5–20% sucrose gradients at 200,000 × g for 4 h at 4°C. For protein-binding experiments, the proteins were incubated together for 10 min at 4°C to allow complexes to form before loading. After centrifugation, fractions were analyzed as described for gel filtration. Each experiment included a gradient of proteins of known S values: cyt c (1.9 S), carbonic anhydrase (2.9 S), BSA (4.6 S), alcohol dehydrogenase (7.4 S), β-amylase (8.9 S), and catalase (11.3 S).
Microtubule Cosedimentation Assay
Taxol-stabilized microtubules were made by incubating tubulin in BRB80K buffer plus 1 mM GTP (Sigma) and 2 μM taxol (Cytoskeleton) at 37°C for 15 min. Microtubules were separated from unpolymerized tubulin by centrifugation at 86,000 × g for 30 min at 25°C and resuspended in BRB80K plus 1 mM GTP and 2 μM taxol. For binding studies, various amounts of microtubules were incubated with 40 μM Bim1 or Bik1 for 10 min at 25°C. Bound protein was separated from unbound protein by centrifugation at 86,000 × g for 30 min at 25°C. Supernatants and pellets were analyzed by SDS-PAGE. Western blotting used anti-α-tubulin antibody DMY1 (Sigma), and anti-Bim1 or anti-Bik1 antibodies (Wolyniak et al., 2006); secondary antibodies were goat anti-rabbit or goat anti-mouse conjugated to HRP (Bio-Rad). Supernatant and pellet band intensities were quantified using ImageJ (http://rsb.info.nih.gov/ij/; NIH, Bethesda, MD).
Localization of Bim1-GFP and Bik1-GFP on Microtubules
A 15 μM 1:20 mixture of rhodamine-labeled to unlabeled tubulin dimers was incubated with sea urchin axonemes fragments in BRB80K at 37°C for 15 min. Microtubules were then incubated with GFP (30 nM), Bim1-GFP (10 nM), Bik1-GFP (100 nM), Bik1-GFP (30 nM) plus Bim1 (10 nM), or Bim1-GFP (10 nM) plus Bik1 (50 nM) at 25°C for 5 min. Mixtures were then fixed in 0.7% glutaraldehyde for 3 min, quenched with 0.1% sodium borohydride for 3 min, and diluted 1:50 for microscopy analysis (van Breugel et al., 2003). Images were acquired using an Axioplan 2 imaging microscope (Zeiss, Thornwood, NY) and Openlab software (Improvision, Lexington, MA). The peak intensities of GFP, Bim1-GFP, or Bik1-GFP along microtubules were determined using the line scan function of ImageJ. No signal was noted for GFP alone on microtubules. Bim1-GFP or Bik1-GFP dots seen at the ends of axonemes were not included in the analysis.
Observation of Microtubule Dynamics
Microtubules were nucleated from sea urchin axoneme fragments and visualized by video-enhanced differential interference contrast (VE-DIC) microscopy as previously described (Walker et al., 1988; Vasquez et al., 1997; Howell et al., 1999). Briefly, ∼10-μl flow chambers were made using double-stick tape to mount coverslips on glass slides. Sea urchin axoneme fragments (Vasquez et al., 1994) were then perfused into the chamber and allowed to adhere to the coverslip for 5 min. Chambers were then perfused with 50 μl PEM plus 0.5% NP40 to remove unbound axonemes. To block nonspecific protein binding to the glass surfaces, 2 mg/ml casein was perfused into the chamber, incubated for 2 min, and then washed out with 50 μl PEM. Tubulin, with or without Bim1 and/or Bik1, in BRB80K containing 1 mM GTP was then perfused into the chamber. Each chamber was warmed on the microscope stage to 35°C to initiate microtubule assembly. Images were converted from super VHS to digital using iDVD (Apple, Cupertino, CA) and imported at a rate of 1 frame/s as individual png files or QuickTime movies. Microtubule lengths were measured using ImageJ.
Measurement of Microtubule Dynamics
Microtubule length was plotted versus time to give a “life-history” plot of each microtubule. From these plots, growth and shrinkage rates were calculated from the slopes by a least-squares regression analysis. Plus and minus ends were assigned based on microtubule elongation velocities, because plus-end elongation rates are faster (Walker et al., 1988, Vasquez et al., 1994). Variations around mean values are given as SDs. Comparisons of statistical significance were done with a two-tailed unpaired t test allowing for unequal variance (Excel; Microsoft, Redmond, WA). To determine the total microtubule length per axoneme end, microtubule lengths were measured at 7–8 min after warming to 35°C.
Transition frequencies were calculated as described previously (Walker et al., 1988; Toso et al., 1993). Catastrophe frequencies were calculated by dividing the number of catastrophes observed by the sum of the total time spent in elongation. Rescue frequencies were calculated by dividing the number of rescues observed by the sum of the total time spent in shortening. SDs for transition frequencies were determined by dividing the catastrophe or rescue frequency by the square root of the number of transitions observed (Walker et al., 1988).
To examine the relationship between microtubule growth rate and catastrophe frequency, we binned individual microtubules according to their growth rates into 0.2-μm/min intervals and calculated the catastrophe frequency for each binned group. Then the average catastrophe frequency was plotted versus the average growth rate for each binned group. This analysis was performed for three conditions: tubulin alone, tubulin plus Bim1, and tubulin plus Bik1. For tubulin alone, we binned microtubule growth rates from experiments using 8.6, 11.5, 13, and 14.4 μM tubulin. For tubulin plus Bim1, we binned microtubule growth rates from experiments using 11.5 μM tubulin with 0.1, 0.5, and 1 μM Bim1. For tubulin plus Bik1, we binned microtubule growth rates from experiments using 11.5 and 14.4 μM tubulin with 0.1, 0.5, and 1 μM Bik1.
RESULTS
Purified Bik1 and Bim1 Form Homodimers That Associate to Form a Tetrameric Complex
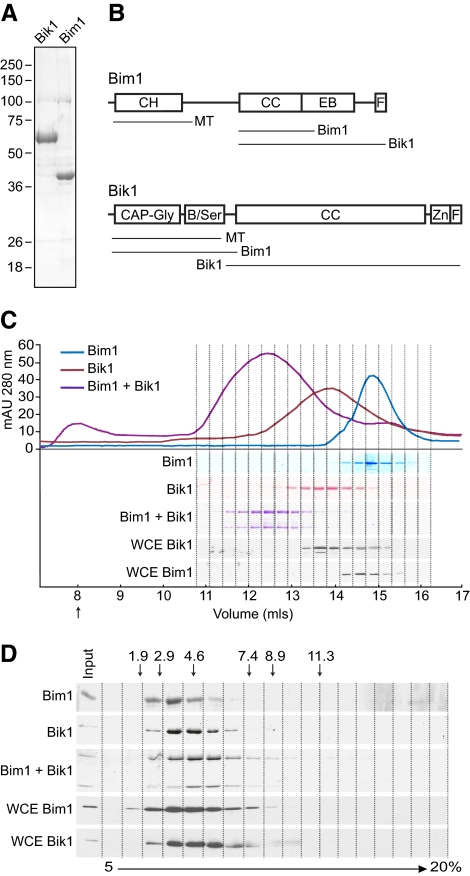
To investigate the biochemical activities of Bik1 and Bim1, we expressed 6xHis-tagged versions of each protein in SF9 cells using a baculovirus system and purified the proteins on Ni-NTA resin. After removal of the 6xHis tag, both proteins migrated on SDS-PAGE at their expected molecular weights (38 kDa for Bim1 and 51 kDa for Bik1) and were >90% pure (Figure 1A).
