Abstract
The goal of this study was to determine the role of the Organic Anion Transporting Polypeptides, OATP1A2, OATP1B1 and OATP1B3 and their genetic variants in the pharmacokinetics of the immunosuppressive drug mycophenolate mofetil (MMF). Using OATP-transfected HEK cells, we measured the uptake of mycophenolic acid (MPA) and its glucuronide (MPAG). MPAG, but not MPA, significantly accumulated in cells expressing OATP1B3 or OATP1B1 (p<0.05). MPA and MPAG pharmacokinetics were significantly influenced by the OATP1B3 polymorphism, 334T>G/699G>A, in 70 renal transplant patients co-treated with tacrolimus or sirolimus but not in 115 patients co-treated with cyclosporine. The decrease in MPA dose-normalized exposure and concomitant increase in the MPAG/MPA metabolic ratio are consistent with reduced enterohepatic cycling in patients carrying the OATP1B3 334G-699A haplotype. Further studies demonstrated that this variant of OATP1B3 exhibited a reduced Vmax in transfected HEK cells, providing functional support of our clinical findings.
Keywords: Cell Line; Genetic Variation; genetics; Humans; Mycophenolic Acid; pharmacokinetics; Organic Anion Transporters; genetics; physiology; Peptides; genetics; physiology; Polymorphism, Genetic; genetics
Keywords: immunosuppressive agents, transplantation, transporters, pharmacogenetics, mycophenolic acid.
Introduction
Mycophenolic acid (MPA), the active moiety of mycophenolate mofetil (MMF), is a non-nephrotoxic, cornerstone immunosuppressant in kidney transplantation. MPA undergoes extensive entero-hepatic recirculation initiated by almost complete hepatic glucuronidation into the inactive MPA-7-O-glucuronide (MPAG) (1) and active biliary excretion of this metabolite by the Multidrug Resistance-associated Protein-2 (MRP2) (2). It is thought that, after MPAG hydrolysis by intestinal flora, the amount of MPA reabsorbed could represent 10 to 60% of total MPA exposure (1). Consequently, inhibitors of MPA entero-hepatic cycling (including cyclosporine) affect MPA levels (3, 4). Although significant associations between polymorphisms in enzymes involved in MPA glucuronidation and MMF pharmacokinetics were described, results differed depending on the post-transplantation period and comedications and could not always be replicated (see supplementary Table 1). The hepatic uptake of MPA and/or MPAG being a prerequisite for MPA entero-hepatic recirculation, it could contribute to the pharmacokinetic variability and drug-drug interactions of MPA-based formulations. There has been only one indirect hint about the implication of the Organic Anion Transporting Polypeptides (OATPs) localized to the sinusoidal membrane of the hepatocytes in the disposition of MMF. Specifically, Miura et al. reported an association between OATP1B3 and OATP1B1 polymorphisms and MPAG and MPA pharmacokinetics in Japanese renal transplants (5, 6).
This study aimed to determine whether MPA and MPAG are substrates of the main OATP expressed in hepatocytes (OATP1B1 and 1B3) or enterocytes (OATP1A2) and the effect of genetic variations in competent transporters on MPA pharmacokinetics in renal transplant recipients receiving MMF combined with different other immunosuppressants, taking into account other SNPs previously reported to affect MPA pharmacokinetics.
Results
Cellular uptake of MPA and MPAG in OATP-expressing cells
After 5 min incubation in cells expressing OATP1A2, 1B1 or 1B3, accumulation of MPA was not affected as compared to that observed in cells transfected with an empty vector (Figure 1a). In contrast, MPAG uptake was significantly and substantially enhanced by OATP1B3 and to a lesser extent by OATP1B1, but not by OATP1A2 (Figure 1b).
Figure 1.
