Positive charges of nascent chain facilitate membrane spanning of a marginally hydrophobic segment, even when separated by 70 residues from the segment. The segment is exposed to the lumen and then slides back into the membrane. They not only fix the hydrophobic segment in the membrane, but exert a much more dynamic action than previously realized.
Abstract
Positively charged amino acid residues are well recognized topology determinants of membrane proteins. They contribute to the stop-translocation of a polypeptide translocating through the translocon and to determine the orientation of signal sequences penetrating the membrane. Here we analyzed the function of these positively charged residues during stop-translocation in vitro. Surprisingly, the positive charges facilitated membrane spanning of a marginally hydrophobic segment, even when separated from the hydrophobic segment by 70 residues. In this case, the hydrophobic segment was exposed to the lumen, and then the downstream positive charges triggered the segment to slide back into the membrane. The marginally hydrophobic segment spanned the membrane, but maintained access to the water environment. The positive charges not only fix the hydrophobic segment in the membrane at its flanking position, but also have a much more dynamic action than previously realized.
INTRODUCTION
Many membrane proteins in the secretory pathway are synthesized by membrane-bound ribosomes and are cotranslationally integrated into the endoplasmic reticulum (ER) membrane. ER targeting and membrane insertion of polypeptide chains are triggered by signal sequences consisting mainly of a hydrophobic sequence (H-segment). Membrane translocation of hydrophilic lumenal domains and membrane integration of transmembrane (TM) segments are mediated by a protein-conducting channel called the translocon. The main part of the translocon comprises a SecY complex in bacteria and a Sec61 complex in eukaryotes (Rapoport, 2007). The x-ray structures of the archaeal and bacterial SecY complex suggest that the translocation pore is surrounded by 10 TM helices of the SecY molecule (Van den Berg et al., 2004; Tsukazaki et al., 2008) and provides an aqueous environment through which a variety of hydrophilic polypeptides can be translocated. The pore appears to open laterally to allow hydrophobic TM segments to exit into the membrane lipid environment (Rapoport, 2007).
At the initial stage of cotranslational translocation and membrane integration, the signal sequence in a nascent polypeptide chain emerging from the ribosome is recognized by the signal recognition particle, which arrests polypeptide chain elongation. The ribosome-nascent chain complex is targeted to the translocon, where the signal sequence is released from the particle by its receptor (Cross et al., 2009). The signal sequence is then recognized by the translocon (Plath et al., 1998), the ribosome docks to the translocon, and either the N-terminal or C-terminal side of the signal sequence is translocated through the translocon pore. At this stage, positive charges tend to resist translocation. When the N-terminus is translocated, type I signal-anchor topology is achieved (Kida et al., 2000, 2005). When the C-terminal portion is translocated, the polypeptide chain, elongating from the ribosome, directly translocates through the translocon tunnel. The signal sequence not only triggers the translocation but also provides a significant translocation motive force that depends on the amino acid sequence (Kida et al., 2009). The geometry around the functioning translocon has been roughly estimated using an in vitro system; 30–40 residues of the nascent chain are included in the ribosome tunnel, 10 residues in an extended conformation span the translocon, and 15 residues of the nascent chain between an oligosaccharyl transferase (OSTase) active site and the membrane (Whitley et al., 1996).
Positive charges have a critical function in determining the orientation of the signal sequence; the more positive side of the H-segment of the signal sequence tends to be retained on the cytoplasmic side, and the other side is translocated though the translocon (High and Dobberstein, 1992; Kida et al., 2006). For example, in the case of a type I signal-anchor sequence, which mediates the translocation of its upstream portion, inserting positive charges into the N-terminal flanking portion and/or deleting positive charges from the C-terminal region causes inversion of the orientation (from N-lumen/C-cytosol to N-cytosol/C-lumen; Sakaguchi, 1997; Goder and Spiess, 2001). In the case of the type I signal-anchor sequence of synaptotagmin II, the downstream positive charges affect the orientation, even when they are separated from the H-segment by 25 residues (Kida et al., 2006).
After the signal sequences trigger translocation of the C-terminal portions (start-translocation), the nascent chain emerging from the ribosome directly enters the translocon. The ongoing polypeptide chain movement through the translocon is then interrupted by a certain segment within the nascent polypeptide (stop-translocation; Yost et al., 1983). The stop-translocation is a critical process for the biogenesis of TM segments of integral membrane proteins. Many TM segments of membrane proteins possess the stop-translocation function (e.g., Ota et al., 1998; Sato et al., 2002). The hydrophobicity of the polypeptide chain is a major determinant of stop-translocation, and the downstream flanking positive charges enhance stop-translocation (Kuroiwa et al., 1990, 1991). Only nine Leu residues are needed to mediate stop-translocation in the ER translocon, whereas more than 19 Ala residues are required to induce a similar extent of stop-translocation (Kuroiwa et al., 1991). A short hydrophobic segment consisting of fewer than 10 Leu residues followed by positive charges can adopt a TM topology, although the TM length does not adopt the width of the lipid bilayer (Jaud et al., 2009). Recent systematic analyses provided a quantitative biological scale of hydrophobicity for membrane insertion of polypeptide chains through the translocon (Hessa et al., 2005, 2007). Simple partitioning of the H-segment of the polypeptide chain into the lipid hydrophobic core appears to determine its membrane insertion. The contribution of positive charges to membrane insertion of the polypeptide chain has also been quantitatively confirmed (Lerch-Bader et al., 2008). Furthermore, the positively charged residues that are 13-residues downstream from the H-segment contribute to the stop-translocation (Lerch-Bader et al., 2008). The functions of the positively charged residues, determination of signal sequence orientation and enhancement of stop-translocation, have been reflected by a statistical rule of membrane topology, i.e., the positive-inside rule of membrane proteins (von Heijne and Gavel, 1988; Sipos and von Heijne, 1993).
In the present study, we systematically evaluated the function of positive charges during the stop-translocation process. Marginal H-segment, which are normally translocated through the membrane, can become TM segment when downstream positive-charge cluster present at distance of up to 70 residues away. During this process, the marginal H-segment is exposed to the ER lumen, but it is eventually retracted to span the membrane. The positive charges cause the H-segment that was translocated into the lumen to slide back into the membrane. Furthermore, such a marginal H-segment spanning the membrane remained in an aqueous environment, suggesting that membrane insertion is not always required for the stop-translocation event. We propose a dynamic process induced by positive charges during membrane integration and folding of membrane proteins.
MATERIALS AND METHODS
Materials
Rough microsomal membrane (RM; Walter and Blobel, 1983) and rabbit reticulocyte lysate (Jackson and Hunt, 1983) were prepared as previously described. RM was treated with EDTA and then with Staphylococcus aureus nuclease as previously described (Walter and Blobel, 1983). Castanospermine (Merck, Tokyo, Japan), the glycosylation acceptor peptide (AP; N-benzoyl-Asn-Leu-Thr-N-methylamide; Quality Controlled Biochemicals, Hopkinton, MA), proteinase K (ProK; Merck), DNA manipulating enzymes (Takara and Toyobo, Tokyo, Japan), (methyl-PEG12)3-PEG4-maleimide (PEGmal; Thermo Scientific, Waltham, MA), and N-ethylmaleimide (NEM; Wako, Tokyo, Japan) were obtained from the indicated sources.