Figure 1.
Bik1 and Bim1 form homodimers that associate to form a tetrameric complex. (A) Coomassie-stained SDS-PAGE gels of ∼5 μg of Bik1 (left lane) and Bim1 (right lane) purified from insect cells. (B) Diagram of Bim1 (top) and Bik1 (bottom) protein domains. Lines below each protein indicate regions mediating microtubule binding, self-interactions, and Bim1-Bik1 interactions in vivo (Miller et al., 2006; Wolyniak et al., 2006; Akhmanova and Steinmetz, 2008). B/Ser, basic, serine-rich region; CH, calponin homology; CC, coiled-coil; CAP-Gly, cytoskeletal-associated protein-glycine–rich; EB, EB1 family homology domain; F, acidic-aromatic motif; Zn, zinc knuckle. (C) Size-exclusion chromatography of purified proteins and yeast whole cell extract (WCE). Chromatograms of purified Bim1 (blue), Bik1 (red), and an equimolar mixture of Bim1 and Bik1 (purple) are shown at top. SDS-PAGE of fractions is shown at bottom. Purified proteins were visualized by Coomassie staining; Bik1 and Bim1 in whole cell extracts were visualized by Western blots using anti-Bim1 or anti-Bik1 antibodies. Arrow indicates the column void volume. (D) Sucrose-gradient sedimentation of purified proteins and yeast whole cell extracts. Fractions from a 5–20% sucrose gradient were analyzed by SDS-PAGE. Purified proteins were visualized by Coomassie staining; Bik1 and Bim1 in whole cell extracts were visualized by Western blots using anti-Bim1 or anti-Bik1 antibodies. Arrows indicate the positions of markers with known S values in the gradient.
We have previously shown that Bim1 and Bik1 self-associate in vivo and that their coiled-coil regions are necessary for mediating this self-interaction (Figure 1B; Wolyniak et al., 2006). To determine the quaternary structure of the recombinant Bik1 and Bim1 proteins, we calculated the molecular weight of each from its Stokes radius and sedimentation coefficient (S; Siegel and Monty, 1966). The Stokes radius was obtained by gel filtration using a Superose 6 column and the S value was obtained by centrifugation through a 5–20% sucrose gradient (Figure 1, C and D; Table 1). Purified Bim1 was calculated to have a molecular weight of 70 kDa, which is close to the predicted molecular weight of 77 kDa for a Bim1 homodimer. This finding agrees with the results of a recent study by Zimniak et al., (2009), which also concludes that Bim1 is a homodimer. Purified Bik1 was calculated to have a molecular weight of 106 kDa, close to the predicted molecular weight of 102 kDa for a Bik1 homodimer. Thus, purified Bim1 and Bik1 appear to exist as homodimers in solution. The early elution of Bim1 and Bik1 and their large calculated Stokes radii determined from size-exclusion chromatography indicate these proteins have elongated rod-like shapes. We determined the Perrin shape parameter for each protein (Bloom et al., 1988), and used the polynomial inversion procedure to calculate an axial ratio for each protein, assuming a prolate ellipsoid (Harding and Colfen, 1995). Bim1was found to have an axial ratio of 13.4, whereas Bik1 had an axial ratio of 23.2.
Table 1.
Physical properties of Bim1 and Bik1
| Protein | Source | Stokes radius (nm) | S value | Calculated MW | Predicted MW |
|---|---|---|---|---|---|
| Bim1 | Pure | 5.2 | 3.2 | 69.7 | 76.7 (dimer) |
| Bim1 | WCE | 5.6 | 3.8 | 84.7 | |
| Bik1 | Pure | 6.7 | 3.8 | 106.3 | 102.2 (dimer) |
| Bik1 | WCE | 6.5 | 4.1 | 112.7 | |
| Bim1+Bik1 | Pure | 8.2 | 5.1 | 170.1 | 178.9 (tetramer) |
| Tubulin | Pure | 4.1 | 5.5 | 96.4 | 110.7 (dimer) |
MW, molecular weight; WCE, whole cell yeast extract.
To compare the physical properties of recombinant Bim1 and Bik1 with endogenous Bim1 and Bik1, yeast extracts were also subjected to gel filtration and sucrose gradient sedimentation, and the fractions analyzed by Western blotting (Figure 1, C and D). We calculated molecular weights of 85 kDa for Bim1 and 113 kDa for Bik1, consistent with these proteins forming homodimers in vivo (Table 1). Previous work found Bim1 in a ∼250-kDa complex in yeast extracts, although it may have existed as a homodimer within this larger complex (Lee et al., 2000).
Bim1 and Bik1 have been shown to interact in vivo and in vitro (Ito et al., 2001; Wolyniak et al., 2006), so we assayed for the formation of a Bim1-Bik1 complex in vitro. Bim1 and Bik1 were mixed in equal molar amounts and then analyzed by gel filtration and sucrose gradient sedimentation. In both conditions, the majority of Bim1 and Bik1 comigrated as a larger complex than either Bim1 or Bik1 alone (Figure 1C and D). The molecular weight of the complex was calculated to be 170 kDa, close to the predicted molecular weight of 179 kDa for two molecules of both Bim1 and Bik1 (Table 1). Thus, Bim1 and Bik1 homodimers associate in solution to form a stable tetrameric complex.
Analysis of Bim1 and Bik1 Binding to Tubulin Subunits and Microtubules
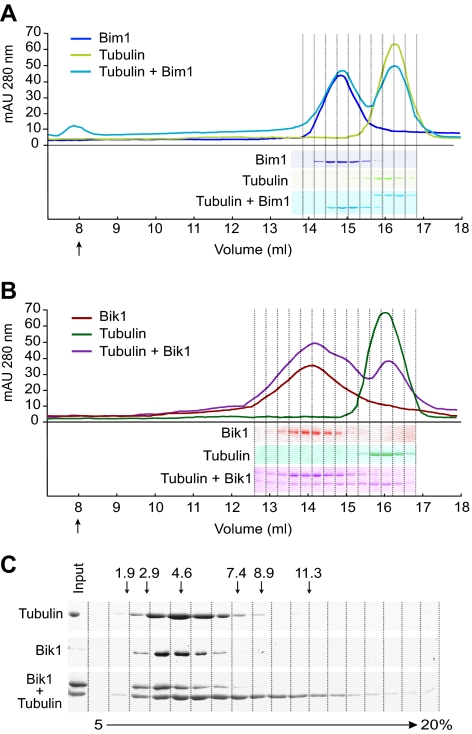
We looked for the formation of Bim1-tubulin and Bik1-tubulin complexes by mixing tubulin dimers with equal molar amounts of Bim1 and Bik1, respectively. The mixtures were then analyzed by gel filtration. In the Bim1-tubulin mixture, each protein eluted at the same position as it did alone, indicating that Bim1 does not bind tubulin dimers (Figure 2A). In the Bik1-tubulin mixture, the majority of Bik1 eluted at the same position as it did alone, although a minor amount of Bik1 migrated as a larger species (Figure 2B). Interestingly, a substantial fraction of the tubulin eluted as a broad peak that overlapped the Bik1 peak. It is unlikely that much of this tubulin is forming a stable complex with Bik1, because the Bik1 peak does not shift and tubulin in the broad peak is not concentrated in the Bik1-containing fractions. Similarly, in sucrose gradient analysis, tubulin in the presence of Bik1 sediments further into the gradient than tubulin alone (Figure 2C). The molecular weight of the largest tubulin species is ∼570 kDa, equivalent to approximately six tubulin subunits. Thus, we conclude that Bik1 binds only weakly, at best, to tubulin. In addition, Bik1 appears to stimulate the polymerization or aggregation of tubulin subunits.
