Abstract
Terreic acid is a metabolite with antibiotic properties produced by the fungus Aspergillus terreus. We found that terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme MurA from E. cloacae and E. coli in-vitro. The crystal structure of the MurA dead-end complex with terreic acid revealed that the quinine ring is covalently attached to the thiol group of Cys115, the molecular target of the antibiotic fosfomycin. Kinetic characterization established that the inactivation requires the presence of substrate UNAG (UDP-N-acetylglucosamine), proceeding with an inactivation rate constant of kinact = 130 M−1s−1. Although the mechanisms of inactivation are similar, fosfomycin is approximately 50 times more potent than terreic acid, and the structural consequences of covalent modification by these two inhibitors are fundamentally different. The MurA-fosfomycin complex exists in the closed enzyme conformation, with the Cys115-fosfomycin adduct buried in the active site. In contrast, the dead-end complex with terreic acid is open, free of UNAG, and has the Cys115-terreic acid adduct solvent–exposed. It appears that terreic acid reacts with Cys115 in the closed, binary state of the enzyme, but that the resulting Cys115-terreic acid adduct imposes steric clashes in the active site. As a consequence, the loop containing Cys115 rearranges, the enzyme opens and UNAG is released. The differential kinetic and structural characteristics of MurA inactivation by terreic acid and fosfomycin reflect the importance of non-covalent binding potential, even for covalent inhibitors, to ensure inactivation efficiency and specificity.
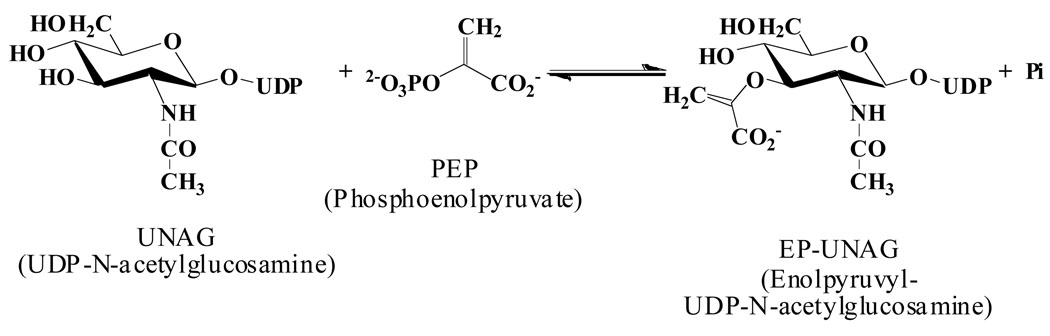
The survival of most bacteria depends on the functionality of the cytosolic enzyme MurA (UDP-N-acetylglucosamine 1-carboxyvinyltransferase, EC 2.5.1.7), which catalyzes the first step in the biosynthesis of the bacterial cell wall (Fig. 1). MurA is an established antibiotic target, but only the natural product fosfomycin [(1R,2S)-1,2-epoxypropyl phosphonic acid], a broad-spectrum bactericidal antibiotic produced by Streptomyces sp., is known to selectively inhibit this enzyme (1). Unfortunately, a growing number of pathogenic bacteria have developed resistance toward fosfomycin through multiple mechanisms (2–4). Hence, there is a critical need for the development of new inhibitors targeting MurA as potential antibiotics in the treatment of a broad range of bacterial infections.
FIGURE 1. Reaction catalyzed by MurA.
The reaction of MurA with substrates UDP-N-acetylglucosamine (UNAG) and phosphoenolpyruvate (PEP) proceeds through an induced-fit mechanism with large conformational changes in the two-domain structure of the enzyme (5, 6). The unliganded “open” enzyme state interacts first with UNAG, forming a binary “closed” state to which the second substrate, PEP, binds. In the course of the open-closed transition, a 12-residue loop containing Cys115 undergoes drastic conformational changes, positioning the Cys115 side chain towards the PEP-binding site. Fosfomycin inactivates MurA by covalent attachment to Cys115 (1, 6–8). The precise role of Cys115 in catalysis is not well understood. Mutating this residue to Asp115 renders the enzyme both active and tolerant to fosfomycin (9), while the Ser115 mutant enzyme is only capable of catalyzing a single-turnover reaction (10). It is unclear if Cys115 participates in the chemical reaction directly, or if the primary role is to facilitate PEP binding and/or product release.
Terreic acid is a metabolite produced by the fungus Aspergillus terreus. The antibiotic properties of terreic acid were recognized more than 60 years ago (11), but its cellular and molecular modes of action remained obscure (12). Chemically, terreic acid is a quinone epoxide, therefore sharing with fosfomycin a potential reactivity towards nucleophiles, such as Cys115 in MurA. This prompted us to study the inhibitory potential of terreic acid toward MurA from E. cloacae and E. coli. We found that terreic acid inactivates MurA by covalently attaching to Cys115. The mechanism of action of terreic acid on MurA is similar to that of fosfomycin, although terreic acid is about 50-fold less potent. The differential inhibitory potency of these inhibitors is reflected by the distinct structural characteristics of the respective dead-end complexes with the enzyme. Implications from these findings for the rational design of novel MurA inhibitors are discussed.