Construction of Model Proteins
In the following DNA construction procedure, DNA fragments obtained by PCR using primers including the appropriate restriction enzyme site (indicated in parentheses) were subcloned into plasmid vectors that had been digested with the restriction enzymes. At each junction, the six bases of the restriction enzyme site were designed to encode two codons. Point mutations were introduced using the method of Kunkel (1985) or the QuikChange procedure (Stratagene, La Jolla CA). All the constructed DNAs were confirmed by DNA sequencing.
Rat serum albumin (RSA) was used as the base for construction (see Figure 1). For reference, the residue numbers of RSA in the database (National Center for Biotechnology Information, NP_599153) are indicated in the figures. The cDNA fragment encoding M1-H319 of RSA (including the Kozak sequence at the 5′-end region and the DNA sequence encoding the Val-stop codon at the 3′-end; XbaI/ApaI) was subcloned into a pRcCMV vector (Invitrogen, Tokyo, Japan, ver. 1.0; XbaI/ApaI). Restriction enzyme sites of NheI and Aor51HI, which encode AS and SA, and the DD-segment were created between V166 and F173 of RSA (Figure 1A). There was no spacer between the two enzyme sites. Synthetic DNA fragments encoding the repeating LA sequences were inserted between these enzyme sites. Glycosylation sites (N67 and N259) were generated at V67 and A259 of RSA by changing V67Q68E69 to NST and A259T260D261 to NST, respectively. Those were 102 residues upstream (−102) and 91 residues downstream (+91) of the LA-segment. The DD segment was exchanged with the KK segment (Figure 1C). Various numbers of positive charges were introduced after the DD segment (Figure 1D). For the 6K-scanning constructs (see Figure 2), the 6K-cluster was inserted into various positions. The distance of the 6K-cluster from the LA-segment is indicated in the parentheses of the cluster name. For standard polypeptides (Figure 2), the termination codons were created at L178, E183, and V188 within the LA8-0K and LA8-6K(10) model proteins. For Cys-scanning constructs (see Figure 3), all of the 19 Cys codons within M1-H319 of RSA were exchanged with Ala codons by the Kunkel method, where at most six primers were included in a single reaction. The upstream glycosylation site (N67), LA-segment, and 6K-cluster were included, and a single Cys codon was created by a point mutation. For glycosylation site–scanning constructs (see Figure 4), a single glycosylation site was created at the indicated position by an amino acid exchange (H164EV to NST, S169ADD to NSTA, L179YY to NST, N186EV to NST, and ES197DK to TNST). The LA8-segment and 6K-cluster were introduced as indicated in the figure. For LAC-segment constructs (see Figure 5), using the Cys-less construct in Figure 3, the indicated LAC-segment, the glycosylation site, and the 6K-cluster were included at the indicated positions. For expression in COS7 cells (see Figure 6), a DNA fragment encoding the model proteins (XbaI/XbaI) was ligated with a pRcCMV-HA expression vector (Kanki et al., 2002), where the HA.11 epitope-tag was C-terminally fused.
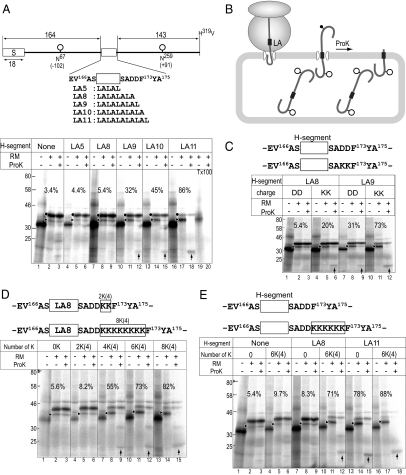
Figure 1.
Positive charges and marginal H-segment cooperate in stop-translocation process. (A and B) Assay for stop-translocation. RSA-based model proteins included a signal peptide (S) at the N-terminus, artificial H-segments consisting of an Leu/Ala repeat sequence in the middle portion (open rectangle), and potential glycosylation sites at N67 and N259 (○). The H-segments were followed by the DD-segment. The model proteins were translated in a cell-free system in the absence or presence of RM. If the product is fully translocated through the membrane, both sites are glycosylated. If the product is stop-translocated at the middle portion, only the upstream site (N67) is glycosylated, and the downstream is left on the cytoplasmic side. Stop-translocation efficiency was estimated with percent (%) of monoglycosylated product among the monoglycosylated and diglycosylated products. Aliquot of the translation product was treated with ProK, which digests the polypeptide on the cytoplasmic side of the membrane. Where indicated, ProK-treatment was performed in the presence of Triton X-100 (Tx100) detergent. ↑, the ProK-resistant fragment. Diglycosylated (•) and monoglycosylated (▶) forms are indicated. The superscripted numbers indicate the residue numbers of the original RSA polypeptide. Stop-translocation clearly depended on the LA-segment length. (C) Positive charges enhanced the stop-translocation. The DD-segment was replaced with two Lys (KK) and the model proteins were subjected to a stop-translocation assay. Monoglycosylated products (▶) and ProK-resistant fragment (↑) are indicated. (D) The positive charges caused stop-translocation depending on their numbers. Various numbers of Lys residues were introduced after the DD-segment. The Lys-clusters are named according to the residue-numbers, and their positions (indicated in parentheses). (E) Cooperation between the marginal H-segment and the 6K-cluster mediated the stop-translocation. The 6K-cluster and/or the H-segment were included in the model proteins as indicated. The 6K-cluster alone does not cause stop-translocation.
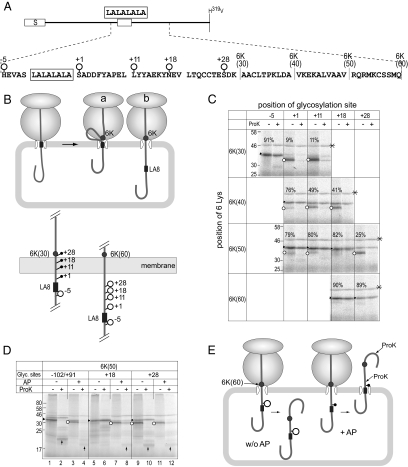
Figure 2.
Distal 6K-cluster induces membrane spanning of the LA8-segment. (A) To examine whether distal downstream 6K-cluster induces stop-translocation, the LA8-segment (rectangle), the 6K-cluster, and glycosylation site were included at the indicated positions of the model proteins. Distances of the 6K-cluster from the LA8-segment are indicated in parentheses. Where indicated, the LA11-segment was included as the H-segment. To generate mobility standards, the termination codon was introduced at the indicated positions (↓). (B) The distal downstream 6K-clusters cause membrane spanning of the LA8-segment. The model proteins were translated in the presence of RM. Aliquots were subjected to ProK treatment. The ProK-resistant fragments (↑) and the monoglycosylated products (▶) are indicated. The experiments were repeated more than three times with each model, and the mean stop-translocation efficiency was calculated and indicated with the SD (bottom panel). (C) Comparison of ProK-resistant fragments with standard polypeptides. Length standards of the LA8-0K and LA8-6K(10) models were translated in the presence of RM. The ProK-resistant fragments were truncated around L179 and E183 and included the 6K(10)-cluster but not the 6K(20)-cluster. (D) Effect of 6K-cluster upstream of the LA8-segment. (E) The downstream 6K-cluster at distance up to 70 residues away can induce membrane spanning of the LA8-segment. ProK degrades only the cytoplasmic segment of a stop-translocated polypeptide. In all cases except the model with 6K(80), the products spanned the membrane by the LA8-segment. Glycosylated potential site (○) and nonglycosylated potential site (shaded dots) are indicated. The model with LA8-6K(80) spanned the membrane via more downstream portion.