Figure 2.
Analysis of Bim1 and Bik1 binding to tubulin heterodimers. Size-exclusion chromatography of (A) Bim1 (blue), tubulin (green), and an equimolar mixture of Bim1 and tubulin (turquoise), and (B) Bik1 (red), tubulin (green), and an equimolar mixture of Bik1 and tubulin (purple). Chromatograms are shown at top and SDS-PAGE of fractions (Coomassie-stained) are shown at bottom. Arrows indicates the column void volumes. (C) Sucrose-gradient sedimentation of Bik1, tubulin, and an equimolar mixture of Bik1 and tubulin. Fractions from a 5–20% sucrose gradient were analyzed by SDS-PAGE, and proteins were visualized by Coomassie staining. Arrows indicate the positions of markers with known S values in the gradient.
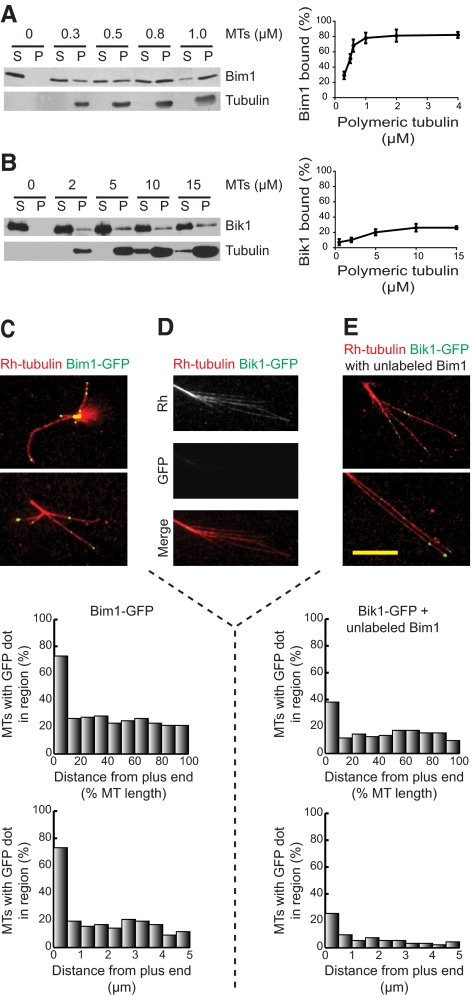
We also measured the abilities of Bim1 or Bik1 to bind assembled microtubules by incubating each with various amounts of taxol-stabilized microtubules. Microtubule bound protein was separated from unbound protein by centrifugation and the supernatant and pellet analyzed by Western blotting. Bim1 bound to microtubules with an apparent dissociation constant, Kd, of 0.5 μM (Figure 3A). In contrast, only small amounts of Bik1 bound microtubules even at much higher tubulin concentrations (Figure 3B).
Figure 3.
Microtubule binding and plus-end localization of Bim1 and Bik1. (A) Bim1 at 40 nM was incubated with various concentrations of taxol-stabilized microtubules. Left, microtubules were pelleted by centrifugation, and the pellet fraction (P) and the supernatant fraction (S) were analyzed by SDS-PAGE and Western blotting using Bim1 and α-tubulin antibodies. Right, band intensities were quantified using ImageJ, and the percentage of Bim1 bound was calculated. Error bars, ±SD. (B) Analysis of Bik1 binding to microtubules was done as described in A, except that higher microtubule concentrations were used. Bik1 was visualized using Bik1 antibody. (C–E) Localization of GFP-labeled proteins on microtubules. (C) Bim1-GFP, (D) Bik1-GFP, or (E) Bik1-GFP and unlabeled Bim1 were incubated with microtubules assembled from sea urchin axonemes in the presence of rhodamine-labeled tubulin. Microtubules are shown in red and GFP-labeled proteins in green. The positions of the Bim1-GFP (C) or Bik1-GFP (E) dots were graphed in two ways: their relative location along the microtubule (top graph) and their absolute distance from the plus end (bottom graph). Bar, 10 μm.
To examine the localization of Bim1 and Bik1 along microtubules, we purified Bim1-GFP and Bik1-GFP. Each of these was incubated with dynamic microtubules assembled from axonemes and then visualized by fluorescence microscopy. Bim1-GFP appeared as dots along microtubules with an average of 2.9 dots per microtubule (Figure 3C). The positions of these dots were analyzed in two ways. First, we divided microtubules into 10 equal-length sections (van Breugel et al., 2003) to determine the relative locations of the Bim1-GFP dots along the microtubule. Second, we measured the absolute distance from the plus end for each dot within 5 μm of the microtubule end. Both calculations showed that 73% of the microtubules had dots at their plus ends, whereas only ∼20% of microtubules had dots in any other region along their length. These results show that Bim1 is an autonomous microtubule end-binding protein, in agreement with Zimniak et al., (2009). In contrast, we observed only faint Bik1-GFP straining along microtubules and axonemes (Figure 3D).
Given that Bim1, but not Bik1, could localize to plus ends, we assayed whether Bik1 could be directed to the microtubule plus end through its interaction with Bim1. When Bik1-GFP was mixed with unlabeled Bim1, Bik1-GFP dots were observed along the microtubule and at the plus-end in a pattern similar to the Bim1-GFP localization (Figure 3E). The average number of Bik1-GFP dots per microtubule was 1.6, and these were enriched at the plus ends. Therefore, Bim1 is able to target Bik1 to microtubule plus ends in vitro. Mixing Bik1 with Bim1-GFP did not substantially alter the plus-end localization of Bim1-GFP (unpublished data).