EXPERIMENTAL PROCEDURES
Materials
Chemicals and reagents were purchased from Sigma Aldrich (St. Louis, MO) and Hampton Research (Aliso Viejo, CA) unless otherwise noted. Terreic acid was obtained from Tocris Bioscience (Ellisville, MO). Cloning and overexpression of E. cloacae MurA and the Cys115Asp mutant enzyme have been described (13). E. coli MurA was sub-cloned from E.coli strain K12 genomic DNA (ATCC, Manassas, VA), inserted into the pET41a vector (Novagen, Merck KGaA, Darmstadt, DE) and overexpressed in E. coli strain BL21(DE3). Purification of MurA was performed as previously described (14). Protein concentration was determined using the Coomassie reagent from BioRad (Hercules, CA) with bovine serum albumin as a standard. Non-linear regression analysis for inhibition kinetics was performed using SigmaPlot (Systat Software, Inc., San Jose, CA).
Inhibition kinetics
MurA activity was assayed in 96-well plates on a Spectra-Max 340PC plate reader (Molecular Devices, Sunnyvale, CA). The amount of inorganic phosphate produced in the forward reaction with UNAG and PEP was determined using malachite green (15). The change in optical density at 650 nm was compared to phosphate standards, and the enzymatic activity was expressed as micromoles of phosphate produced per minute of reaction per milligram of enzyme (U/mg). All inactivation studies were performed in the absence of reducing agents such as dithiothreitol (DTT) or β-mercaptoethanol. MurA (5.0 µM) was first incubated with varying concentrations of UNAG and terreic acid or fosfomycin; at time intervals aliquots (10 uL) were assayed for the MurA residual activity. The assay mixture (100 µl) contained 50 mM HEPES (pH 7.5), 0.5 µM MurA, 1 mM PEP and 1 mM UNAG. Control experiments were performed under the same conditions. Residual activity was plotted as a function of incubation time (t), with data fit to equation (1), where kobs is the observed first order rate constant of inactivation at a single concentration of inhibitor and UNAG. Data sets were evaluated by plotting kobs values vs. inhibitor concentration (I) and fitting the data to equation (2), where kinact equals the inactivation rate constant at a single UNAG concentration. The overall inactivation constant (k*inact) was determined by fitting data to equation (3), where Kd(S1) is the dissociation constant of the MurA-UNAG complex. IC50 values were determined by fitting data to equation (4), where A is the relative activity remaining, [I] is the concentration of inhibitor, and n is the Hill slope.
| Equation(1) |
| Equation(2) |
| Equation(3) |
| Equation(4) |
Protein crystallography
Crystallization was performed at 19°C using the hanging drop vapor-diffusion method. Crystals of the MurA-terreic acid complex were grown from a solution of 10 mg/mL E. cloacae MurA in the presence of 2.5 mM UNAG and 2.5 mM terreic acid with 50 mM HEPES/NaOH (pH 7.5), and 15% (v/v) polyethylene glycol 400; no cryo-protectant was added for data collection. Crystals of the MurA-fosfomycin complex were grown from a solution of 50 mg/mL E. cloacae MurA in the presence of 2.5 mM UNAG and 2.5 mM fosfomycin, with 12.5 mM MES/NaOH (pH 6.2), 25 mM Na/K phosphate buffer, and 6% (w/v) polyethylene glycol 20,000; for data collection, 20% (v/v) glycerol was added as a cryoprotectant. X-ray diffraction data were recorded at −180°C using the rotation method on single flash-frozen crystals [detector: Rigaku HTC image plate; rotating anode: Rigaku Micro-Max 007-HF (CuKα, focused by mirror optics)]. Data were processed with XDS (16) or HKL2000 (HKL Research, Inc., Charlottesville, VA). The structures were determined by molecular replacement using CNS (17). For the MurA-terreic acid complex, the coordinates of E. cloacae unliganded MurA type II ((18); PDB entry 1EJC) served as a search model; the MurA-UNAG-fosfomycin complex was solved using the coordinates of the E. coli MurA-fosfomycin complex ((10); PDB entry 1UAE). Refinement cycles were performed using data to the highest resolution with no sigma cut-off applied. Several rounds of minimization, simulated annealing (2500 K starting temperature) and restrained individual B-factor refinement were carried out. Model building was performed with O (19). Data collection and refinement statistics are summarized in Table 1. Figures 3–5 were produced with Pymol (DeLano Scientific, Palo Alto, CA).
TABLE 1.
Summary of data collection and structure refinement for the dead-end complexes of E. cloacace MurA with terreic acid and fosfomycin*.
| MurA-terreic acid | MurA-fosfomycin | |
|---|---|---|
| Data collection | ||
| Space group | P43212 | P212121 |
| Unit cell dimensions (Å) | a=b=115.3 c=277.4 | a=64.4 b=61.8 c=134.4 |
| α=β=γ=90° | α=β=γ=90° | |
| Resolution range (Å) | 12-2.25 (2.3-2.25) | 20-1.75 (1.79-1.75) |
| Unique reflections | 88863 (5612) | 46694 (3050) |
| Completeness (%) | 99.7 (100.0) | 99.4 (98.9) |
| I/σI | 23.4 (27.9) | 11.7 (2.2) |
| Rmergeb (%) | 4.9 (17.5) | 8.4 (35.4) |
| Structure refinement | ||
| Protein atoms | 4×3,154 | 3,151 |
| Average B-factor (Å2) | 30.5 | 20.2 |
| Inhibitor atoms | 4×11 at Cys115 | 8 at Cys115 |
| Average B-factor (Å2) | 31.5 | 15.6 |
| UNAG atoms | n/a | 39 |
| Average B-factor (Å2) | 16.9 | |
| Solvent molecules | 526 | 523 |
| average B-factor (Å2) | 30.1 | 30.9 |
| rmsda bonds (Å) | 0.011 | 0.012 |
| rmsd angles (°) | 1.6 | 1.6 |
| Rcrystc (%) | 19.6 | 16.7 |
| Rfreed (%) | 25.4 | 19.9 |
| Cross-validated estimated coordinate error: | ||
| From Luzzati plot (Å) | 0.35 | 0.20 |
| From SigmaA (Å) | 0.29 | 0.17 |
Values in parentheses refer to the highest resolution shell.