Figure 3.
Downstream of the LA8-segment is exposed to the cytoplasmic side. (A) Cysteine scanning model protein to assess membrane topology. Cys-less model proteins included one glycosylation site (N67) and either the LA8-segment or LA11-segment. A single Cys residue and the 6K-cluster were included at the indicated positions. (B) Position dependency of PEGmal reaction. The models were translated in the presence of RM and then subjected to PEGmal treatment. Where indicated, RM was not included in the reaction (w/o RM). The PEGylated form (←), glycosylated form (▶), and nonglycosylated form (○) are indicated. The experiments were repeated two times with each model and the mean PEGylation efficiency was plotted. (C) Reactivity to PEGmal and sensitivity to ProK digestion demonstrate that the LA8-segment spanned the membrane as the LA11-segment.
Figure 4.
LA8-segment is exposed to the lumen. (A) Glycosylation site scanning model protein to probe accessibility for OSTase in the lumen. Glycosylation site was created at one of the indicated positions (○) and 6K-cluster was inserted at one of the indicated sites. (B) Possible topologies of the LA8-segment during stop-translocation; the LA8-segment is statically retained at the membrane translocon (a) or is exposed to the lumen (b) before the 6K-cluster exerts its function. To address these possibilities, potential glycosylation site was scanned. Their glycosylation status of the 6K(30) and 6K(60) models assessed in C are schematically shown. Glycosylated potential site (○) and nonglycosylated potential site (shaded dots) are indicated. (C) The full-length model proteins were expressed in the presence of RM. Aliquots were treated with ProK. Monoglycosylated (▶) and nonglycosylated forms (○) are indicated. Percent monoglycosylation is indicated in the panel. Asterisks denote nonspecific product. (D) Effect of glycosylation inhibition on the translocation. The model proteins included 6K-cluster at position (+50) and potential glycosylation sites at indicated position. Translation of each full-length model protein was performed in the presence or absence of AP. Monoglycosylated forms (▶), nonglycosylated forms (○), and ProK-resistant fragments (↑) are indicated. In these experiments, a high concentration of ProK (400 μg/ml) was used. The product monoglycosylated at position (−102) was ProK-sensitive, whereas that monoglycosylated at position (+18) was completely ProK-resistant. The latter model became ProK-sensitive when the glycosylation was suppressed with AP. (E) The sugar chain at position (+18) induced full translocation. Inhibition of the glycosylation by AP caused membrane spanning of the LA8-segment. Glycosylated potential site (○) and nonglycosylated potential site (shaded dots) are indicated. Sugar chain keeps the LA8-segment in the lumen and prevents cooperation of the LA8-segment and the 6K-cluster to cause forward movement.
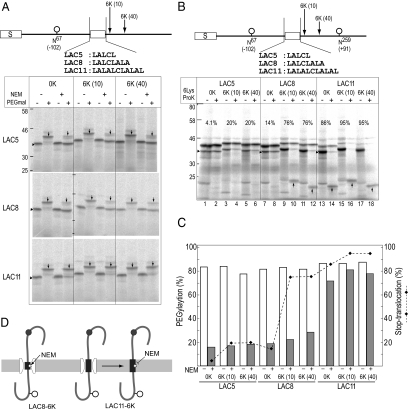
Figure 5.
LA8-segment spans membrane but exists in aqueous environment. (A) To assess water environment of H-segment, a single Cys residue, one glycosylation site (N67), and the 6K-cluster were included at the indicated positions of the Cys-less model protein. After translation in the presence of RM, an aliquot was incubated with NEM under native conditions, where SH-group in an aqueous environment reacts with NEM, and then treated with PEGmal under SDS-denaturing conditions. Only the SH-group integrated in membrane lipid is PEGylated. PEGylated products (↓) and monoglycosylated forms (▶) are indicated. (B) To assess stop-translocation efficiency of the LAC-segments, glycosylation sites were incorporated in the indicated positions. Monoglycosylated products (▶) and the ProK-resistant fragments (↑) are indicated. Stop-translocation percentages are indicated (%). (C) Quantification of PEGylation (%; □, NEM −;  , NEM +) and stop-translocation percent (%; ♦). (D) The LAC8-segment spanned the membrane depending on the 6K-cluster, but the Cys residue (☆) remained in a water-accessible environment, where the SH-group reacted with NEM and thus did not react with PEGmal. In contrast, the LAC11-segment was in a hydrophobic membrane environment, where the Cys residue did not react with NEM and thus reacted with PEGmal reagent.
, NEM +) and stop-translocation percent (%; ♦). (D) The LAC8-segment spanned the membrane depending on the 6K-cluster, but the Cys residue (☆) remained in a water-accessible environment, where the SH-group reacted with NEM and thus did not react with PEGmal. In contrast, the LAC11-segment was in a hydrophobic membrane environment, where the Cys residue did not react with NEM and thus reacted with PEGmal reagent.
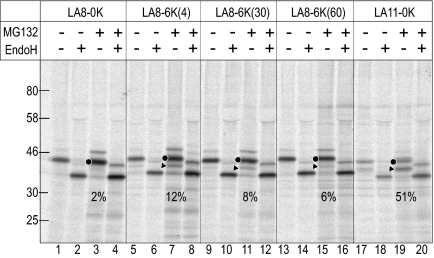
Figure 6.
Function of LA8 and 6K segments in vivo. The HA-tagged model proteins were expressed in COS7 cells and pulse-labeled for 30 min in the absence or presence of the proteasome inhibitor, MG132. The proteins were immunoprecipitated with rabbit polyclonal anti-HA antibodies and analyzed by SDS-PAGE and subsequent image analysis. Where indicated, aliquots were treated with EndoH. Diglycosylated (•) and monoglycosylated (▶) forms are indicated. Percent stop-translocation is shown in each panel (%).
Cell-Free Transcription and Translation
Cell-free transcription and translation were performed essentially as previously described (Sakaguchi et al., 1992), except that translation reactions with RM contained 20 μg/ml castanospermine to prevent heterogeneity of the products due to trimming of the sugar chain. Plasmid DNA was linearized with XhoI and then transcribed with T7-RNA polymerase. The RNA was translated in the reticulocyte lysate cell-free system for 1 h at 30°C in either the absence or presence of RM. The translation reaction included 110 mM potassium acetate (KOAc), 0.8 mM magnesium acetate (Mg[OAc]2), 32% reticulocyte lysate, 20 μg/ml castanospermine, and 15.5 kBq/μl [35S]EXPRESS protein-labeling mix (Perkin Elmer-Cetus, Norwalk, CT). Where indicated, the AP (80 μg/ml) dissolved in DMSO was included. After the translation reaction, aliquots were treated with 200 μg/ml ProK for 1 h on ice. Radiolabeled polypeptide chains were analyzed by SDS-PAGE, visualized with imaging analyzer (BAS1800; Fuji Film, Tokyo, Japan), and quantified using Image Gauge software (Fuji Film). Stop-translocation percentage was estimated as the ratio of the monoglycosylated product among the monoglycosylated and diglycosylated products. In the cases of models with short H-segment, trace amount of monoglycosylated form was ProK-resistant due to glycosylation efficiency. The extent (p) of ProK- resistance among the monoglycosylated one was quantified and (1 − p) value was used to correct the stop-translocation extent.