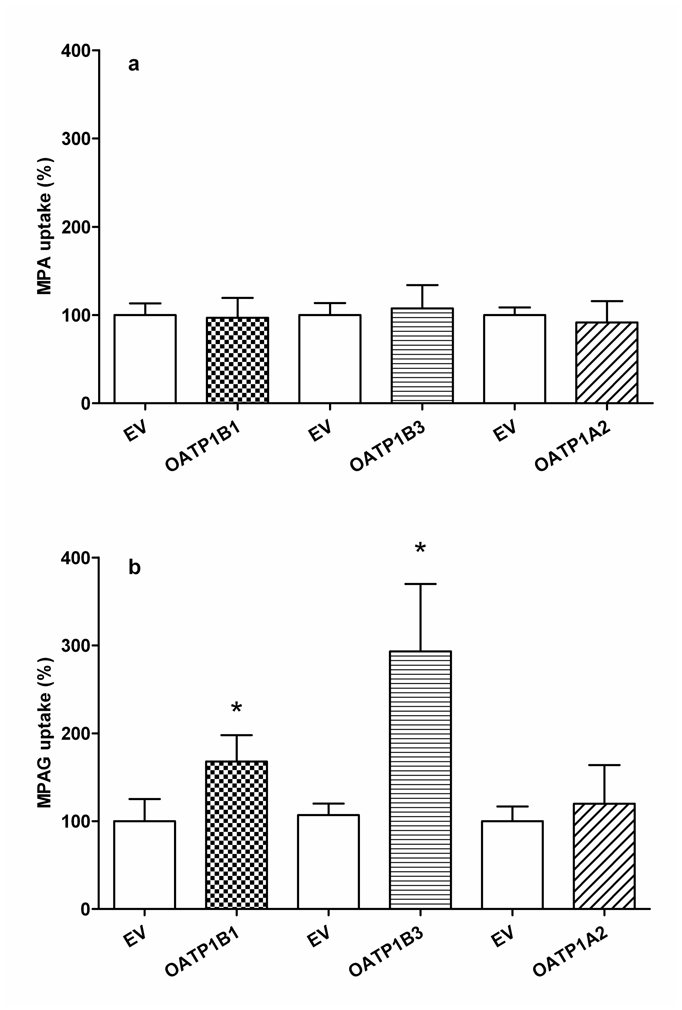
Mycophenolic acid (MPA; a) and mycophenolic acid phenyl-glucuronide (MPAG; b) (20μM) uptake into HEK293T cells transiently transfected with OATP1B1, 1B3 and 1A2. Empty vector (EV) transfected cells served as control. Data are mean ± SD of three independent experiments performed in triplicate. Uptake time was 5 min. (*p<0.05 for comparison with control cells).
Experiments with radiolabeled model substrates confirmed the activity of each transporter (see supplementary data).
Identification and frequency distribution of patient genotypes
Our clinical pharmacogenetic investigation was primarily based on our aforementioned in-vitro results and thus aimed to assess the effect of SLCO1B3 and SLCO1B1 polymorphisms on MMF pharmacokinetics. In addition, patients were genotyped for polymorphisms in UGT1A9, UGT1A8 and ABCC2, previously reported to significantly affect MPA pharmacokinetics.
SNPs to be investigated in SLCO genes were selected from the literature and from the SOPHIE (Study of Pharmacogenetics in Ethnically Diverse Populations) database (http://pharmacogenetics.ucsf.edu/index.html). Two frequent coding, non-synonymous SNPs in complete linkage disequilibrium were reported in SLCO1B3 (T334G, G699A) (7). In the Caucasian group of the SOPHIE cohort one additional common non-synonymous SNP was found in SLCO1B3 (G767C) with allele frequency of 0.18, and three in SLCO1B1 (A388G, C463A and T521C) with allelic frequencies of 0.44, 0.16 and 0.17, respectively.
No linkage disequilibrium was observed between the SLCO1B3 T334G and G767C SNPs. In contrast, for SLCO1B1, the A388G SNP showed a strong linkage disequilibrium with the C463A (D′=l; r2=0.21) and the T521C (D′=0.79; r2=0.17) SNPs, while there was no linkage between the C463A and the T521C SNPs. This lead to six SLCO1B1 haplotypes: A-C-T (0.53), G-C-C (known as SLCO1B1*15; 0.15), G-C-T (0.14), G-A-T (0.13), A-C-C (0.021), and G-A-C (0.004).
All genotypes were in Hardy-Weinberg equilibrium. Their distributions were compared to those observed in the HapMap Caucasian panel (www.hapmap.org; Release 27, Feb 09), except for UGT1A9 C-440T, T98C and SLCO1B3 C767G, not reported in HapMap. No significant difference was found, except for ABCC2 C-24T (p=0.01592) whose allelic distribution was however not significantly different from that reported in the literature for Caucasians (8, 9).