Rmerge = 100 × ΣhΣi |Ihi - Ih| / ΣhiIhi where h are unique reflection indices.
r.m.s.d. = root mean square deviation from ideal values.
Rcryst = 100 × Σ | Fobs-Fmodel | / Σ̣Fobs where Fobs and Fmodel are observed and calculated structure factor amplitudes, respectively.
Rfree is Rcryst calculated for randomly chosen unique reflections, which were excluded from the refinement (1063 for MurA-terreic acid and 1450 for MurA-fosfomycin).
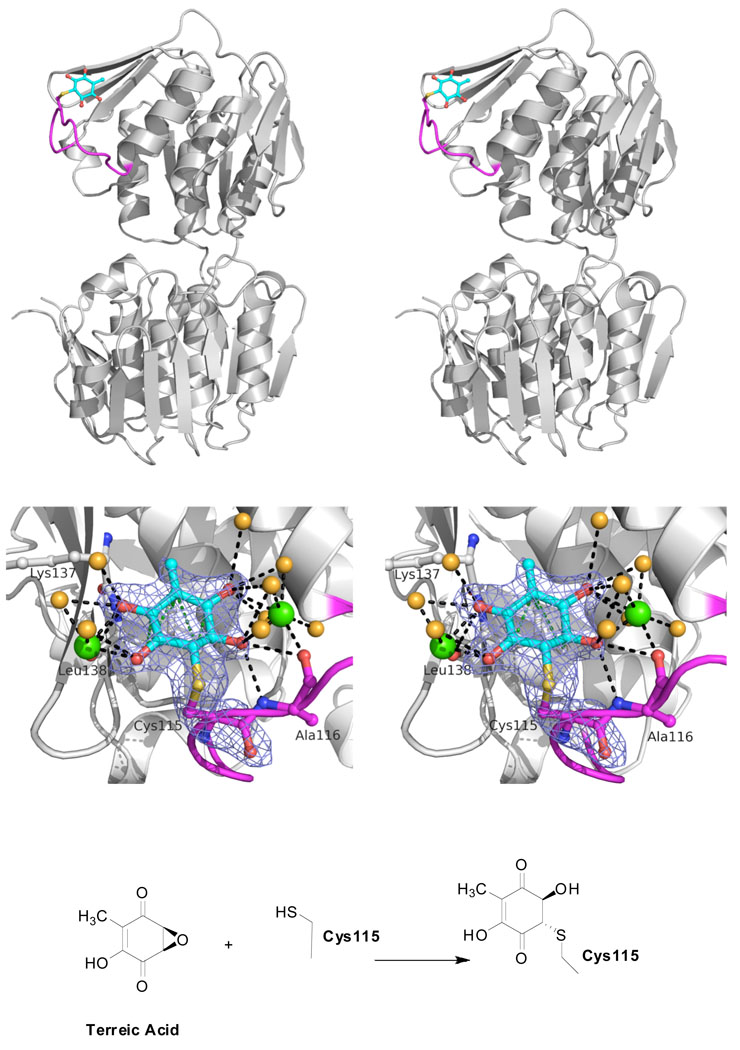
FIGURE 3. Crystal structure of the dead-end complex of E. cloacae MurA with terreic acid.
(Top, stereoview) The overall structure of MurA inactivated by terreic acid exists in an open conformation and is free of UNAG. The loop hosting Cys115 is shown in magenta; the covalently bound terreic acid molecule (cyan) is largely solvent–exposed.
(Middle, stereoview) A detailed view of the Cys115-terreic acid adduct reveals multiple polar interactions (d ≤ 3.3 Å, black dotted lines) with water molecules (orange spheres) and two Ca2+ ions from the crystallization solution (green spheres); hydrophobic interactions (d ≤ 3.8 Å, green dotted lines) exist with Leu138. The blue-colored mesh represents the Fo-Fc difference electron density map (at 2.25 Å resolution and contoured at 3σ), omitting the modified Cys115 residue during the refinement.
(Bottom) Proposed chemical reaction of terreic acid with Cys115.