NEM and PEGmal Reaction Experiments
The translation reaction was performed in the presence of RM for 1 h at 30°C and was terminated with 2 mM cycloheximide. For a direct PEGmal reaction, 1 μl of 100 mM PEGmal dissolved in DMSO was added to the translation mixture (9 μl) to the final concentration of 10 mM PEGmal and incubated at 30°C for 60 min. Then, 20 μl SDS-PAGE sample buffer containing 100 mM DTT was added and incubated for 10 min at 30°C to quench the reaction. After incubation for 5 min at 95°C, the sample was subjected to SDS-PAGE. For sequential reactions with NEM and PEGmal, 0.6 μl of either NEM (100 mM), which was freshly dissolved in 10 mM Tris/Cl (pH 8.0) immediately before use, or the buffer only was added to the translation reaction mixture (9 μl) to 6 mM NEM. After incubation for 60 min at 15°C, the reaction was quenched with 30 mM DTT for 10 min at 15°C. Then, 500 μl of dilution buffer (30 mM HEPES, pH 7.4, 150 mM KOAc, and 2 mM Mg[OAc]2) was added to the mixture, and the mixture was centrifuged for 5 min at 104,000 × gav (S100AT4 rotor, 50,000 rpm; Hitachi, Tokyo, Japan) at 4°C. The obtained membrane precipitates were dissolved in 9 μl of SDS-reducing buffer (50 mM Tris/Cl, pH 8.5, 2% SDS, 2 mM Tris[2-carboxyethyl]phosphine hydrochloride), and heated at 95°C for 5 min. Then, 1.0 μl of PEGmal solution (dissolved in DMSO at 50 mM) was added to a final concentration of 5 mM and incubated for 60 min at 37°C. The reaction was quenched and subjected to SDS-PAGE as described above.
Expression and Pulse Labeling in COS7 Cells
The expression plasmids were transfected into COS7 cells using FuGene 6 reagent (Roche Chemical, Indianapolis, IN). After culture for 24 h, cells were pulse-labeled with 7.25 MBq/ml [35S]EXPRESS protein-labeling mix for 30 min in the absence or presence of the proteasome inhibitor, MG132 (Sigma-Aldrich, Tokyo, Japan). Cells were scraped, heat-treated in 1% SDS buffer, and disrupted by brief sonication. The model proteins were immunoprecipitated with rabbit polyclonal anti-hemagglutinin (HA) antibodies (Covance, Richmond, CA) as previously described (Kanki et al., 2002). Aliquots were treated with endoglycosidase H (EndoH; New England Biolabs, Ipswich, MA) to confirm the glycosylation. The proteins were analyzed by SDS-PAGE and subsequent image analysis.
RESULTS
Positive Charge Mediates Stop-Translocation
For quantitative analysis of stop-translocation through the ER translocon, we designed model proteins containing RSA as a polypeptide backbone, various H-segments in the middle portion, and N-glycosylation sites upstream (Asn67) and downstream (Asn259) of the H-segment (Figure 1A). Each glycosylation site was located 102 residues upstream (−102) and 91 residues downstream (+91) of the H-segment. The H-segment comprised repeated Leu-Ala sequences of various lengths. Translocation of the model is initiated by the N-terminal signal peptide of RSA (Oda et al., 1990). If the polypeptide was completely translocated, both potential glycosylation sites would be glycosylated in the ER lumen (Figure 1B). If the translocation was stopped between the two glycosylation sites, only the upstream site would be glycosylated. ProK treatment degrades the polypeptide chain retained on the cytoplasmic side, but not the translocated regions. When the models were expressed in a reticulocyte lysate cell-free system, single major bands of 32 kDa were observed. When synthesized in the presence of RM, larger forms of 35 and 38 kDa were observed, depending on the length of the H-segment (Figure 1A, ▶ and •). Models with a shorter H-segment (none, LA5, and LA8) gave mainly the 38-kDa form and some of the 35-kDa form. Both of the larger forms were converted to the 32-kDa form by EndoH treatment (data not shown), indicating that the 38- and 35-kDa forms were the diglycosylated and monoglycosylated forms, respectively. With a longer H-segment (LA9 and LA10), there was an increase in the monoglycosylated form. The LA11 model gave the monoglycosylated form as the major product. The percentage of the monoglycosylated form among the monoglycosylated and diglycosylated forms was used to estimate the percent stop-translocation.
When the products were treated with ProK, the diglycosylated form was not degraded, whereas the monoglycosylated forms were largely degraded and ProK-resistant fragments with higher mobility were observed (Figure 1A, ProK + lanes; ↑). ProK-resistant bands were not observed in the presence of the mild detergent Triton X-100 (Figure 1A, lane 20). The diglycosylated product was in the lumen and was protected from ProK degradation by the membrane, whereas the monoglycosylated forms left their C-terminal halves on the cytosolic side. Relative to the monoglycosylated form, the signals of the ProK-resistant fragments were faint because the upstream region of the H-segment contained only one methionine, whereas the downstream region contained four methionines. Taking these methionine numbers into account, the yields of the ProK-resistant fragment were quantitatively reasonable. The ProK-resistant fragment is a good indication of the membrane topology, where the inserted H-segment spans the membrane. Trace amounts of the monoglycosylated forms of the models with the shorter H-segment were resistant to ProK, indicating that they were translocated and became ProK-resistant, but glycosylation was not complete. The ProK-resistance of the monoglycosylated form was quantified and used to compensate the percent stop-translocation (see Materials and Methods for details).
To examine the effect of charged residues in this system, the DD segment two residues downstream of the H-segment was exchanged with two Lys (KK) residues (Figure 1C). The KK residues increased the percent stop-translocation. More ProK-resistant fragments were observed with an increase in KK-induced stop-translocation. Various numbers of Lys residues were then inserted just after the DD segment (Figure 1D). Here, the Lys-clusters were denoted according to the residue number and position; e.g., 2K(4), where 2K indicates 2 Lys residues and (4) indicates 4 residues downstream of the H-segment. Stop-translocation clearly increased depending on the number of inserted Lys residues.
Neither the 6K(4)-cluster nor the LA8-segment alone had remarkable stop-translocation activity (Figure 1E, lanes 5 and 8). When the two were combined, however, there was remarkable stop-translocation activity (Figure 1E, lane 11), indicating that the H-segment and 6K-cluster cooperated to induce efficient stop-translocation. The LA11-segment by itself showed high percent stop-translocation, and adding the 6K(4)-cluster further increased stop-translocation. The ProK-resistant fragments generated from the LA8-6K(4) and LA11 models had similar mobilities, indicating that these models similarly spanned the membrane at the H-segment.