Influence of comedications on MPA pharmacokinetics
As the nature of the immunosuppressant coadministered with MMF influences MPA pharmacokinetics, this effect was evaluated before the pharmacogenetic association study. Since no difference in MMF dose (1416±528 vs 1304±478 mg; p=0.3633), MPA dose-normalized (dn) AUC0-12h (64.6±34.7 vs 66.0±29.7 mg.h/l/g, p=0.6401), dn-C0 (4.4±3.5 vs 3.9±2.5 mg/l/g, p=0.6958) and dn-Cmax, (20.1±9.4 vs 27.2±21 mg/l/g, p=0.2674) or MPAG dn-AUC0-9h (1011±595 vs 1183±611 mg.h/l/g; p=0.1220), dn-C0 (99±75 vs 108±59 mg/l/g, p=0.3436) and dn-Cmax (152±84 vs 177±95 mg/l/g, p=0.2059) was observed between patients receiving the drug in combination with sirolimus or tacrolimus, the two sub-groups of patients were combined for the pharmacogenetic study. In contrast, patients co-treated with cyclosporine were studied independently as they received significantly higher doses of MMF (1890±679mg) and had lower MPA dn-AUC0-12h(44.3±20.8 mg.h/l/g) as compared to patients on sirolimus or tacrolimus (p<0.0001).
SLCO genotypes and MMF pharmacokinetics
In patients receiving MMF in combination with cyclosporine (n=115), no significant association between SLCO1B3 or 1B1 genotypes or haplotypes and MPA dn-AUC0-12h was found (Table 1).
Table 1.
Mycophenolic acid (MPA) dose-normalized (dn) AUC0-12h according to SLCO, UGT or ABCC2 Single Nucleotide Polymorphisms (SNPs) in patients co-treated with cyclosporine
| Gene | SNP | Patients genotypes (n) | MPA dn-AUC0-12h (mg.h/l)/g | P values |
|---|---|---|---|---|
| SLCO1B3 | T334G | TT (n=6) | 42.7±18.4 | 0.4747 |
| TG (n=32) | 48.6±22.9 | |||
| GG (n=76) | 42.7±20.2 | |||
| G767C | GG (n= 79) | 47.1±23.2 | 0.0469 | |
| GC (n= 36) | 38.1±12.4 | |||
| SLCO1B1 | T521C | TT (n=75) | 44.8±20.7 | 0.7507 |
| TC (n=36) | 43.7±22.1 | |||
| CC (n=4) | 38.6±10.9 | |||
| A388G | AA (n=37) | 45.5±17.1 | 0.2800 | |
| AG (n=52) | 44.1±22.2 | |||
| GG (n=26) | 42.8±23.3 | |||
| C463A | CC (n=83) | 43.6±18.3 | 0.9717 | |
| CA (n=30) | 46.3±27.0 | |||
| AA (n=2) | 42.8±25.0 | |||
| UGT1A8 | C518G | CC (n=59) | 43.7±18.9 | 0.9066 |
| CG (n=50) | 44.1±22.1 | |||
| GG (n=6) | 51.4±29.3 | |||
| UGT1A9 | C-440T | CC (n=69) | 40.8±15.3 | 0.2721 |
| CT (n=39) | 49.2±27.1 | |||
| TT (n=7) | 50.7±24.0 | |||
| T-275A and/or C-2152T* | TTandCC(n=101) | 44.1±21.2 | 0.4498 | |
| TAandCT or CC(n=12) | 47.8±19.1 | |||
| AA and TT (n=2) | 31.8±7.9 | |||
| T98C | TT (n=105) | 43.7±21.1 | 0.0634 | |
| TC (n=10) | 50.7±17.1 | |||
| ABCC2 | C-24T | CC (n=68) | 40.2±14.5 | 0.0087 |
| CT (n=35) | 53.6±26.0 | |||
| TT (n=12) | 39.9±26.7 | |||
SNPs in strong linkage disequilibrium: D′=1; r2=0.87
In 70 patients co-treated with sirolimus or tacrolimus, MPAG dn-AUC0-9h showed a trend to a gradual decrease with the number of SLCO1B3 T334 alleles (Table 2). Although not significant, important differences in MPAG dose-normalized levels were observed at all the sampling time points (Figure 2b). The metabolic ratio of MPAG to MPA showed a very significant decrease in carriers of the SLCO1B3 T334 allele (p=0.0001) (Figure 2c).
Table 2.