Bim1 Promotes and Bik1 Inhibits Microtubule Assembly In Vitro
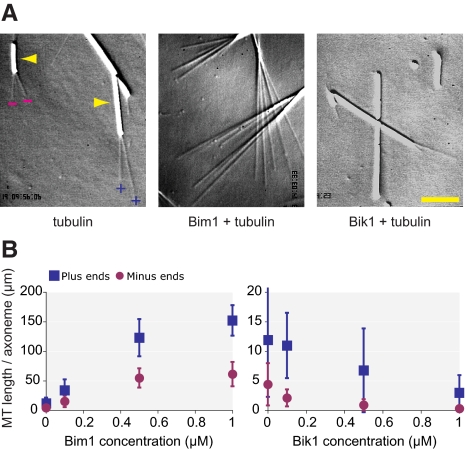
We next examined the effects of Bim1 and Bik1 on the assembly of microtubules in vitro. Microtubules were nucleated from sea urchin axonemes in 11.5 μM tubulin alone or in the presence of 0.1, 0.5, and 1.0 μM Bim1 or Bik1. Microtubules were visualized using VE-DIC microscopy. Because axonemes contain parallel bundles of uniformly oriented microtubules, microtubules extending from one end are plus-ended, whereas microtubules extending from the other end are minus-ended. Thus, it was possible to examine the assembly of plus- and minus-ended microtubules independently.
Bim1 increased the length and number of both plus- and minus-ended microtubules (Figure 4A). To quantify this effect, we calculated the total microtubule length per axoneme end by multiplying the average number of microtubules per end by their average length. Axonemes in 11.5 μM tubulin alone contained 11.9 ± 9.6 μm of total microtubule length at their plus ends and 4.4 ± 3.6 μm of total microtubule length at their minus ends (Figure 4B and Supplementary Table 1). Addition of 0.1 μM Bim1 increased total microtubule length 2.8-fold at plus ends and 3.5-fold at minus ends; addition of 1 μM Bim1 increased total microtubule length by 12.8-fold at plus ends and 13.9-fold minus ends (Figure 4B and Supplementary Table 1). Thus, Bim1 promotes microtubule assembly. In contrast to Bim1, Bik1 decreased the length and number of both plus- and minus-ended microtubules (Figure 4, A and B). Addition of 0.1 μM Bik1 decreased total microtubule length by 8% at plus ends and 52% at minus ends; addition of 1 μM Bik1 decreased total microtubule length by 4.0-fold at plus ends and 14.2-fold at minus ends (Figure 4B and Supplementary Table 2). Thus, Bik1 inhibits microtubule assembly.
Figure 4.
Bim1 promotes and Bik1 inhibits microtubule assembly in vitro. (A) Sea urchin axonemes were incubated with 11.5 μM tubulin alone (left) and in the presence of 1 μM Bim1 (center) or 1 μM Bik1 (right). Microtubules were visualized using VE-DIC microscopy. Axonemes are the larger rod-shaped structures indicated by the arrowheads in the left panel. Microtubules nucleated from the plus end or minus end of the axoneme are indicated by (+) and (−) symbols, respectively. Scale bar, 5 μM. (B) To quantify the effects of Bim1 (left) and Bik1 (right) on microtubule assembly, the total microtubule length per axoneme was calculated by multiplying the average length of microtubules by the average number of microtubules per axoneme. Error bars, ±SD.
Individual Effects of Bim1 and Bik1 on Microtubule Dynamics
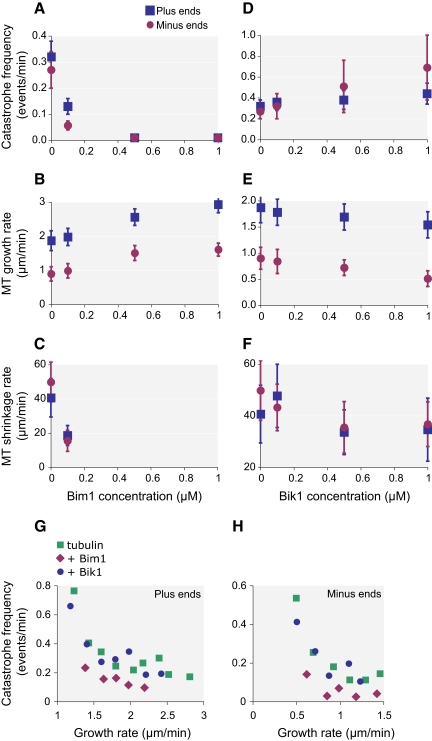
To determine the mechanisms by which Bim1 and Bik1 influence microtubule assembly, we examined the effects of these proteins on microtubule dynamics. Microtubules dynamics are defined by four parameters: the rates of microtubule growth and shrinkage, and the frequencies of catastrophes (transitions from growing to shrinking) and rescues (transitions from shrinking to growing). To determine how Bim1 and Bik1 affect these parameters, we observed microtubules over time using VE-DIC microscopy.
The most substantial effect of Bim1 is on catastrophe frequency. The addition of 0.1 μM Bim1 decreased the frequency of catastrophes 2.5-fold at plus ends and 5.6-fold at minus ends (Figure 5A and Supplementary Table 1). We did not observe any catastrophes at higher Bim1 concentrations, indicating that catastrophe frequencies were reduced at both plus and minus ends >20-fold in the presence of 0.5 μM Bim1 and >40-fold in the presence of 1 μM Bim1. Under the assembly conditions used, rescues are rare events with 11.5 μM tubulin alone; no rescue events were observed at plus ends and only two rescue events (1.96 events/min) were seen at minus ends (Supplementary Table 1). We did observe a higher frequency of rescues in 0.1 μM Bim1 with 0.51 events/min at plus ends, and 3.33 events/min at minus ends. We could not calculate a rescue frequency at higher Bim1 concentrations because of the absence of shrinking microtubules.
Figure 5.
Effects of Bim1 and Bik1 on microtubule dynamics. (A–F) Sea urchin axonemes were incubated with 11.5 μM tubulin in the presence of varying concentrations of Bim1 or Bik1. Individual microtubules were visualized using VE-DIC microscopy and their lengths measured over time. From this data, we calculated microtubule growth and shrinkage rates, and catastrophe frequencies; rescues were rare (see Supplementary Tables 1 and 2). Plots show the effects of increasing concentrations of Bim1 (A–C) or Bik1 (D–F) on these parameters. Error bars, ±SD. (G and H) Catastrophe frequencies were plotted with respect to microtubule growth rates for microtubule plus ends (G) and minus ends (H). See Materials and Methods for details. Green, tubulin alone; purple, Bim1 plus tubulin; blue, Bik1 plus tubulin.
In addition, Bim1 significantly increased microtubule growth rates and lowered shrinkage rates (Figure 5, B and C, Supplementary Table 1). Bim1 at 1 μM increased plus- and minus-end growth rates by 57 and 83%, respectively. Bim1 at 0.1 μM lowered plus- and minus-end shrinkage rates by 2.5- and 3.5-fold, respectively. We could not calculate shrinkage rates at higher Bim1 concentrations because shrinking microtubules were not observed. Overall, these results demonstrate that Bim1 decreases the catastrophe frequency and shrinkage rate and increases the growth rate and rescue frequency.