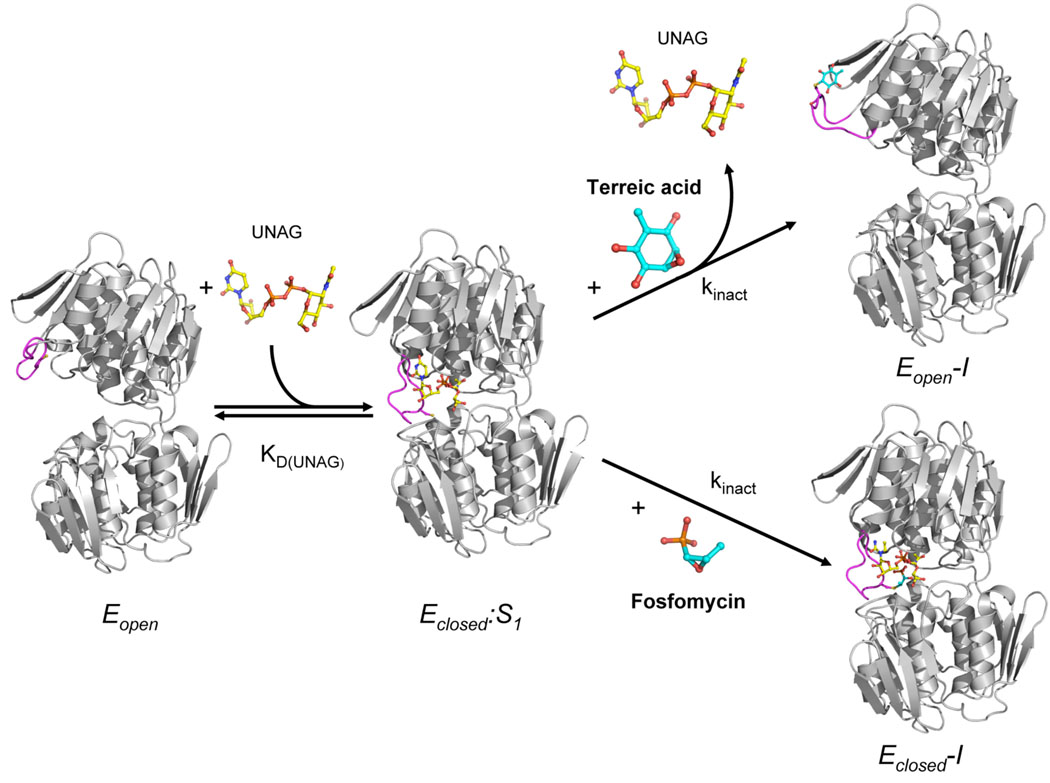
FIGURE 5. The molecular modes of action of terreic acid and fosfomycin on E. cloacae MurA.
In its unliganded state, MurA exists in an open conformation (Eopen) with Cys115 solvent–exposed. Binding of the substrate UNAG (S1) induces the structural transition to the closed binary state (Eclosed:S1). Both terreic acid and fosfomycin interact with Cys115 in this binary complex. The reaction of terreic acid with Cys115 forces the enzyme to open and to release UNAG, leading to an open dead-end complex (Eopen-I). By contrast, fosfomycin interaction with the binary complex renders the overall conformation of MurA unchanged, and the dead-end complex exists as the closed enzyme state with UNAG still bound (Eclosed-I). The loop containing Cys115 is shown in magenta, UNAG in yellow, and the inhibitors in cyan.
RESULTS
Inactivation of MurA by terreic acid
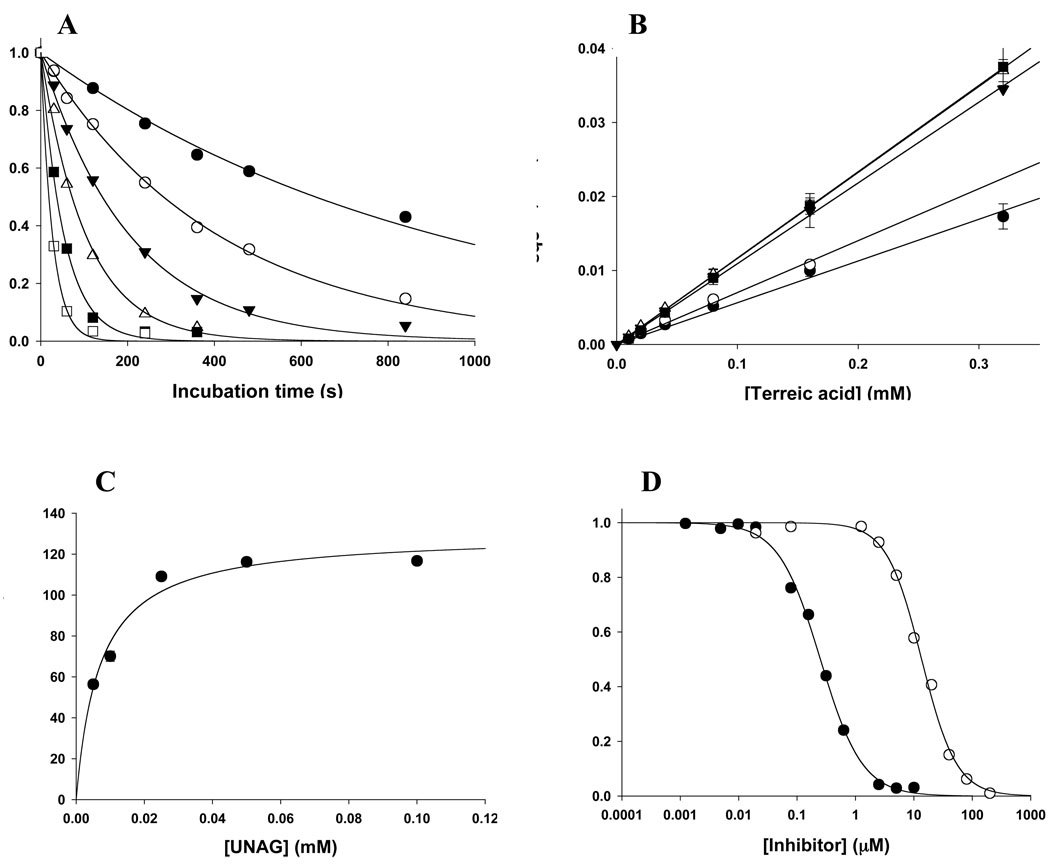
Initial studies established that terreic acid inhibits MurA in a time-dependent manner (Fig. 2 and Figs. S1-S6 of the supplemental material). The inhibition proceeds single-exponentially as a function of incubation time (Fig. 2A). The linear relationship between the first-order inactivation rate constants and terreic acid (Fig. 2B) indicates a single-step interaction between enzyme and inhibitor, without the formation of a rapidly reversible enzyme-inhibitor complex. This type of inactivation mechanism is typical of group-specific reagents, which often display non-selective reactivity toward a large number of proteins. Remarkably, however, the activity of terreic acid toward MurA strictly depends on the presence of UNAG (Fig. 2C), suggesting that inactivation requires the enzyme to interact with the substrate first. Kinetic characterization with varying concentrations of UNAG yielded an overall inactivation rate constant of kinact = 130 M−1s−1. The reaction of fosfomycin with MurA also depends on the presence of UNAG (8, 14). However, fosfomycin is about 50 times more potent than terreic acid when assayed under identical conditions (Fig. 2D). The Cys115Asp mutant MurA, a naturally occurring fosfomycin-resistant variant found in bacteria such as M. tuberculosis (2, 9), is not inhibited by terreic acid (data not shown). Taken together, the kinetic characteristics suggest that terreic acid covalently reacts with MurA through residue Cys115, the molecular target of fosfomycin. Notably, the presence DTT in the assay mixture substantially reduced the inhibitory potency of terreic acid towards MurA. Further examination revealed that incubation of 0.1 mM terreic acid with 2 mM DTT, glutathione or β-mercaptoethanol for 1 hour, prior to addition of MurA, abolished inhibitory activity. By contrast, overnight incubation with 2 mM L-cysteine did not affect the activity of terreic acid. It appears that terreic acid readily reacts with thiol groups of strong reducing agents, but not of free cysteine. The reaction of terreic acid with reducing agents is likely to proceed through the same chemical mechanism that leads to the covalent adduct with the nucleophilic Cys115 side chain of MurA, as outlined below (Fig. 3).
FIGURE 2. Inactivation of E. cloacae MurA by terreic acid.
(A) Time-dependent loss of MurA activity in the presence of 0.05 mM UNAG. The terreic acid concentration was 0.01 mM (•), 0.02 mM (○), 0.04 mM (▼), 0.08 mM (△), 0.16 mM (■), and 0.32 mM (□). Data were fit to equation 1.
(B) Replot of the observed first-order rate constants of inactivation (kobs) vs. terreic acid at varying UNAG concentrations [0.005 mM (•), 0.01 mM (○), 0.025 mM (▼), 0.05mM (△), 0.10 mM (■)]. Data were fit to equation 2.