Pharmacokinetic parameters of mycophenolic acid (MPA) and its glucuronide (MPAG) according to SLCO Single Nucleotide Polymorphisms (SNPs) in patients co-treated with tacrolimus or sirolimus
| Gene | SNP | Genotypes (n) | MPA dn-C0 (mg/l)/g | MPA dn-Cmax (mg/l)/g | MPA Tmax (min) | MPA dn-AUC0-12h (mg.h/l)/g | MPAG dn-C0 (mg/l) | MPAG dn-Cmax (mg/l) | MPAG Tmax (min) | MPAG dn-AUC0-9h (mg.h/l) |
|---|---|---|---|---|---|---|---|---|---|---|
| SLCO1B3 | T334G | TT (n= 2) | 3.4±0.6 | 21.4±10.5 | 65.0±35.3 | 73.9±30.2 | 79.6±42.7 | 111.6±32.8 | 90.0±42.4 | 621.7±235.3 |
| TG (n=20) | 5.3±2.8 | 28.6±10.7 | 71.5±111.6 | 82.6±32.6 | 91.0±51.0 | 137.4±54.0 | 114.5±112.1 | 964.8±457.6 | ||
| GG (n=48) | 3.7±3.1 | 20.6±16.7 | 69.6±86.7 | 57.5±30.2 | 108.8±74.7 | 173.7±98.7 | 123.3±75.6 | 1146.2±644.2 | ||
| p values | 0.0413 | 0.0012 | 0.8113 | 0.0040 | 0.6296 | 0.3664 | 0.3297 | 0.2477 | ||
| p values (TT+TG vs GG) | 0.0150 | 0.0004 | 0.8233 | 0.0009 | 0.3774 | 0.1947 | 0.1364 | 0.2394 | ||
| G767C | GG (n=49) | 4.4±3.2 | 23.5±13.2 | 63.5±74.5 | 68.5±33.4 | 103.9±72.1 | 157.2±82.7 | 117.9±88.1 | 1042.6±524.5 | |
| GC (n=21) | 3.7±2.7 | 21.5±19.8 | 85.2±126.0 | 57.4±30.0 | 100.9±60.2 | 171.7±101.7 | 124.3±83.5 | 1165.1±741.9 | ||
| p values | 0.3153 | 0.1644 | 0.9183 | 0.2234 | 0.9365 | 0.5010 | 0.6570 | 0.4769 | ||
| SLCO1B1 | T521C | TT (n=47) | 3.6±2.3 | 22.1±17.0 | 64.0±75.7 | 60.0±30.0 | 112.3±76.3 | 170.0±96.7 | 116.6±84.1 | 1137.9±639.8 |
| TC (n=23) | 5.3±4.1 | 24.6±11.6 | 82.2±121.0 | 75.8±35.6 | 84.8±44.7 | 144.3±66.6 | 126.5±91.8 | 959.7±482.2 | ||
| p values | 0.0942 | 0.0902 | 0.6753 | 0.0534 | 0.1168 | 0.2685 | 0.9695 | 0.2578 | ||
| A388G | AA (n=19) | 4.0±2.5 | 18.5±9.0 | 80.0±115.2 | 58.2±30.4 | 123.9±93.7 | 167.8±95.9 | 117.4±114.9 | 1079.4±545.2 | |
| AG (n=43) | 4.0±3.3 | 23.7±16.3 | 71.6±89.9 | 65.9±33.0 | 91.0±55.7 | 156.6±87.1 | 127.4±75.5 | 1054.0±634.9 | ||
| GG (n=8) | 5.2±3.1 | 29.4±20.5 | 37.5±16.7 | 77.9±34.8 | 118.9±52.3 | 173.2±85.5 | 85.0±58.1 | 1164.6±542.6 | ||
| p values | 0.4849 | 0.1966 | 0.1858 | 0.2692 | 0.0883 | 0.6920 | 0.2154 | 0.5720 | ||
| C463A | CC (n=52) | 3.9±2.3 | 23.3±15.1 | 74.8±106.4 | 65.7±33.0 | 96.7±70.2 | 154.2±86.1 | 119.2±90.5 | 1015.1±562.9 | |
| CA (n=17) | 4.6±4.6 | 22.1±16.9 | 58.2±21.9 | 63.5±33.2 | 119.5±62.6 | 185.2±96.2 | 127.6±72.3 | 1291.0±672.6 | ||
| AA (n=1) | 8.6 | 18.3 | 20.0 | 62.9 | 137.0 | 145.6 | 20.0 | 824.9 | ||
| p values | 0.2799 | 0.7794 | 0.1598 | 0.9049 | 0.1393 | 0.3723 | 0.1249 | 0.1680 | ||
dn: dose-normalized
Figure 2.