Bik1 also had a substantial effect on catastrophe frequency but in the opposite direction from Bim1. In the presence of 1 μM Bik1, catastrophe frequencies rose by 38% at plus ends and 2.4-fold at minus ends (Figure 5D and Supplementary Table 2). Growth rates decreased by 18% at plus ends and 43% at minus ends, but there was no significant change in shrinkage rates (Figure 5, E and F, Supplementary Table 2). We did not observe a significant increase in rescue events in the presence of Bik1 (Supplementary Table 2). Because Bik1 stimulated catastrophes in 11.5 μM tubulin, we also examined its effect in 14.4 μM tubulin, a tubulin concentration at which catastrophes are rarely observed (Supplementary Table 3). The addition of 0.1 μM Bik1 to 14.4 μM tubulin increased the catastrophe frequency >8-fold at plus ends and >4-fold at minus ends, resulting in frequencies similar to those observed in 11.5 μM tubulin alone. In summary, these results indicate that Bik1 decreases growth rates and increases catastrophe frequency.
The Effect of Bim1, But Not Bik1, on Catastrophe Frequency Is Independent of Its Effect on Growth Rates
Catastrophe frequency is inversely related to growth rate (Drechsel et al., 1992; van Breugel et al., 2003), so the effects of Bim1 and Bik1 on catastrophe frequencies could be an indirect effect of their abilities to increase and decrease growth rates, respectively. To test this possibility, we examined the relationship between microtubule growth rate and catastrophe frequency for three conditions: tubulin alone, tubulin plus Bim1, and tubulin plus Bik1. For each condition, we binned microtubules according to their growth rates and calculated the corresponding catastrophe frequencies. We then plotted average catastrophe frequency versus average growth rate for each binned group. As expected for tubulin alone, catastrophe frequency decreases as growth rate increases for both plus and minus ends (Figure 5, G and H). In the presence of Bik1, the catastrophe frequency at each growth rate is nearly the same as for tubulin alone, indicating that Bik1 likely promotes catastrophe frequency by inhibiting growth rate. However, in the presence of Bim1, the catastrophe frequency at each growth rate is lower than for tubulin alone. Thus, the reduction in catastrophe frequencies by Bim1 is not due solely to its effects on growth rates.
The Combined Effects of Bim1 and Bik1 on Microtubule Dynamics
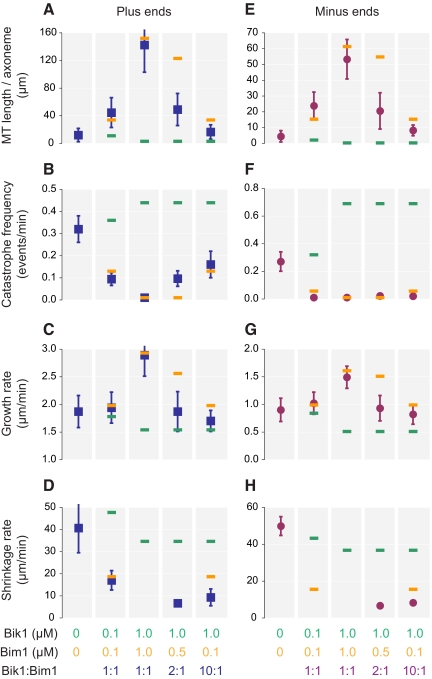
To determine the effects of the Bim1-Bik1 complex on microtubule dynamics, we combined Bim1 and Bik1 in equimolar amounts (0.1 and 1 μM each). Overall, this complex affected microtubule assembly in nearly the same way as Bim1 alone (Figure 6 and Supplementary Table 4). This result is more apparent at the higher concentrations of protein (1 μM) because the differences between the effects of the individual proteins at 1 μM are greater. The addition of both proteins at 1 μM increased total microtubule length 12.0-fold at plus ends and 12.1-fold at minus ends compared with tubulin alone (Figure 6, A and E). We did not observe any catastrophes at 1 μM Bim1 and Bik1, indicating that catastrophe frequencies were reduced >48-fold at plus ends and >29-fold at minus ends (Figure 6, B and F). Growth rates increased 50% at plus ends and 70% at minus ends (Figure 6, C and G). All of these parameters are very close to those obtained with 1 μM Bim1 alone and substantially different from those produced by 1 μM Bik1. Lack of shrinking microtubules prevented measuring shrinkage rates at 1 μM (Figure 6, D and H). However, at 0.1 μM Bim1-Bik1 complex, shrinkage rate at plus ends is the same as for 0.1 μM Bim1 alone.
Figure 6.
Combined effects of Bim1 and Bik1 on microtubule dynamics. (A–H) Sea urchin axonemes were incubated with 11.5 μM tubulin in the presence of varying concentrations of both Bim1 and Bik1. Parameters of microtubule dynamic instability were determined as in Figure 5 (see Supplemental Table 4). Error bars, ±SD. For comparison, the orange and green bars indicate the individual effects of Bim1 (orange) or Bik1 (green) at the concentration indicated, as previously shown in Figures 4B and 5, A–F.
To see whether Bim1 maintained this dominant effect at higher Bik1 to Bim1 ratios, we kept the Bik1 concentration at 1 μM and decreased the concentration of Bim1 to 0.5 or 0.1 μM. At 0.1 μM Bim1 and 1.0 μM Bik1, the assay should contain 0.1 μM Bim1-Bik1 complex and 0.9 μM Bik1, or a 9:1 ratio of free Bik1 to Bim1-Bik1 complex. This excess Bik1 did lead to decreases in total microtubule length and microtubule growth rate relative to 0.1 μM Bim1-Bik1 complex alone; the values for these parameters were approximately midway between those for 1.0 μM Bik1 and 0.1 μM Bim1. However, catastrophe frequency changed very little, remaining close the value for 0.1 μM Bim1 alone. Thus, even though it is out-numbered by Bik1 10:1, Bim1 is still the major influence on microtubule catastrophe frequency.
DISCUSSION
We have purified Bim1 and Bik1, the sole S. cerevisiae members of the EB1 and CLIP-170 families of proteins, respectively. Both proteins form elongated homodimeric molecules, similar to EB1 and CLIP-170 (Pierre et al., 1992; Scheel et al., 1999; Lansbergen et al., 2004; Honnappa et al., 2005; Slep et al., 2005; Akhmanova and Steinmetz, 2008). The conservation of this structure from yeast to humans underscores the importance of this arrangement in function. When Bik1 and Bim1 are mixed in a 1:1 ratio, they form a stable tetrameric complex comprised of one Bik1 homodimer and one Bim1 homodimer. Here we discuss the effects of Bim1, Bik1, and the Bim1-Bik1 complex on microtubule dynamics in vitro.
Bim1 Activity In Vitro
We found that Bim1 stimulates microtubule polymerization in vitro, a result that is in line with previous reports showing that EB1-family proteins are promoters of microtubule polymerization in a variety of systems (Tirnauer and Bierer, 2000). Promotion of microtubule polymerization can occur by increasing microtubule growth rates or rescue frequencies or by decreasing shortening rates or catastrophe frequencies. Previous work has examined the effects of EB1 (Manna et al., 2008; Vitre et al., 2008), EB3 (Komarova et al., 2009), and the Schizosaccharomyces pombe EB protein, Mal3 (Bieling et al., 2007; des Georges et al., 2008) on these parameters of microtubule assembly in vitro. Although Bim1 shares some of the properties of these EB1-family proteins, in several respects its effects on microtubule dynamics are quite different.