(C) Replot of the second-order inactivation rate constants (kinact) vs. UNAG concentration. Data were fit to equation 3, yielding k*inact = 130 ± 5.5 M−1s−1 and Kd(UNAG) = 7 ± 1 µM.
(D) Dose-response curves for inhibition of MurA by fosfomycin (•) and terreic acid (○). The enzyme was preincubated with inhibitor and 0.1mM UNAG for 8 minutes before the reaction was started by addition of 1mM PEP. Data were fit to equation 4, yielding IC50 values of 0.25 ± 0.01 µM for fosfomycin and 14 ± 0.6 µM for terreic acid.
Molecular mode of action of terreic acid
In an attempt to understand the inactivation of MurA at the molecular level, the E. cloacae enzyme was crystallized in the presence of UNAG and terreic acid, with the structure determined at 2.25 Å resolution (Table 1). The asymmetric unit consists of four MurA molecules arranged into two dimers. The dimeric state is likely a crystallization artifact, since the enzyme is active as a monomer, and it is well known that MurA tends to crystallize in diverse oligomeric states depending on the ligand present. In solution, terreic acid does not induce oligomerization of MurA as judged by size exclusion chromatography (data not shown). Unexpectedly, the MurA-terreic acid dead-end complex exists in an open, UNAG-free enzyme state (Fig. 3). Terreic acid is covalently bound to Cys115 with full occupancy in all four MurA molecules of the asymmetric unit. The Cys115-terreic acid adduct is largely solvent–exposed; multiple polar interactions with water molecules and two Ca2+ ions are observed, which stem from the crystallization solution. The presence of CaCl2 did not influence the activity of terreic acid toward MurA (data not shown). In addition, hydrogen bonding interactions are formed with the main chain atoms of residues Ala116, Lys137 and Leu138. The opposite side of the terreic acid quinone ring interacts predominantly with the side chain of Leu138 through van-der-Waals forces. The loop around the modified Cys115 residue adopts a distinct, well-defined conformation, different from any other known MurA structure.
In contrast to the structural changes induced by the reaction with terreic acid, inactivation by fosfomycin renders the binary complex with UNAG intact, as shown previously for the enzymes from E. coli (6) and H. influenza (20). We obtained X-ray grade crystals of the MurA-terreic acid complex for only the enzyme from E. cloacae, but not from E. coli. In order to genuinely compare the molecular mode of action of both inhibitors on the identical enzyme (the amino acid sequences of E. coli and E. cloacae MurA share 93.6 % identity and 96.2 % similarity), we determined the crystal structure of the E. cloacae enzyme, inactivated by fosfomycin, at 1.75 Å resolution (Table 1, Fig. 4). In the course of these studies, we also obtained high-quality crystals of the E. coli MurA-fosfomycin complex and re-determined this structure at 1.7 Å resolution (Table S1). The Cys115-fosfomycin adduct in the dead-end complexes of both E. cloacae and E. coli MurA extends into the PEP-binding site, where it is held in place through multiple hydrogen bonding and electrostatic interactions with the side chains of Arg397, Arg120 and Lys22. Fosfomycin, therefore, exploits the binding potential of the highly charged and narrow PEP-binding site, whereas terreic acid lacks this functionality. It appears that the covalent reaction with terreic acid induces large conformational changes in the loop that hosts Cys115, presumably as a result of steric clashes between the Cys115-terreic acid adduct and the active site (Fig. 5). As a consequence, the enzyme opens, UNAG is released and the loop around Cys115 undergoes structural rearrangement.