Mean ± SEM of dose normalized concentration-time profiles of mycophenolic acid (MPA; a), MPA-phenyl glucuronide (MPAG; b) and distribution of MPA to MPAG metabolic ratio (MPAG/MPA AUC0-9h ratio; c) as a function SLCO1B3 T334G genotype in patients co-treated with tacrolimus or sirolimus. In panel c, the lines represent the mean values. *p<0.05; **p<0.005
In these patients, MPA dose-normalized exposure was significantly different depending on SLCO1B3 T334G genotype (Table 2): the SLCO1B3 T334 allele resulted in significantly higher MPA dose-normalized levels at every concentration-time points, except T20min (p=0.0636) and T120min (p=0.0975) (Figure 2a). Consequently, carriers of a least one SLCO1B3 T334 allele (n=22) had 1.42-fold higher (IC95%: 1.39–1.45; p=0.0003) MPA dn-AUC0-12h and a 1.38-fold higher (IC95%: 1.32–1.49; p=0.0010) dn-Cmax as compared to carriers of the 334GG genotype (n=48). There was no difference in Tmax (Table 2). MPA AUC4-9h was studied as a surrogate for enterohepatic cycling: patients heterozygous or homozygous for the SLCO1B 3 T334 allele (n=22) had significantly higher MPA dn-AUC4-9h than other patients (n=48; p=0.0027). However, they also had higher dn-AUC0-4h (p=0.0008), resulting in unchanged AUC4-9h/AUC0-9h ratio (p=0.4945).
The SLCO1B3 T334 allele was associated with a similar increase in MPA dn-AUC0-12h (p=0.0074 and p=0.0651, respectively) and decrease in MPAG/MPA metabolic ratio (p=0.0517 and 0.0002, respectively) in the subgroups of patients co-treated with sirolimus (n=42) and tacrolimus (n=28).
There was no association between the SLCO1B3 T334G genotype and MMF daily dose, categorized as low (≤1g), intermediate (=1.5g) or standard/high (≥2g) (p=0.1080). However, a significant association was observed between this genotype and MPA AUC0-12h categorized as below, in, or above the generally admitted therapeutic range (30–60 mg.h/l; p=0.03548). One third of the patients with the 334 TT or TG genotype (n=7/22) had MPA AUC0-12h higher than the admitted therapeutic range, as compared to only 8% of patients of the 334GG genotype (n=4/48). Conversely, in carriers of the 334GG genotype, 40% (n=19/48) had MPA AUC0-12h below the therapeutic range as compared to 23% (n=5/22) patients of the TT or TG genotype. There was no association of the second SLCO1B3 SNP tested (G767C) with any of the exposure indices.
Despite a trend to higher MPA dn-AUC0-12h in heterozygous carriers of the SLCO1B1 T521C SNP as compared to homozygous TT521 carriers (p=0.0534), this SNP was not associated with MPAG exposure (Table 2). The other SLCO1B1 SNP tested (A388G and C463A) did not show any association with MPA or MPAG pharmacokinetics (Table 2).
Thesias® haplotype analysis revealed an association between the SLCO1B1*15 allele and MPA dn-AUC0-12h (p=0.0246) or MPAG/MPA ratio (p=0.0267), but not with MPAG dn-AUC0-9h.
No statistical interaction between the effect of SLCO1B3 T334G genotype and SLCO1B1*15 allele was found using ANOVA, but the effect of the SLCO1B1*15 allele did not remain significant regarding MPA dn-AUC0-12h (p=0.0553) and only showed a trend regarding MPAG/MPA ratio (p=0.0302, NS after Bonferroni correction). In contrast, the effect of SLCO1B3 T334G genotype remained highly significant (p values of 0.0008 and 0.0043 for MPA dn-AUC0-12h and MPAG/MPA ratio, respectively).
UGT and ABCC2 genotypes and MMF pharmacokinetics
In patients co-treated with cyclosporine, no influence of UGT1A8 and 1A9 polymorphisms was observed on MPA dn-AUC0-12h (Table 1). There was a trend towards an association between the ABCC2 C-24T SNP and MPA dn-AUC0-12h, however, without apparent additivity: heterozygotes for this SNP showed higher MPA dn-AUC0-12h than homozygotes for the reference or the variant genotype.
No significant association of ABCC2, UGT1A8 or UGT1A9 genotypes with MPAG exposure was found in patients co-treated with tacrolimus or sirolimus (Table 3). Regarding MPA pharmacokinetics, a trend to lower MPA dn-Cmax was found in two patients heterozygous for the UGT1A9 T98C genotype as compared to patients of the reference genotype (p=0.0344; NS after Bonferroni correction). However, no association between this polymorphism and other MPA (or MPAG) exposure indices or with MPAG/MPA ratio was found (Table 3).
Table 3.