Long-Range Effect of the 6K-Cluster
To examine the effect of the distance between the H-segment and the 6K-cluster, the cluster was scanned at 10-residue intervals (Figure 2A). Surprisingly, all of the downstream 6K-clusters induced stop-translocation (Figure 2B). All of the monoglycosylated forms were sensitive to ProK, whereas the diglycosylated forms were fully resistant. All of the models, except LA8-6K(80), generated ProK-resistant fragments of ∼20 kDa, which was also observed with the LA11–6K(4) model, indicating that the LA8-segments of those models spanned the membrane as the LA11-segment. The model with the 6K(10)-cluster gave larger ProK-resistant fragment than that with the 6K(20)-cluster, suggesting that the ProK cleaved between L178 and V188. Comparison with standard polypeptides allowed us to estimate the ProK-cleavage site (Figure 2C). The ProK-resistant fragment of the LA8-6K(20) model protein was slightly larger than that of the LA8-0K model protein cleaved at L178 and smaller than that cleaved at E183. The ProK-resistant fragment of the LA8-6K(10) model protein was slightly smaller than that of the LA8-6K(10) model cleaved at E183. The 6K-cluster was included in the ProK-resistant fragment of the 6K(10) model, but not in that of the 6K(20) model (Figure 2E). Thus, in all cases except the LA8-8K(80) model, ProK cleaved the model polypeptide around L179-E183, irrespective of the 6K-cluster position. The same experiments were performed with an Arg-cluster and essentially the same results were obtained; the Arg-cluster 60 residues downstream of the LA8-segment induced membrane spanning of the LA8-segment (data not shown). On the other hand, the model protein with the 6K(80)-cluster, in which the 6K-cluster was separated by 80 residues from the LA8-segment, gave a larger ProK-resistant fragment of 28 kDa. It is highly likely that the polypeptide chain spanned the membrane because of a segment downstream of the LA8-segment (Figure 2E). In contrast to the downstream 6K-clusters, the 6K-cluster inserted upstream of the H-segment (6K[-5] and 6K[-10]) caused little or a much smaller increase in the percent stop-translocation (Figure 2D). These findings indicate that the 6K-clusters within the 70 residues downstream of the LA8-segment caused the LA8-segment to span the membrane (Figure 2E).
LA8-Segment Spans the Membrane
To assess the TM topology of the LA8-segment using different criteria, we performed a chemical modification experiment in which the SH-group of the Cys residue was modified with a membrane-impermeable SH-reagent, PEGmal. For this purpose, all Cys residues in the model protein were mutated to Ala to make a Cys-less model polypeptide; a single Cys residue was then created downstream of the LA8-segment by point mutation and the 6K-cluster was inserted (Figure 3A). To easily identify the PEG-modified form, the second glycosylation site was not included. These Cys-mutations had no effect on the stop-translocation behavior (data not shown). When the LA8-0K series was translated in the absence of RM, PEGmal treatment caused the products to shift up by 2 kDa (Figure 3B, LA8-0K w/o RM), indicating that the introduced Cys residues could efficiently react with PEGmal. In the presence of RM, the LA8-0K series models showed low reactivity (Figure 3B, LA8-0K), consistent with the fact that the majority of synthesized polypeptides were sequestered in the ER lumen. The models with the LA11-segment, which had efficient stop-translocation activity, had remarkable reactivity with PEGmal, depending on the Cys position. Because the LA11-segment spanned the membrane, the PEGmal reactivity profile was used as a reference for an authentic TM segment. The PEGmal reactivity of the LA8-6K(40) and LA8-6K(50) models were position-dependent, similar to that of the LA11 model, indicating that the LA8-segment spanned the membrane in the presence of the downstream 6K-cluster in a manner similar to that of the LA11-segment (Figure 3C).
LA8-Segment Is Temporarily Exposed to the Lumen
The ribosome tunnel includes a nascent polypeptide chain of at most 40 amino acid residues (Matlack and Walter, 1995). The 6K-clusters more than 40 residues downstream of the LA8-segment are not yet synthesized when the LA8-segment reaches the translocon pore (Figure 4B). When the 6K-cluster reaches the translocon, the LA8-segment can be exposed to the lumen. We next examined whether the LA8-segment is released into the ER lumen during the positive charge–mediated stop-translocation process (Figure 4). A single glycosylation site was created around the LA8-segment (Figure 4, A and B). The full-length models were expressed in the presence of RM. In the case of the LA8-6K(30) model, position (−5) was highly glycosylated (Figure 4C, ▶), whereas position (+1) was only slightly glycosylated (Figure 4C, ○). Based on a previous report that a potential glycosylation site separated by 15 residues from the membrane is accessible to OSTase (Nilsson and von Heijne, 1993; Popov et al., 1997), glycosylation of position (−5) indicated full exposure of the LA8-segment to the lumen. Position (+1) of the LA8-6K(40) model was highly glycosylated. Position (+11) and position (+18) of the LA8-6K(50) model were accessible to OSTase, whereas the position (+28) glycosylation level was low. Furthermore, when the 6K-cluster was further shifted downstream by 10 residues, even position (+28) became glycosylated, as observed with the LA8-6K(60) model. These results indicate that the LA8-segment and its downstream sequence were exposed to the lumen and accessible to OSTase (Figure 4B, bottom panel). The extent of the exposure depended on the position of the 6K-cluster.
Unexpectedly, the products glycosylated at flanking or downstream potential sites were fully ProK-resistant in this assay (Figure 4C, ▶), indicating that they were fully translocated into the lumen even in the presence of both the LA8-segment and the 6K-cluster. In contrast, the products glycosylated at position (−102) were ProK-sensitive (Figures 2B and 4D, lanes 1 and 2, ▶). It is highly possible that the sugar chain flanking or downstream of the LA8-segment prevented the LA8-segment from sliding back into the membrane, whereas that in the distal upstream position did not. To examine the contribution of the sugar chain, translation was performed in the presence of AP, which inhibits glycosylation by OSTase (Geetha-Habib et al., 1990; Figure 4D). Glycosylation of the LA8-6K(50) model with the potential sites (−102/+91) was largely suppressed (Figure 4D, lanes 3 and 4, ○). The product was largely ProK-sensitive and a nonglycosylated ProK-resistant fragment was observed (lane 4, ↑). The stop-translocation behavior of this model was not affected by AP. Similarly, the model with the potential site at position (+18) was largely degraded by ProK in the presence of AP, and the ProK-resistant fragment was not glycosylated (Figure 4D, lanes 7 and 8), although it was efficiently glycosylated and entirely resistant to ProK in the absence of AP (Figure 4D, lanes 5 and 6). The full translocation of this model was strongly inhibited by AP and the product spanned the membrane. We thus concluded that the sugar chain at position (+18) induced full translocation. The model with potential site (+28) was glycosylated as much as the model with potential sites (−102/+91) was diglycosylated, indicating that the models were stop-translocated to a similar extent. The sugar chain at position (+18) promoted translocation of the polypeptide chain, whereas the sugar chain at position (−102) did not. The sugar chain at position (+18) prevented the LA8-segment from sliding back into the membrane and mediated the forward movement in a position-dependent manner (Figure 4E). These results suggested that sugar chains at specific positions can modulate the membrane topology.
LA8-Segment Spans Membrane But Remains in an Aqueous Environment
The LA8-segment spanned the membrane depending on the downstream positive charges. We next examined whether the LA8-segment existed in the hydrophobic environment of the membrane or in the water accessible environment. We utilized the chemical reaction of the SH-group with maleimide whose reactivity depends on the water environment (Koide et al., 2007). Using the Cys-less model protein, a single Cys residue was created in the H-segment, the second glycosylation site was silenced, and the 6K-cluster was inserted as indicated (Figure 5A). After translation in the presence of RM, the product was treated with the SH-reagent NEM, which is membrane permeable and does not react with the SH-group in a lipid environment. After the NEM reaction, membranes were solubilized with SDS and then the residual SH-group was reacted with PEGmal. In this sequential treatment, only the SH-group in the membrane lipid reacted with the second PEGmal reagent. In the absence of the first NEM reaction, all the models efficiently reacted with PEGmal and their molecular weight increased by 2 kDa (Figure 5A, ↓). The PEGmal reactivity of the LAC5- and LAC8-models was greatly reduced by the preceding NEM treatment (Figure 5, A and C, NEM + lanes), indicating that the LAC5- and LAC8-segments were in a water-accessible environment. On the other hand, the PEGmal reactivity of the LAC11-model was little affected by the preceding NEM treatment, indicating that the SH-group of the LAC11-segment was in a lipid environment.