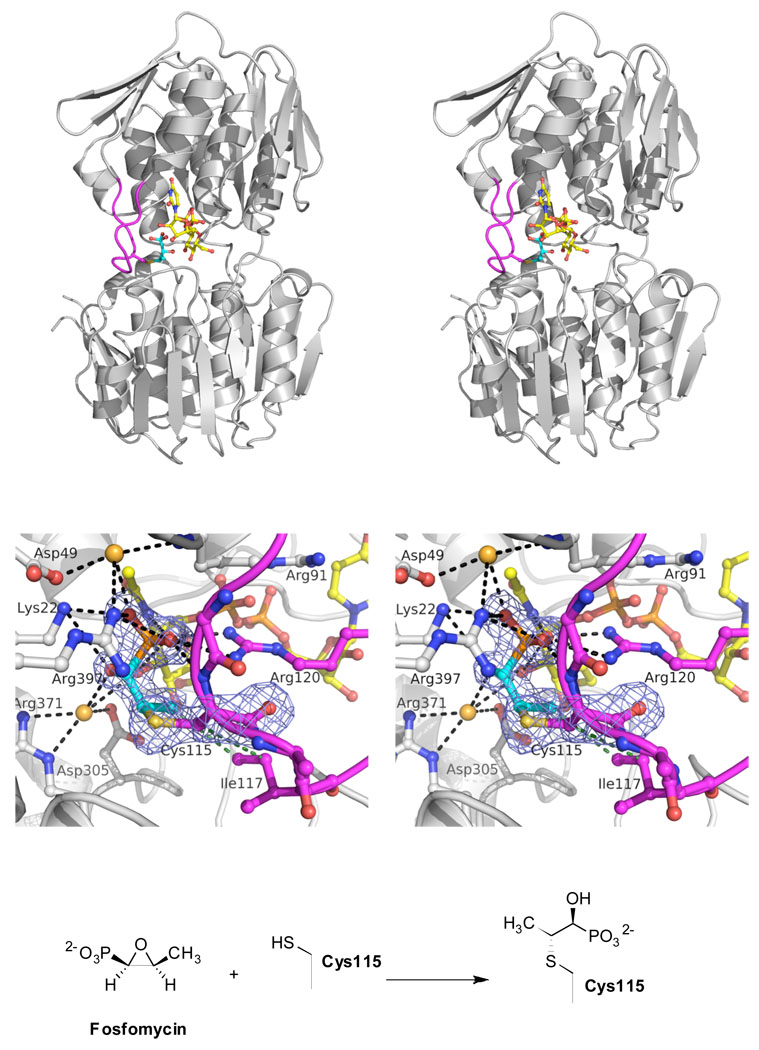
FIGURE 4. Crystal structure of the dead-end complex of E. cloacae MurA with fosfomycin.
(Top, stereoview) The overall structure of MurA inactivated by fosfomycin exists in a closed conformation with UNAG bound. The loop hosting Cys115 is shown in magenta; the covalently bound fosfomycin molecule (cyan) occupies the putative PEP-binding site.
(Middle, stereoview) A detailed view of the Cys115-fosfomycin adduct reveals multiple polar and electrostatic interactions (d ≤ 3.5 Å, black dotted lines) with active site residues, water molecules and the UNAG molecule. Hydrophobic interactions (d ≤ 3.8 Å, green dotted lines) exist with Ile117. The blue-colored mesh represents the Fo-Fc difference electron density map (at 1.75 Å resolution and contoured at 3σ), omitting the Cys115-fosfomycin adduct during the refinement.
(Bottom) Chemical reaction of fosfomycin with Cys115.
DISCUSSION
Despite the critical importance of MurA for bacterial survival, and thus the potential for development of novel antibacterial therapeutics, the only antibiotic known to specifically target this enzyme is fosfomycin. The finding that terreic acid inactivates MurA through a mechanism of action similar to that of fosfomycin indicates that Cys115 is accessible to covalent attack by a broader range of electrophilic compounds than previously envisioned. Although the terreic acid molecule differs considerably from fosfomycin with respect to size and charge, it appears to be small enough to collide with and chemically modify Cys115. Taken together, the data demonstrate that chemical modification of Cys115 by terreic acid or fosfomycin leads to differential structural states of the enzyme. The closed state of the MurA-fosfomycin complex indicates the complementarity between the bound inhibitor and the active site, explaining the superiority of fosfomycin with respect to potency and selectivity. The open state of the MurA-terreic acid complex renders the inhibitor largely solvent–exposed; consequently, less binding energy is generated by the enzyme. It appears that the differential kinetic and structural characteristics of MurA inactivation by terreic acid and fosfomycin reflect the importance of non-covalent binding potential, even for covalent inhibitors, to ensure inactivation efficiency and specificity.