Pharmacokinetic parameters of mycophenolic acid (MPA) and its glucuronide (MPAG) according to UGT or ABCC2 Single Nucleotide Polymorphisms (SNPs) in patients co-treated with tacrolimus or sirolimus
| Gene | SNP | Patients genotypes (n) | MPA dn-C0 (mg/l)/g | MPA dn-Cmax (mg/l)/g | MPA Tmax (min) | MPA dn-AUC0-12h (mg.h/l)/g | MPAG dn-C0 (mg/l) | MPAG dn- Cmax (mg/l) | MPAG Tmax (min) | MPAG dn-AUC0-9h (mg.h/l) |
|---|---|---|---|---|---|---|---|---|---|---|
| UGT1A9 | C-440T (T-331C) | CC (n=39) | 4.1±3.7 | 22.0±18.3 | 75.6±93.8 | 63.1±35.0 | 108.5±88.3 | 171.9±104.2 | 130.0±95.1 | 1143.5±692.2 |
| TC (n=27) | 4.1±2.3 | 23.3±11.3 | 68.5±97.4 | 68.7±30.1 | 95.0±50.6 | 144.7±59.0 | 111.5±76.5 | 974.3±429.7 | ||
| TT (n=4) | 4.4±2.1 | 29.0±6.7 | 25.0±10.0 | 61.5±17.7 | 102.2±46.6 | 174.7±86.4 | 77.5±35.0 | 1163.1±559.7 | ||
| p values | 0.5049 | 0.0753 | 0.0330 | 0.5080 | 0.7807 | 0.7867 | 0.3018 | 0.6777 | ||
| T-275A or C-2152T* | TT(CC) (n=63) | 4.3±3.1 | 23.3±15.8 | 71.9±97.2 | 65.7±30.4 | 105.8±69.8 | 163.4±90.4 | 118.4±88.9 | 1090.6±605.2 | |
| TA (CT) (n=7) | 2.8±2.8 | 19.3±11.7 | 52.8±22.1 | 60.3±51.2 | 79.7±50.6 | 144.9±69.9 | 132.8±59.6 | 978.2±523.4 | ||
| p values | 0.0912 | 0.3325 | 0.8469 | 0.1799 | 0.5119 | 0.9143 | 0.2858 | 0.9143 | ||
| T98C | TT (n=68) | 4.1±3.0 | 23.3±15.4 | 70.1±93.8 | 66.2±32.5 | 103.7±68.9 | 163.4.2±88.7 | 119.0±87.2 | 1089.2±599.4 | |
| TC (n=2) | 6.1±6.5 | 8.9±3.1 | 65.0±35.3 | 30.9±8.7 | 78.9±38.3 | 98.0±51.0 | 150.0±42.4 | 745.6±371.3 | ||
| p values | 0.5671 | 0.0344 | 0.5957 | 0.0570 | 0.6764 | 0.2172 | 0.2358 | 0.4175 | ||
| UGT1A8 | C518G | CC (n=40) | 4.2±3.2 | 25.4±18.1 | 69.0±94.5 | 66.8±29.6 | 105.1±75.5 | 166.8±95.4 | 109.7±73.8 | 1066.9±587.5 |
| CG (n=26) | 4.5±3.0 | 21.0±9.8 | 73.5±97.8 | 67.9±36.8 | 105.1±60.9 | 160.7±41.9 | 139.6±105.6 | 1040.4±643.9 | ||
| GG (n=4) | 1.1±0.4 | 10.1±6.0 | 57.5±28.7 | 30.9±8.3 | 68.4±30.5 | 114.2±41.8 | 92.5±37.7 | 807.7±263.5 | ||
| p values | 0.0130 | 0.0368 | 0.4694 | 0.0171 | 0.5331 | 0.5350 | 0.5786 | 0.6701 | ||
| ABCC2 | C-24T | CC (n=48) | 4.1±3.2 | 22.7±17.6 | 60.4±51.6 | 64.1±33.0 | 98.4±72.5 | 158.1±93.7 | 127.1±78.0 | 683.1±299.7 |
| CT (n=19) | 4.7±3.0 | 23.0±9.8 | 96.8±157.5 | 67.4±33.6 | 112.8±60.1 | 165.1±75.9 | 103.15±110.05 | 1107.0±648.1 | ||
| TT (n=3) | 2.6±1.2 | 26.2±4.7 | 53.3±11.5 | 68.0±27.7 | 116.2±49.4 | 193.9±95.4 | 110.0±17.3 | 1341.8±577.8 | ||
| p values | 0.5066 | 0.1804 | 0.7746 | 0.8393 | 0.3411 | 0.2371 | 0.0913 | 0.3835 | ||
SNPs in complete linkage disequilibrium
The UGT1A8 C518G SNP tended to be associated with MPA dn-C0 (p=0.0130), Cmax (p=0.0368) and dn-AUC0-12h (p=0.0171; Table 3) but not with MPAG exposure indices (Table 3) or MPAG/MPA ratio. Finally, patients homozygous for the UGT1A9 C-440T variant genotype (n=4) had significantly lower MPA Tmax than patients heterozygous or homozygous for the reference allele, but similar MPA or MPAG exposure indices.