The contribution of the LAC-segments to stop-translocation was examined using models with two glycosylation sites (Figure 5B). The LAC5-model was efficiently diglycosylated. Percent stop-translocation was low but was significantly increased by the 6K-cluster (Figure 5, B and C). The diglycosylated forms were resistant to ProK and no ProK-resistant smaller fragments were observed. The LAC5-segment was translocated into the lumen and reactive with NEM and thus showed low reactivity with PEGmal. On the other hand, the stop-translocation of the model protein with the LAC8 segment was greatly enhanced by the 6K(10)- and 6K(40)-clusters (Figure 5, B and C), and the ProK-resistant fragment was significantly visible only in the presence of the 6K-cluster. We concluded that the LAC8-segment spanned the membrane but remained in the water-accessible environment. In contrast, the LAC11 segment stopped translocation by itself, irrespective of the 6K-cluster (Figure 5, B and C). The Cys in the LAC11 segment showed high reactivity with PEGmal despite the preceding NEM treatment, indicating that it was integrated in a fully hydrophobic environment.
Function of LA8 and 6K Segments In Vivo
To examine behavior of the model proteins in vivo, the model proteins were C-terminally HA-tagged, transiently expressed in the COS7 cells, and pulse-labeled for 30 min in the absence or presence of a proteasome inhibitor, MG132. The models were immunoprecipitated with anti-HA antibodies (Figure 6). The LA8-0K model was little monoglycosylated, but largely diglycosylated, indicating that LA8-0K model was fully translocated into ER lumen. On the other hand, significant amount of the monoglycosylated forms were observed with the LA8-6K(4), LA8-6K(30), and LA8-6K(60) model proteins in the presence of MG132 (Figure 6, lanes 7, 11, and 15). The diglycosylated forms and monoglycosylated forms were confirmed by EndoH treatment. The LA11-segment induced 50% stop-translocation in vivo, suggesting that the behaviors of the H-segment somewhat differ in vitro and in vivo; the LA11-segment is more easily translocated through translocon in the cells than in vitro. Even though the monoglycosylated forms of the LA8-6K model proteins, which are retained in the water environment of translocon, should be extremely unstable, the significant accumulation of the monoglycosylated forms were observed. These results demonstrated that the sequence elements, the LA8-segment and the 6K-cluster at distal downstream positions, can cooperate and induce the membrane spanning of the product in vivo.
DISCUSSION
Positively charged amino acid residues have long been recognized as topology determinants of membrane proteins. Here we demonstrated that these residues have more dynamic actions during stop-translocation of nascent polypeptide chains than previously recognized. The residues mediate membrane spanning of a marginal H-segment, not only at its downstream flanking region but also at distal downstream positions. In the latter case, the marginal H-segment is exposed to the lumen, and the positive charges allow the segment to slide back to the membrane. From more than 60 residues downstream, the cluster of positive charges induces the TM topology of the marginal H-segment (LA8-segment), as observed with the LA8-6K(60) model protein (Figure 7). This unexpected stop-translocation process includes the following dynamic events. At most, 40 residues of the nascent polypeptide chain fit inside the ribosome tunnel. When the LA8-segment enters the translocon during the ongoing cotranslational translocation, the 6K-cluster is not yet synthesized (A). A polypeptide chain of 10 residues in an expanded conformation as well as a polypeptide chain of 20 residues in an α-helical conformation spans the membrane (Mingarro et al., 2000). When the 6K-cluster is synthesized in the ribosome, the LA8-segment can pass through the translocon into the lumen (B1) or remains in the translocon (B2). Then, the polypeptide chain elongates and the position (+28) glycosylation site, which is 28 residues downstream of the LA8-segment, is accessible to OSTase (C1). Although the LA8-segment might decrease the translocation rate and remain at the translocon (C2), it is not statically retained in the membrane, but is exposed to the lumen for a significant period of time (C1). For access to OSTase, the potential site should be at least 15 residues away from the membrane (Nilsson and von Heijne, 1993; Popov et al., 1997). Thus, more than 42 residues after the LA8-segment should be exposed to the lumen, although the conformational details are unknown. At this stage, the 6K-cluster should be near the ribosome exit or the translocon entrance. Despite such a large exposure, the LA8-segment eventually spans the membrane (D).
Figure 7.
Possible geometry of the LA8-6K(60) model protein during stop-translocation. (A) When the LA8-segment is at the translocon during cotranslational translocation, the 6K(60)-cluster is not yet synthesized. (B1 and B2) When the 6K-cluster is synthesized in the ribosome, the LA8-segment can be exposed to the lumen or retain in the translocon pore. (C1 and C2) The polypeptide chain elongates and the position (+28) is accessible to OSTase (C1). At this stage, the 6K-cluster is near the ribosome-translocon interface. Although the LA8-segment might be retained in the translocon (C2), at least 43 residues after the LA8-segment are exposed to the lumen for a significant period of time (C1). The extent of exposure to the lumen depends on the position of the 6K-cluster. The 6K-cluster triggers the accommodation of the LA8-segment in the translocon (C2). Possibly, the 6K-cluster transiently arrests the cotranslational translocation and allow the nascent chain to move in and out of translocon. Likely, the 6K-cluster and LA8-segment are sensed simultaneously by translocon. The cluster stabilize the slide backed LA8-segment in a TM topology. (D) Finally, the LA8-segment spans the membrane to allow the downstream portion to be accessible to ProK and PEGmal. The segment remains in water environment despite of TM topology. Numbers indicate the number of amino acid residues in the indicated region. Conformations of polypeptide chains are not taken into account.
Positive charges enhance the stop-translocation of the polypeptide chain translocating through the translocon as well as determine the orientation of the signal sequences being inserted into the translocon. These topogenic functions are reflected by the positive-inside rule (von Heijne and Gavel, 1988; Sipos and von Heijne, 1993), and have been demonstrated in numerous experiments (e.g., Kuroiwa et al., 1991; Kida et al., 2006; Lerch-Bader et al., 2008). These functions can be simply explained by their inhibitory effect on polypeptide chain movement through the translocon. Our finding indicates that the resistance not only fixes the cytoplasmic portion of the TM segment on the membrane, but also allows the polypeptide chain to slide back to the translocon from the lumen. The geometry of the H-segment and positive charges on the membrane leads us to propose sequential actions of the stop-translocation effectors, the H-segment and the positive charges. Initial insertion of the polypeptide chain into the translocon is driven by the motive force of the signal sequence (Kida et al., 2009). After the initiation of cotranslational translocation, chain elongation from the ribosome is probably a major motive force for the constitutive vectorial movement of the polypeptide chain (Figure 7A). The marginal H-segment is insufficient to interrupt the ongoing polypeptide chain movement in the presence of the elongation motive force and passes through the translocon (B1). The extent of the exposure in the lumen depends on the position of the positive charges (C1). When the positive charges reach the membrane, they should tentatively arrest the elongation-driven movement (C1). While the forward movement is arrested, the upstream polypeptide chain might be able to move back-and-forth via Brownian motion (Figure 7, C1 and C2). During such dynamic movement, the translocon pore scans the polypeptide chain and accommodates the marginal H-segment on balance (D). At this stage, the positive charges and the H-segment should be simultaneously sensed by the translocon. The charges might somehow increase the stability of the H-segment in the translocon to cause membrane spanning of the H-segment. Alternatively, the positive charges might increase the affinity of the translocon for the marginal H-segment. It is likely that the positive charges exert their unique function via electrostatic interaction with translocon subunits, rRNA, and/or membrane lipid. In the absence of the LA8-segment, there is no stable position of our model protein within a certain upstream window, and thus the positive charges would be eventually translocated by the Brownian motion of the chain. In the case of the model with the 6K(80)-cluster, a different segment just upstream of the 6K(80) should be accommodated in the translocon instead of the LA8-segment (Figure 2).