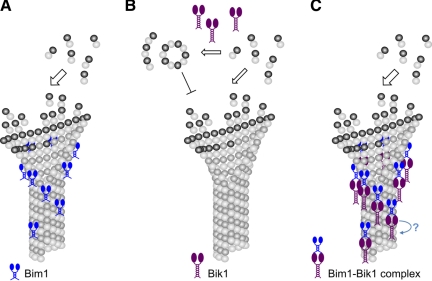
Bim1 stimulates microtubule growth rate by less than twofold at 1 μM. This modest stimulation of growth rate is similar to that of other EB1-family proteins, which have either no significant effect or cause up to a several-fold increase in growth rates, depending on the study (Bieling et al., 2007, 2008; Manna et al., 2008; Vitre et al., 2008; Dixit et al., 2009; Komarova et al., 2009). Bim1, like EB1 (Niethammer et al., 2007; Slep and Vale, 2007), does not bind tubulin. Therefore, Bim1 does not act as a microtubule polymerase like the XMAP215 family of proteins, which bind tubulin dimers and add them to growing plus ends (Al-Bassam et al., 2006; Kerssemakers et al., 2006; Brouhard et al., 2008). Instead, Bim1 has an intrinsic affinity for the plus end and may stimulate microtubule growth by a mechanism that has been proposed for other EB1-family proteins, stabilizing the lateral associations between protofilaments that promotes the growth of tubulin sheets (Figure 7A; Sandblad et al., 2006; des Georges et al., 2008; Vitre et al., 2008).
Figure 7.
Working model for the effects of Bim1, Bik1, and the Bim1-Bik1 complex on microtubule dynamics. (A) Bim1 binding to microtubule plus ends may stabilize lateral associations between protofilaments and promote growth. A similar activity could inhibit catastrophes by preventing the protofilament peeling that accompanies microtubule depolymerization. (B) Bik1 promotes the oligomerization of tubulin subunits. Assuming that these tubulin oligomers cannot be incorporated into microtubules, their formation will lower the effective tubulin concentration, inhibiting microtubule growth and increasing catastrophe frequency. (C) In the Bim1-Bik1 complex, the N-terminal CH domain of Bim1 is free to bind microtubule plus ends and affects microtubule dynamics in much the same way as Bim1 alone. On the other hand, the N-terminal CAP-Gly domain of Bik1 is bound to the C-terminal tail of Bim1 and is unable to interact with tubulin. It is possible that in vivo Bim1 is used to recruit Bik1 to the microtubule plus end, and an additional factor is needed to bring about the transfer of Bik1 from Bim1 to the microtubule (blue arrow).
Bim1 also promotes microtubule polymerization by dramatically inhibiting catastrophe frequencies, an effect that is too large to be only a secondary consequence of Bim1's ability to stimulate growth rate. This effect could also be due to the protein's ability to stabilize lateral associations between protofilaments and prevent the transition to protofilament peeling that accompanies microtubule depolymerization. In contrast, other EB1-family proteins have been shown to stimulate catastrophes in vitro (Bieling et al., 2007; Vitre et al., 2008; Komarova et al., 2009) or, in one study, to have only a small inhibitory effect (Manna et al., 2008). Vitre et al., (2008) proposed that EB1's ability to induce sheet closure by sealing the seam at growing plus ends makes microtubules more prone to catastrophe events. Thus, either Bim1 does not induce sheet closure or EB1 stimulates catastrophes by some other mechanism.
Like EB1, Bim1 also decreases microtubule shrinkage rates and increases rescue frequencies. EB1 does not associate with the plus ends of shrinking microtubules (Mimori-Kiyosue et al., 2000), so its effects on shrinkage rates and rescue frequencies have been attributed to its ability to bind along the microtubule lattice (Manna et al., 2008; Vitre et al., 2008). However, Bim1 suppresses shrinkage rates at a low Bim1-dimer to tubulin-dimer ratio of 1:230, so it is unlikely that this effect is mediated solely by uniform binding along the microtubule lattice. Perhaps, Bim1 inhibits microtubule shrinkage and stimulates rescues by acting directly at the plus end to stabilize lateral associations in the shrinking microtubule. Bim1 does associate with the plus ends of shrinking microtubules in vivo (Wolyniak et al., 2006); however, Zimniak et al., (2009) report that Bim1 is not observed on depolymerizing microtubules in vitro.
Bik1 Activity In Vitro
We found that Bik1 inhibits microtubule polymerization in vitro by slowing growth rates and, consequently, promoting catastrophes. This result is distinctly different from that obtained with CLIP-170, although these studies utilized only the N-terminal part of CLIP-170 that contains the tubulin/microtubule binding CAP-Gly domains. The N-terminal portion of CLIP-170 promoted microtubule polymerization (Arnal et al., 2004; Gupta et al., 2009) and stimulated rescues (Arnal et al., 2004). Bik1 could act directly at plus ends to specifically slow growth rates, which would be a novel activity, but the fact that we do not observe Bik1 binding to microtubule ends argues against this model. Alternatively, Bik1 could bind and sequester tubulin subunits, like Op18/Stathmin (Cassimeris, 2002), slowing growth by effectively lowering the free tubulin concentration. The affinity of Bik1 for tubulin was not sufficiently high to measure by gel filtration binding studies, but these results do not rule out a direct interaction of these proteins in solution. We did see oligomerization of tubulin in the presence of Bik1, indicating that Bik1 does interact with tubulin at least transiently to influence tubulin polymerization. If these oligomers cannot be incorporated into microtubules, then their formation will lower the effective tubulin concentration and inhibit microtubule growth, as we observed in the presence of Bik1 (Figure 7B). Oligomerization has been reported previously for tubulin in the presence of the N-terminal domain of CLIP-170, but these oligomers are proposed to stimulate, rather than inhibit, microtubule growth (Diamantopoulos et al., 1999; Arnal et al., 2004). Overall, our results show that Bik1 inhibits microtubule growth, but its mechanism of action is still unclear.
In yeast cells, loss of either Bik1 or Bim1 increases the amount of time microtubules spend in a paused state, neither growing nor shrinking to a significant degree, as judged by light microscopy. Because pausing occurs infrequently in our in vitro system (Walker et al., 1988), we could not determine whether Bik1 or Bim1 affects pausing in vitro.