Transport activity of OATP1B3 reference and variant proteins
The effect of the SLCO1B3 variant found to be associated with MPA clinical pharmacokinetics was investigated in-vitro.
The uptake of MPAG into cells expressing the OATP1B3 variant protein (i.e., 334G-699A haplotype) was markedly lower than in cells expressing the reference sequence. OATP1B3 variant protein showed a similar Km (122±109 vs 114±69.8 μM) with the reference OATP1B3 but a reduced Vmax (313±188 vs 508±205 pmol/min/mg protein, respectively) (Figure 3).
Figure 3.
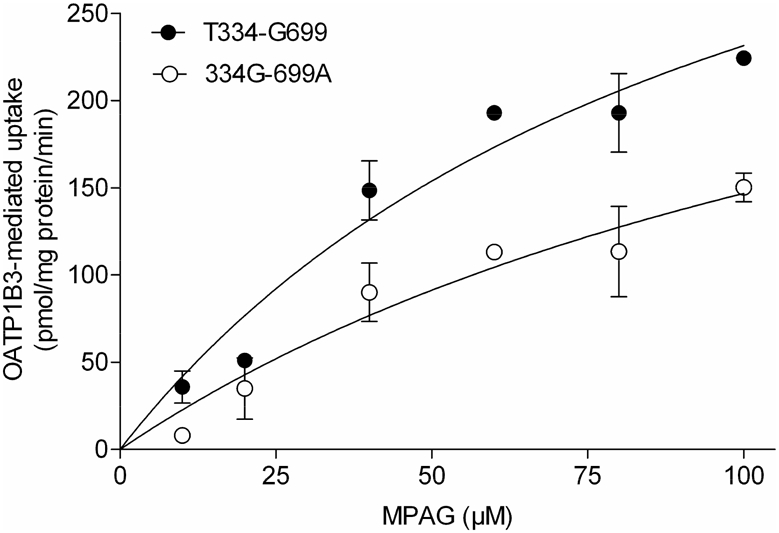
Kinetics of mycophenolic acid phenyl-glucuronide (MPAG) transport by reference (T334-G699) and variant (334G-699A OATP1B3. HEK293 cells transiently expressing either the OATP-reference or the OATP-variant proteins were incubated in the presence of increasing concentrations of MPAG for 10 min. Data were fitted to the Michaelis–Menten equation after subtracting background uptake. Data points represent the mean ±SD from n=3 experiments.
Discussion
This study provides direct evidence that MPAG is a substrate of the liver-specific uptake transporter OATP1B3 and that the pharmacokinetics of MPA is significantly influenced by the common SLCO1B3 T334G polymorphism of this gene in renal transplant patients receiving MMF in a cyclosporine-free immunosuppressive regimen. Reduced transport of MPAG was observed in cells expressing this particular OATP1B3 variant, which is consistent with the trend towards a gene-dose decrease in MPAG exposure with the number of variant alleles observed in these patients. We found a significant decrease in MPA dn-Cmax and dn-AUC0-12h in carriers of the 334G allele. Based on our in-vitro findings and on the change in MPAG exposure in transplant patients, we suggest that this is secondary to reduced MPAG hepatic uptake, hence reduced re-absorption of MPA via the entero-hepatic cycling.
Entero-hepatic cycling is indeed a major feature of MPA pharmacokinetics and its contribution to MPA exposure is well-known. MPAG is extensively produced in the liver, partly excreted into the bile and substantially hydrolysed to MPA in the small intestine, leading to MPA re-absorption estimated to contribute 10 to 60% to total MPA exposure (1). It is generally thought that MPAG biliary excretion occurs just after its production in the liver. Here we suggest that, additionally, the active transport of circulating MPAG into hepatocytes substantially contributes to MPA enterohepatic cycling and then in MPA systemic exposure. Indeed, MPAG plasma concentrations are by far higher than those of MPA and this study strongly suggests that MPAG is actively taken up by the hepatocyte. The trend towards higher MPAG plasma levels in patients carrying the SLCO1B3 G-allele is consistent with our in-vitro findings and supports the hypothesis that reduced OATP1B3 activity would decrease MPAG hepatic uptake. Nonetheless, the change in MPAG levels was not significant, maybe because renal elimination is the main disposition pathway for this metabolite. Thus, despite reducing MPA re-circulation, the decrease in MPAG biliary excretion might have a limited effect on MPAG overall elimination.