The chemical reactivity of terreic acid against reducing agents such as DTT suggests covalent reactions with nucleophilic residues of other proteins as well. However, only one other enzyme to date, Bruton's tyrosine kinase (Btk), has been reported as a target of terreic acid, which obstructs the interaction of the pleckstrin homology (PH) domain of Btk with protein kinase C in mast cells and other immune cells (21). The mechanism of action of terreic acid on Btk has not yet been determined, but it is conceivable that one or more of the five cysteine residues of the PH domain of Btk is subject to covalent modification by terreic acid in a manner similar to Cys115 in MurA.
Recently, it was reported that other natural products with antibiotic activity, such as the sesquiterpene lactone cnicin (22) and tuliposides (23), inhibit MurA in vitro as well. While the mechanism of action of tuliposides on MurA is based on circumstantial evidence, the interaction of MurA with cnicin has been studied in greater detail. Based on X-ray crystallographic data, it was concluded that MurA catalyzes an unusual “anti-Michael” 1,3-addition to form a UNAG-cnicin adduct that blocks the active site (24). However, with the exception of fosfomycin, it has never been demonstrated that the antibacterial activity of in vitro MurA inhibitors is actually caused by inhibition of MurA in the cell. For example, recent studies from our laboratory indicate that the antibacterial properties of terreic acid and fosfomycin are fundamentally different, suggesting that MurA is not the primary target of terreic acid in E. coli (Schönbrunn, unpublished). We hypothesize that highly reactive compounds with antibiotic activity, such as terreic acid, cnicins or tuliposides, predominantly react with proteins on the outside of the bacterial cell membrane, instead of entering the cytoplasm to inactivate MurA. Inhibitors designed to selectively target Cys115 in MurA should exhibit only latent electrophilicity, as does fosfomycin, in order to reduce unspecific interactions with other proteins.
It is alarming that, almost 50 years after the discovery of fosfomycin, no other MurA-specific antibiotic agents have been identified. Reversible high-affinity inhibitors with new molecular modes of action could present more desirable candidates for the rational design of therapeutics. Unfortunately, initial high-throughput screening (HTS) efforts in this regard did not reveal prospective pharmacophores suitable for further development. For example, derivatives of sulfonoxy-anthranilic acid, discovered by HTS, were shown to reversibly inhibit MurA by binding to the open form of the enzyme, largely through van-der-Waals interactions (25). However, these inhibitors are relatively weak (Ki = 16 µM), presumably because they lack the functionality to induce the enzyme’s structural transition to the closed state. The conformational flexibility of MurA and the highly charged and polar character of the active site impose challenges for drug design that are difficult to overcome by conventional HTS and molecular modeling technology. Therefore, it seems that intense screening efforts using new, diverse HTS compound libraries and more sophisticated computational approaches are necessary in order to enable the design of new chemical entities as prospective lead molecules with therapeutic potential. Experimentally determined structures of new MurA-ligand complexes, such as the MurA-terreic acid complex, will be instrumental in the understanding and subsequent exploitation of the unique structural features of this enzyme for the design of new inhibitors with novel mechanisms of action.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Moffitt Structural Biology Core for use of the X-ray crystallography facility. We thank Frank Marsilio and Donna J. Lubbers from the Schönbrunn laboratory for help with enzyme purification and preparation of the manuscript.
The abbreviations used are
- PEP
phosphoenolpyruvate
- UNAG
UDP-N-acetylglucosamine
Footnotes
This work was supported by the National Institutes of Health (NIH) Grant 5R01GM070633.
The atomic coordinates and structure factors (codes 3KQA, 3KR6 and 3LTH) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
SUPPORTING INFORMATION
Crystallographic data statistics for the dead-end complex of E. coli MurA with fosfomycin and additional kinetic data for the inactivation of MurA by terreic acid and fosfomycin are provided as supplementary material (http://pubs.acs.org).
REFERENCES
- 1.Kahan FM, Kahan JS, Cassidy PJ, Kropp H. The Mechanism of Action of Fosfomycin (Phosphonomycin) Ann. N. Y. Acad. Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 2.De Smet KA, Kempsell KE, Gallagher A, Duncan K, Young DB. Alteration of a Single Amino Acid Residue Reverses Fosfomycin Resistance of Recombinant Mura from Mycobacterium Tuberculosis. Microbiology. 1999;145(Pt 11):3177–3184. doi: 10.1099/00221287-145-11-3177. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Lobo JM, Ortiz JM. Tn2921, a Transposon Encoding Fosfomycin Resistance. J Bacteriol. 1982;151:477–479. doi: 10.1128/jb.151.1.477-479.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navas J, Garcia-Lobo JM, Leon J, Ortiz JM. Structual and Functional Analyses of the Fosfomycin Resistance Transposon Tn2921. J Bacteriol. 1985;162:1061–1067. doi: 10.1128/jb.162.3.1061-1067.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schonbrunn E, Sack S, Eschenburg S, Perrakis A, Krekel F, Amrhein N, Mandelkow E. Crystal Structure of UDP-N-Acetylglucosamine Enolpyruvyltransferase, the Target of the Antibiotic Fosfomycin. Structure. 1996;4:1065–1075. doi: 10.1016/s0969-2126(96)00113-x. [DOI] [PubMed] [Google Scholar]