Bim1-Bik1 Complex Activity in Vitro
The Bim1-Bik1 complex associates with microtubules and binds preferentially to microtubule ends, similar to Bim1 alone. Because Bik1 alone does not localize to plus ends, we conclude that Bim1 can target Bik1 to microtubule ends in the absence of other proteins. Similarly, EB1 is necessary and sufficient for the plus-end tracking by CLIP-170 in vitro (Bieling et al., 2008; Dixit et al., 2009). CLIP-170-family proteins contain one or two N-terminal CAP-Gly domains. CAP-Gly domains interact with EEY/F motifs (Honnappa et al., 2006; Mishima et al., 2007; Weisbrich et al., 2007), which are present at the C-terminus of both EB1/Bim1 and α-tubulin. EEY/F motifs are also found at the C-terminus of CLIP-170 members, although this motif is less apparent in Bik1, and have been implicated in the auto-regulation of CLIP-170 (Miller et al., 2006; Akhmanova and Steinmetz, 2008). Interestingly, in vivo, the majority of Bik1 localization to astral microtubule plus ends depends on the kinesin motor, Kip2, and not on Bim1 (Carvalho et al., 2004). However, in the presence of a mutant α-tubulin that lacks the EEY/F tail, Bik1 binding to the plus end decreases several-fold (Badin-Larcon et al., 2004), and the residual Bik1 localization to plus ends is dependent on Bim1 (Caudron et al., 2008). Overall, these results indicate that Bim1 can localize Bik1 to astral microtubule plus ends in vivo, consistent our in vitro results, but that this may not be the primarily mechanism of Bik1 localization on astral microtubules. Bim1 may be responsible for Bik1 localization to spindle microtubule plus ends, as Bik1 localizes to spindle microtubules in the absence of Kip2 (Carvalho et al., 2004) and Kip2 is not present on spindle microtubules (Huyett et al., 1998; Miller et al., 1998).
Interestingly, the effect of the Bik1-Bim1 complex on microtubule dynamics appears to be identical to that of Bim1 alone. The simplest conclusion is that the Bim1-Bik1 interaction does not alter Bim1's activity but completely inhibits Bik1's activity. As noted above, previous evidence suggests that the N-terminal CAP-Gly domain of Bik1 interacts with the C-terminal tail of Bim1 (Honnappa et al., 2006; Wolyniak et al., 2006). Because both EB1-family and CLIP-170-family proteins associate with microtubules through their N-terminal domains: the calponin homology (CH) domain and CAP-Gly domain, respectively (Akhmanova and Steinmetz, 2008), one might expect that this arrangement of Bim1 and Bik1 would strongly inhibit Bik1's activity while leaving Bim1's activity largely unchanged (Figure 7C).
On the other hand, EB1 has been shown to be autoinhibited by interactions of its C-terminal tail with its N-terminal microtubule-binding domain (Hayashi et al., 2005). Binding of p150glued, another CAP-Gly containing protein, to the C-terminus of EB1, or deletion of the EB1 C-terminus, enhances EB1 binding and activity at microtubule ends (Hayashi et al., 2005; Manna et al., 2008). Although Bik1 does bind to the C-terminus of Bim1 (Wolyniak et al., 2006), we saw no increase of Bim1 activity in the presence of Bik1. These results indicate that Bim1, unlike EB1, does not exist in an autoinhibited state. A similar conclusion was reached by Zimniak et al., (2009).
Given that the Bim1-Bik1 complex behaves much like Bim1 alone, what is the value of this interaction? Perhaps its role is simply to suppress the activity of Bik1. Because Bik1 has only one CAP-Gly domain, it is likely that Bim1-bound Bik1 will not be able to simultaneously interact with the microtubule. In this way, Bim1 could buffer Bik1 activity at plus ends. Alternatively, formation of the Bim1-Bik1 complex may be the first step in a Kip2-independent mechanism for recruiting Bik1 to the microtubule end. Clearly, binding of Bik1 to Bim1 brings Bik1 into close proximity to the microtubule plus end. A second step would be the dissociation of Bik1 from Bim1, freeing its CAP-Gly domain to interact with the microtubule plus end directly. This transfer of Bik1 from Bim1 to the microtubule may require an additional factor, which could, for example, phosphorylate one or both proteins and weaken their interaction. Any such factor will be lacking in our in vitro assay, which may explain why we do not see any Bik1 activity in the presence of equal amounts of Bim1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gary Isaacs and Lee Kraus for advice and the use of their FPLC and Beth Lalonde and James Hodek for comments on the manuscript. This work was supported by National Institutes of Health (NIH) Grants GM40479 (T.C.H.) and GM058025 (L.C.). K.B.H. was supported in part by predoctoral training grants from the NIH and the U.S. Department of Education.
Abbreviations used:
- +TIP
plus-end tracking protein
- VE-DIC
video-enhanced differential interference contrast.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-02-0083) on April 14, 2010.
REFERENCES
- Akhmanova A., Hoogenraad C. C. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol. 2005;1:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Steinmetz M. O. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008;4:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J., van Breugel M., Harrison S. C., Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 2006;7:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnal I., Heichette C., Diamantopoulos G. S., Chretien D. CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr. Biol. 2004;23:2086–2095. doi: 10.1016/j.cub.2004.11.055. [DOI] [PubMed] [Google Scholar]
- Badin-Larcon A. C., Boscheron C., Soleilhac J. M., Piel M., Mann C., Denarier E., Fourest-Lieuvin A., Lafanechere L., Bornens M., Job D. Suppression of nuclear oscillations in Saccharomyces cerevisiae expressing Glu tubulin. Proc. Natl. Acad. Sci. USA. 2004;15:5577–5582. doi: 10.1073/pnas.0307917101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin V., Styles C. A., Fink G. R. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J. Cell Biol. 1990;6(Pt 1):2573–2586. doi: 10.1083/jcb.111.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Kandels-Lewis S., Telley I. A., van Dijk J., Janke C., Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 2008;7:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E. L., Sandblad L., Dogterom M., Brunner D., Surrey T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;7172:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- Bloom G. S., Wagner M. C., Pfister K. K., Brady S. T. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 1988;9:3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J., Hyman A. A. XMAP215 is a processive microtubule polymerase. Cell. 2008;1:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Gupta M. L., Jr, Hoyt M. A., Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 2002;1:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- Caudron F., Andrieux A., Job D., Boscheron C. A new role for kinesin-directed transport of Bik1p (CLIP-170) in Saccharomyces cerevisiae. J. Cell Sci. 2008;121(Pt 9):1506–1513. doi: 10.1242/jcs.023374. [DOI] [PubMed] [Google Scholar]
- des Georges A., Katsuki M., Drummond D. R., Osei M., Cross R. A., Amos L. A. Mal3, the Schizosaccharomyces pombe homolog of EB1, changes the microtubule lattice. Nat. Struct. Mol. Biol. 2008;10:1102–1108. doi: 10.1038/nsmb.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Mitchison T. J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Diamantopoulos G. S., Perez F., Goodson H. V., Batelier G., Melki R., Kreis T. E., Rickard J. E. Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J. Cell Biol. 1999;1:99–112. doi: 10.1083/jcb.144.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Barnett B., Lazarus J. E., Tokito M., Goldman Y. E., Holzbaur E. L. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc. Natl. Acad. Sci. USA. 2009;2:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel D. N., Hyman A. A., Cobb M. H., Kirschner M. W. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol. Biol. Cell. 1992;10:1141–1154. doi: 10.1091/mbc.3.10.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M. K., et al. The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J. Cell Biol. 2008;1:91–100. doi: 10.1083/jcb.200710164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K. K., Paulson B. A., Folker E. S., Charlebois B., Hunt A. J., Goodson H. V. Minimal plus-end tracking unit of the cytoplasmic linker protein CLIP-170. J. Biol. Chem. 2009;11:6735–6742. doi: 10.1074/jbc.M807675200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S. E., Colfen H. Inversion formulae for ellipsoid of revolution macromolecular shape functions. Anal. Biochem. 1995;1:131–142. doi: 10.1006/abio.1995.1324. [DOI] [PubMed] [Google Scholar]