We initially hypothesized an alternative possibility that the LA8-segment is statically retained in the translocon and waits for the action of the subsequent positive charges. This, however, is not the case; the LA8-segment is exposed to the lumen for a specific period of time. Glycosylation at position (+28) indicated that a large portion of the downstream polypeptide chain of the LA8-segment was exposed to the lumen. Furthermore, efficient glycosylation at positions (−5) and (+1) indicates that the entire LA8-segment was exposed to the lumen. Adding a sugar chain to certain positions caused full translocation, even in the presence of the 6K-cluster and the H-segment. A sugar chain distal upstream of the H-segment, however, did not have such an effect. It is highly likely that the sugar chain caused steric hindrance, prevented the H-segment from sliding back into the translocon, and blocked the cooperation between the 6K-cluster and the LA8-segment. The sugar chain can therefore be thought of as a position-specific ratchet.
Our systematically designed model proteins demonstrated the unexpected effect of distal positive charges and the long range cooperation between the H-segment and downstream positive charges, which induce the polypeptide chain to slide back into the translocon from the ER lumen. The extent of such a back sliding motion is likely dependent on the experimental system as well as various conditions; e.g., chaperone activity, translocation rate, and folding efficiency. When the same constructs were examined in cultured cells, the 6K-cluster 4, 30, and 60 residues downstream of the LA8-segment caused significant stop-translocation, although to a lesser extent than in the cell-free system (Figure 6). The stop-translocation efficiency of the LA11-segment in cultured cells was also lower than that in the cell-free system. The long-range cooperation is likely to be operative to varying degrees in vivo and in vitro. Anyway, our finding implies that the translocon can sense structural information in a translocating polypeptide chain spreading over 60 residues in vitro and in vivo. Chaperone binding and folding of the polypeptide chain in the lumen should likely prevent the sliding back to the membrane. The findings from the LA8-6K(80) model protein suggest this possibility; the segment between positions (+70) and (+80) might cause specific folding and/or binding to a lumenal chaperone. We thus propose that when the positive charges transiently arrest the forward translocation, backsliding may occur up to a point where something obstructs further movement and that this may or may not be the marginal H-segment. If there is no significant stable position, the polypeptide chain should eventually be translocated into the lumen. Studies of such backsliding determinants are in progress in our laboratory.
We observed an intermediate state of membrane insertion in which a marginal H-segment spanned the membrane depending on the downstream 6K-cluster but still existed in a water accessible environment. The model with the 6K(80)-cluster spanned the membrane via a nonhydrophobic segment upstream of the cluster. The physicochemical nature required for stop-translocation does not seem to be identical to that required for membrane insertion. Actually, the H-segment alone is sufficient for stop-translocation if it is significantly hydrophobic. The membrane partition of the polypeptide chain is a simple determinant of membrane insertion. A segment consisting of only 9 Leu residues stopped ongoing translocation, whereas 18 Ala residues were required for similar stop-translocation (Kuroiwa et al., 1991). The hydrophobic nature reflects the biological hydrophobicity scale of each amino acid residue, which is estimated by quantitative analysis of translocon-mediated membrane insertion (Hessa et al., 2007). The second stop-transfer effecter of the positive charge strongly contributes to stop-translocation processes but contributes little to membrane insertion. The stop-transfer step must then be recognized as one of several sequential steps required for membrane insertion. In the case of single-spanning membrane proteins, the marginal H-segment cannot be normally included in TM-segment because it should be retained in the translocon and extremely unstable. Consequently, the TM segments of native single-pass proteins are usually fairly hydrophobic. On the other hand, in the case of multispanning membrane protein biogenesis, the marginal H-segment might be tentatively retained in the translocon or its surrounding sites (Sadlish et al., 2005) and then assembled with other hydrophobic TM segments. The assembly forms hydrophobic TM bundles and is laterally released into the membrane (Skach, 2009). The large water environment of the translocon would mediate such a dynamic assembly (Borel and Simon, 1996; Alder and Johnson, 2004; Kida et al., 2007). The unexpectedly flexible nature of the multimeric translocon pores, through which two translocating polypeptide chains can be accommodated, may be key to the assembly (Kida et al., 2007; Skach, 2007).
The findings of the present study suggest that long-range retrograde movement induced by distal downstream positive charges should be considered during membrane protein integration and that the presence of a sugar chain at a critical position has drastic effects on the final membrane topology of multispanning membrane protein. The N-terminal signal-anchor sequence can be reoriented by its downstream TM segment, and such reorientation is prevented by glycosylation of the segment initially exposed to the lumen (Goder et al., 1999). Translocation intermediates of secretory proteins whose C-terminus is retained in the ribosome can be released from the RM into the cytosol. The release is also prevented by glycosylation in the lumen (Ooi and Weiss, 1992). A specific TM segment (TM2) of the multispanning membrane protein, aquaporin-1, is exposed to the lumen once and then reoriented into the membrane via protein folding (Skach, 2009). A membrane embedded region of Band3 protein is exposed to the lumen where it becomes accessible to OSTase once and is then inserted into the membrane (Kanki et al., 2002). It has also been suggested that a nascent chain TM segment can be drawn back from the translocon during protein synthesis (Daniel et al., 2008). The sugar chain functions as a ratchet in the ER lumen to retain the polypeptide segment during such dynamic movement of the nascent chain. We propose that the unexpectedly wide-range distributions and combinations of sugar chains, positive charges, and marginal H-segments are critical structural determinants of multispanning membrane proteins.
ACKNOWLEDGMENTS
We thank Mrs. Y. Sakaguchi and M. Tsukuda for excellent technical assistance. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Global COE program; the Sumitomo Foundation; and the Hyogo Science and Technology Association.
Abbreviations used:
- AP
glycosylation acceptor peptide
- EndoH
endoglycosidase H
- ER
endoplasmic reticulum
- H-segment
hydrophobic segment
- NEM
N-ethylmaleimide
- OSTase
oligosaccharyl transferase
- PEGmal
polyethylene glycol-maleimide
- ProK
proteinase K
- RM
rough microsomal membrane
- RSA
rat serum albumin
- TM
transmembrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-12-1060) on April 28, 2010.