- 6.Skarzynski T, Mistry A, Wonacott A, Hutchinson SE, Kelly VA, Duncan K. Structure of UDP-N-Acetylglucosamine Enolpyruvyl Transferase, an Enzyme Essential for the Synthesis of Bacterial Peptidoglycan, Complexed with Substrate UDP-N-Acetylglucosamine and the Drug Fosfomycin. Structure. 1996;4:1465–1474. doi: 10.1016/s0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]
- 7.Wanke C, Amrhein N. Evidence That the Reaction of the UDP-N-Acetylglucosamine 1-Carboxyvinyltransferase Proceeds through the O-Phosphothioketal of Pyruvic Acid Bound to Cys115 of the Enzyme. Eur J Biochem. 1993;218:861–870. doi: 10.1111/j.1432-1033.1993.tb18442.x. [DOI] [PubMed] [Google Scholar]
- 8.Marquardt JL, Brown ED, Lane WS, Haley TM, Ichikawa Y, Wong CH, Walsh CT. Kinetics, Stoichiometry, and Identification of the Reactive Thiolate in the Inactivation of UDP-Glcnac Enolpyruvoyl Transferase by the Antibiotic Fosfomycin. Biochemistry. 1994;33:10646–10651. doi: 10.1021/bi00201a011. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, Lees WJ, Kempsell KE, Lane WS, Duncan K, Walsh CT. Characterization of a Cys115 to Asp Substitution in the Escherichia Coli Cell Wall Biosynthetic Enzyme UDP-Glcnac Enolpyruvyl Transferase (Mura) That Confers Resistance to Inactivation by the Antibiotic Fosfomycin. Biochemistry. 1996;35:4923–4928. doi: 10.1021/bi952937w. [DOI] [PubMed] [Google Scholar]
- 10.Eschenburg S, Priestman M, Schonbrunn E. Evidence That the Fosfomycin Target Cys115 in UDP-N-Acetylglucosamine Enolpyruvyl Transferase (Mura) Is Essential for Product Release. J. Biol. Chem. 2005;280:3757–3763. doi: 10.1074/jbc.M411325200. [DOI] [PubMed] [Google Scholar]
- 11.Sheehan JC, Lawson WB, Gaul RJ. The Structure of Terreic Acid. J. Am. Chem. Soc. 1958;80 [Google Scholar]
- 12.Yamamoto H, Moriyama K, Jinnouchi H, Yagishita K. Studies on Terreic Acid. Jpn J Antibiot. 1980;33:320–328. [PubMed] [Google Scholar]
- 13.Wanke C, Falchetto R, Amrhein N. The UDP-N-Acetylglucosamine 1-Carboxyvinyl-Transferase of Enterobacter Cloacae. Molecular Cloning, Sequencing of the Gene and Overexpression of the Enzyme. FEBS Letters. 1992;301:271–276. doi: 10.1016/0014-5793(92)80255-f. [DOI] [PubMed] [Google Scholar]
- 14.Schonbrunn E, Eschenburg S, Krekel F, Luger K, Amrhein N. Role of the Loop Containing Residue 115 in the Induced-Fit Mechanism of the Bacterial Cell Wall Biosynthetic Enzyme Mura. Biochemistry. 2000;39:2164–2173. doi: 10.1021/bi991091j. [DOI] [PubMed] [Google Scholar]
- 15.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An Improved Assay for Nanomole Amounts of Inorganic Phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W. Automatic Procession of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constrants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 17.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Crystallogr Sec D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Eschenburg S, Schonbrunn E. Comparative X-Ray Analysis of the Un-Liganded Fosfomycin-Target Mura. Proteins. 2000;40:290–298. doi: 10.1002/(sici)1097-0134(20000801)40:2<290::aid-prot90>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 19.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron Density Maps and the Location of Errors in These Models. Acta Crystallogr Sec A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Yoon HJ, Lee SJ, Mikami B, Park HJ, Yoo J, Suh SW. Crystal Structure of UDP-N-Acetylglucosamine Enolpyruvyl Transferase from Haemophilus influenzae in Complex with UDP-N-Acetylglucosamine and Fosfomycin. Proteins. 2008;71:1032–1037. doi: 10.1002/prot.21959. [DOI] [PubMed] [Google Scholar]
- 21.Kawakami Y, Hartman SE, Kinoshita E, Suzuki H, Kitaura J, Yao L, Inagaki N, Franco A, Hata D, Maeda-Yamamoto M, Fukamachi H, Nagai H, Kawakami T. Terreic Acid, a Quinone Epoxide Inhibitor of Bruton's Tyrosine Kinase. Proc Natl Acad Sci U S A. 1999;96:2227–2232. doi: 10.1073/pnas.96.5.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachelier A, Mayer R, Klein CD. Sesquiterpene Lactones Are Potent and Irreversible Inhibitors of the Antibacterial Target Enzyme Mura. Bioorg Med Chem Lett. 2006;16:5605–5609. doi: 10.1016/j.bmcl.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Shigetomi K, Shoji K, Mitsuhashi S, Ubukata M. The Antibacterial Properties of 6-Tuliposide B. Synthesis of 6-Tuliposide B Analogues and Structure-Activity Relationship. Phytochemistry. 2010;71:312–324. doi: 10.1016/j.phytochem.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Steinbach A, Scheidig AJ, Klein CD. The Unusual Binding Mode of Cnicin to the Antibacterial Target Enzyme Mura Revealed by X-Ray Crystallography. J Med Chem. 2008;51:5143–5147. doi: 10.1021/jm800609p. [DOI] [PubMed] [Google Scholar]
- 25.Eschenburg S, Priestman MA, Abdul-Latif FA, Delachaume C, Fassy F, Schönbrunn E. A Novel Inhibitor That Suspends the Induced-Fit Mechanism of UDP-N-Acetylglucosamine Enolpyruvyl Transferase (MurA) J Biol Chem. 2005;280:14070–14075. doi: 10.1074/jbc.M414412200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.