- Hayashi I., Wilde A., Mal T. K., Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol. Cell. 2005;4:449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- Honnappa S., John C. M., Kostrewa D., Winkler F. K., Steinmetz M. O. Structural insights into the EB1-APC interaction. EMBO J. 2005;2:261–269. doi: 10.1038/sj.emboj.7600529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Okhrimenko O., Jaussi R., Jawhari H., Jelesarov I., Winkler F. K., Steinmetz M. O. Key interaction modes of dynamic +TIP networks. Mol. Cell. 2006;5:663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Howard J., Hyman A. A. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 2007;1:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Howell B., Larsson N., Gullberg M., Cassimeris L. Dissociation of the tubulin-sequestering and microtubule catastrophe-promoting activities of oncoprotein 18/stathmin. Mol. Biol. Cell. 1999;1:105–118. doi: 10.1091/mbc.10.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyett A., Kahana J., Silver P., Zeng X., Saunders W. S. The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J. Cell Sci. 1998;111(Pt 3):295–301. doi: 10.1242/jcs.111.3.295. [DOI] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y., Huffaker T. C. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 2003;3:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;8:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerssemakers J. W., Munteanu E. L., Laan L., Noetzel T. L., Janson M. E., Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;7103:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- Komarova Y., et al. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 2009;5:691–706. doi: 10.1083/jcb.200807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Akhmanova A. Microtubule plus end: a hub of cellular activities. Traffic. 2006;5:499–507. doi: 10.1111/j.1600-0854.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- Lansbergen G., et al. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J. Cell Biol. 2004;7:1003–1014. doi: 10.1083/jcb.200402082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent T. C., Killander J. A theory of gel filtration and its experimental verification. J. Chromatogr. 1964;14:317–330. [Google Scholar]
- Lee L., Tirnauer J. S., Li J., Schuyler S. C., Liu J. Y., Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;5461:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Manna T., Honnappa S., Steinmetz M. O., Wilson L. Suppression of microtubule dynamic instability by the +TIP protein EB1 and its modulation by the CAP-Gly domain of p150glued. Biochemistry. 2008;2:779–786. doi: 10.1021/bi701912g. [DOI] [PubMed] [Google Scholar]
- Miller R. K., D'Silva S., Moore J. K., Goodson H. V. The CLIP-170 orthologue Bik1p and positioning the mitotic spindle in yeast. Curr. Top. Dev. Biol. 2006:49–87. doi: 10.1016/S0070-2153(06)76002-1. [DOI] [PubMed] [Google Scholar]
- Miller R. K., Heller K. K., Frisen L., Wallack D. L., Loayza D., Gammie A. E., Rose M. D. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell. 1998;8:2051–2068. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Shiina N., Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 2000;14:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Mishima M., Maesaki R., Kasa M., Watanabe T., Fukata M., Kaibuchi K., Hakoshima T. Structural basis for tubulin recognition by cytoplasmic linker protein 170 and its autoinhibition. Proc. Natl. Acad. Sci. USA. 2007;25:10346–10351. doi: 10.1073/pnas.0703876104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. B., Sagot I., Manning A. L., Xu Y., Eck M. J., Pellman D., Goode B. L. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 2004;2:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer P., Kronja I., Kandels-Lewis S., Rybina S., Bastiaens P., Karsenti E. Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biol. 2007;2:e29. doi: 10.1371/journal.pbio.0050029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre P., Scheel J., Rickard J. E., Kreis T. E. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;6:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- Porath J. Some recently developed fractionation procedures and their application to peptide and protein hormones. Pure Appl. Chem. 1963;6:233–244. [Google Scholar]
- Sandblad L., Busch K. E., Tittmann P., Gross H., Brunner D., Hoenger A. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006;7:1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Scheel J., Pierre P., Rickard J. E., Diamantopoulos G. S., Valetti C., van der Goot F. G., Haner M., Aebi U., Kreis T. E. Purification and analysis of authentic CLIP-170 and recombinant fragments. J. Biol. Chem. 1999;36:25883–25891. doi: 10.1074/jbc.274.36.25883. [DOI] [PubMed] [Google Scholar]
- Schuyler S. C., Pellman D. Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell. 2001;4:421–424. doi: 10.1016/s0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- Schwartz K., Richards K., Botstein D. BIM1 encodes a microtubule-binding protein in yeast. Mol. Biol. Cell. 1997;12:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel L. M., Monty K. J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 1966;2:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Slep K. C., Rogers S. L., Elliott S. L., Ohkura H., Kolodziej P. A., Vale R. D. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J. Cell Biol. 2005;4:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep K. C., Vale R. D. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell. 2007;6:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Doheny K. F., Hieter P., Kopski K. M., Huffaker T. C., Hyman A. A. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc. Natl. Acad. Sci. USA. 1995;26:12026–12030. doi: 10.1073/pnas.92.26.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J. S., Bierer B. E. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J. Cell Biol. 2000;4:761–766. doi: 10.1083/jcb.149.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J. S., O'Toole E., Berrueta L., Bierer B. E., Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 1999;5:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso R. J., Jordan M. A., Farrell K. W., Matsumoto B., Wilson L. Kinetic stabilization of microtubule dynamic instability in vitro by vinblastine. Biochemistry. 1993;5:1285–1293. doi: 10.1021/bi00056a013. [DOI] [PubMed] [Google Scholar]
- van Breugel M., Drechsel D., Hyman A. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J. Cell Biol. 2003;2:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez R. J., Gard D. L., Cassimeris L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol. 1994;4:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez R. J., Howell B., Yvon A. M., Wadsworth P., Cassimeris L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell. 1997;6:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B., Coquelle F. M., Heichette C., Garnier C., Chretien D., Arnal I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 2008;4:415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- Walker R. A., O'Brien E. T., Pryer N. K., Soboeiro M. F., Voter W. A., Erickson H. P., Salmon E. D. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 1988;4:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrich A., Honnappa S., Jaussi R., Okhrimenko O., Frey D., Jelesarov I., Akhmanova A., Steinmetz M. O. Structure-function relationship of CAP-Gly domains. Nat. Struct. Mol. Biol. 2007;10:959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- Wolyniak M. J., Blake-Hodek K., Kosco K., Hwang E., You L., Huffaker T. C. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell. 2006;6:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., Westermann S. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 2009;3:379–391. doi: 10.1083/jcb.200901036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.