REFERENCES
- Alder N. N., Johnson A. E. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J. Biol. Chem. 2004;279:22787–22790. doi: 10.1074/jbc.R400002200. [DOI] [PubMed] [Google Scholar]
- Borel A. C., Simon S. M. Biogenesis of polytopic membrane proteins: membrane segments assemble within translocation channels prior to membrane integration. Cell. 1996;85:379–389. doi: 10.1016/s0092-8674(00)81116-2. [DOI] [PubMed] [Google Scholar]
- Cross B. C., Sinning I., Luirink J., High S. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- Daniel C. J., Conti B., Johnson A. E., Skach W. R. Control of translocation through the Sec61 translocon by nascent polypeptide structure within the ribosome. J. Biol. Chem. 2008;283:20864–20873. doi: 10.1074/jbc.M803517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha-Habib M., Park H. R., Lennarz W. J. In vivo N-glycosylation and fate of Asn-X-Ser/Thr tripeptides. J. Biol. Chem. 1990;265:13655–13660. [PubMed] [Google Scholar]
- Goder V., Bieri C., Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J. Cell Biol. 1999;147:257–266. doi: 10.1083/jcb.147.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V., Spiess M. Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 2001;504:87–93. doi: 10.1016/s0014-5793(01)02712-0. [DOI] [PubMed] [Google Scholar]
- Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- High S., Dobberstein B. Mechanisms that determine the transmembrane disposition of proteins. Curr. Opin. Cell Biol. 1992;4:581–586. doi: 10.1016/0955-0674(92)90075-n. [DOI] [PubMed] [Google Scholar]
- Jackson R. J., Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Jaud S., Fernández-Vidal M., Nilsson I., Meindl-Beinker N. M., Hübner N. C., Tobias D. J., von Heijne G., White S. H. Insertion of short transmembrane helices by the Sec61 translocon. Proc. Natl. Acad. Sci. USA. 2009;106:11588–11593. doi: 10.1073/pnas.0900638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Sakaguchi M., Kitamura A., Sato T., Mihara K., Hamasaki N. The tenth membrane region of band 3 is initially exposed to the luminal side of the endoplasmic reticulum and then integrated into a partially folded band 3 intermediate. Biochemistry. 2002;41:13973–13981. doi: 10.1021/bi026619q. [DOI] [PubMed] [Google Scholar]
- Kida Y., Mihara K., Sakaguchi M. Translocation of a long amino-terminal domain through ER membrane by following signal-anchor sequence. EMBO J. 2005;24:3202–3213. doi: 10.1038/sj.emboj.7600788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y., Morimoto F., Mihara K., Sakaguchi M. Function of positive charges following signal-anchor sequences during translocation of the N-terminal domain. J. Biol. Chem. 2006;281:1152–1158. doi: 10.1074/jbc.M506613200. [DOI] [PubMed] [Google Scholar]
- Kida Y., Morimoto F., Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J. Cell Biol. 2007;179:1441–1452. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y., Morimoto F., Sakaguchi M. Signal-anchor sequence provides motive force for polypeptide-chain translocation through the endoplasmic reticulum membrane. J. Biol. Chem. 2009;284:2861–2866. doi: 10.1074/jbc.M808020200. [DOI] [PubMed] [Google Scholar]
- Kida Y., Sakaguchi M., Fukuda M., Mikoshiba K., Mihara K. Membrane topogenesis of a type I signal-anchor protein, mouse synaptotagmin II, on the endoplasmic reticulum. J. Cell Biol. 2000;150:719–730. doi: 10.1083/jcb.150.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide K., Maegawa S., Ito K., Akiyama Y. Environment of the active site region of RseP, an Escherichia coli regulated intramembrane proteolysis protease, assessed by site-directed cysteine alkylation. J. Biol. Chem. 2007;282:4553–4560. doi: 10.1074/jbc.M607339200. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T., Sakaguchi M., Mihara K., Omura T. Structural requirements for interruption of protein translocation across rough endoplasmic reticulum membrane. J. Biochem. 1990;108:829–834. doi: 10.1093/oxfordjournals.jbchem.a123288. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T., Sakaguchi M., Mihara K., Omura T. Systematic analysis of stop-transfer sequence for microsomal membrane. J. Biol. Chem. 1991;266:9251–9255. [PubMed] [Google Scholar]
- Lerch-Bader M., Lundin C., Kim H., Nilsson I., von Heijne G. Contribution of positively charged flanking residues to the insertion of transmembrane helices into the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2008;105:4127–4132. doi: 10.1073/pnas.0711580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack K.E.S., Walter P. The 70 carboxyl-terminal amino acids of nascent secretory proteins are protected from proteolysis by the ribosome and the protein translocation apparatus of the endoplasmic reticulum membrane. J. Biol. Chem. 1995;270:6170–6180. doi: 10.1074/jbc.270.11.6170. [DOI] [PubMed] [Google Scholar]
- Mingarro I., Nilsson I., Whitley P., von Heijne G. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 2000;1:3. doi: 10.1186/1471-2121-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I. M., von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- Oda K., Fujiwara T., Ogata S., Ikehara Y. Production of propeptide-directed antibody: its application to the purification of proalbumin and analysis of proalbumin processing. J. Biochem. 1990;108:549–553. doi: 10.1093/oxfordjournals.jbchem.a123240. [DOI] [PubMed] [Google Scholar]
- Ooi C. E., Weiss J. Bidirectional movement of a nascent polypeptide across microsomal membranes reveals requirement for vectorial translocation of proteins. Cell. 1992;71:87–96. doi: 10.1016/0092-8674(92)90268-h. [DOI] [PubMed] [Google Scholar]
- Ota K., Sakaguchi M., Hamasaki N., Mihara K. Assessment of topogenic functions of anticipated transmembrane segments of human band 3. J. Biol. Chem. 1998;273:28286–28291. doi: 10.1074/jbc.273.43.28286. [DOI] [PubMed] [Google Scholar]
- Plath K., Mothes W., Wilkinson B. M., Stirling C. J., Rapoport T. A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Popov M., Tam L. Y., Li J., Reithmeier R.A.F. Mapping the ends of transmembrane segments in a polytopic membrane protein. J. Biol. Chem. 1997;272:18325–18332. doi: 10.1074/jbc.272.29.18325. [DOI] [PubMed] [Google Scholar]
- Rapoport T. A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Sadlish H., Pitonzo D., Johnson A. E., Skach W. R. Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M. Mutational analysis of signal-anchor and stop-transfer sequences in membrane proteins. In: von Heijne G., editor. Membrane protein assembly. Austin, TX.: R. G. Landes Company; 1997. pp. 135–150. [Google Scholar]
- Sakaguchi M., Hachiya N., Mihara K., Omura T. Mitochondrial porin can be translocated across both endoplasmic reticulum and mitochondrial membranes. J. Biochem. 1992;112:243–248. doi: 10.1093/oxfordjournals.jbchem.a123884. [DOI] [PubMed] [Google Scholar]
- Sato Y., Sakaguchi M., Goshima S., Nakamura T., Uozumi N. Integration of Shaker-type K+ channel, KAT1, into the endoplasmic reticulum membrane: synergistic insertion of voltage-sensing segments, S3–S4 and independent insertion of pore-forming segments, S5-P-S6. Proc. Natl. Acad. Sci. USA. 2002;99:60–65. doi: 10.1073/pnas.012399799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos L., von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur. J. Biochem. 1993;213:1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- Skach W. R. The expanding role of the ER translocon in membrane protein folding. J. Cell Biol. 2007;179:1333–1335. doi: 10.1083/jcb.200711107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach W. R. Cellular mechanisms of membrane protein folding. Nat. Struct. Mol. Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T., et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B., Clemons W. M., Jr, Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- von Heijne G., Gavel Y. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Preparation of microsomal membranes for co-translational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- Whitley P., Nilsson I. M., von Heijne G. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 1996;271:6241–6244. doi: 10.1074/jbc.271.11.6241. [DOI] [PubMed] [Google Scholar]
- Yost C. S., Hedgpeth J., Lingappa V. R. A stop transfer sequence confers predictable transmembrane orientation to a previously secreted protein in cell-free systems. Cell. 1983;34:759–766. doi: 10.1016/0092-8674(83)90532-9. [DOI] [PubMed] [Google Scholar]