We did not observe any association between this SNP and MPA exposure in 115 patients co-treated with cyclosporine, which might be due to the inhibitory effect of cyclosporine on MPA biliary excretion. Inhibition of MRP2 by cyclosporine will indeed reduce MPA recirculation, whatever the activity of OATP1B3 is. Looking at the means ± SD in the three genotype groups (Table 1) as well as to the p value obtained, it seems that the number of patients in this comedication subgroup was sufficient to exclude any clinically relevant influence of this particular SNP on MPA exposure in patients receiving cyclosporine. In contrast, increasing the number of patients cotreated with tacrolimus or sirolimus might have allowed us to investigate a potential additive gene-dose effect.
Human OATP1B3 is mainly expressed in hepatocytes where it mediates, together with OATP1B1, the uptake of a number of drugs or endogenous compounds from blood to hepatocytes (10). In this study, we showed that cell expression of either transporter resulted in the facilitated uptake of the main metabolite of MPA (MPAG). However, OATP1B1 activity with respect to MPAG was relatively modest as compared to that of OATP1B3, suggesting limited implication of OATP1B1 in MPAG uptake in-vivo as compared to OATP1B3. The finding that none of the OATP1B1 polymorphisms or haplotypes tested in this study was significantly and markedly associated with MPA or MPAG pharmacokinetics supports this hypothesis.
We did not observe any difference between HEK293T native and OATP-expressing cells regarding the uptake of the parent drug (MPA). We cannot totally exclude that MPA is a substrate of OATP. Indeed, passive diffusion (or transport via some unknown transporter in HEK) apparently plays a much higher role in MPA than MPAG cellular transport, with a net uptake rate approximately 3-fold higher for MPA than MPAG in HEK293T native cells (0.021 vs 0.008 pmol/mg/min). This may mask the active transport of the drug (if any) in our cellular model.
OATP1A2 is highly expressed in the intestine, the kidney, in cholangiocytes and at the blood-brain barrier (10, 11) where it is supposed to play an important role in drug absorption and drug-drug interactions (11). We observed no difference between native and OATP1A2-expressing HEK293T cells regarding MPA or MPAG uptake, suggesting that these compounds are not substrates of OATP1A2.
The first study investigating the functional consequences of SLCO1B3 polymorphisms showed in stably transfected MDCKII and HEK293 cells that variant OATP1B3 proteins resulting from either of the two coding non-synonymous SNPs T334G (S122A) and G699A (M233I) had similar basolateral membrane localization with the reference protein (12). They did not find differences in uptake activities between these proteins using several model substrates (Estrone-3-sulfate, bromosulfophthalein and 17-β-glucuronosyl estradiol). However, the authors did not investigate the consequence of the combined amino acid substitutions resulting from these two SNPs (in complete linkage disequilibrium). Hamada et al. found that COS-7 cells transfected with the variant SLCO1B3 haplotype (334G-699A) had a significantly reduced ability to uptake testosterone as compared to cells transfected with the reference SLCO1B3 gene or a sequence containing either the 334G or the 699A SNP (7). In the present study, our SLCO1B3 plasmid integrated simultaneously the two SNPs and we obtained results in line with those of Hamada et al, with a decreased uptake of MPAG in cells transfected with the 334G-699A variant as compared to the reference sequence. Our clinical results are compatible with decreased activity of the OATP1B3 variant protein but surprisingly different from those recently published for the same drug in Japanese patients. Miura el al. described higher dn-AUC6-12 (regarded a marker of MPA recirculation) in patients carrying the 334GG (or 699AA) genotype as compared to carriers of the 334TT genotype (6). Based on this pharmacokinetic observation, the authors proposed that MPA uptake into hepatocytes and excretion into bile might be increased in patients with SLCO1B3 334GG genotype. This is in contradiction with previous (12) and the present in-vitro evidence that the 334G allele reduces OATP1B3 activity. Also, the present results do not support this hypothesis as MPA is not a substrate of OATP1B3 in-vitro, and yet we found a lower dose-normalized MPA exposure in carriers of the 334GG genotype. It should be noted that Miura et al. recently reported, in accordance with our hypothesis, that patients homozygous for the SLCO1B3 334G-699A allele had significantly higher MPAG dn-AUC0-12 than patients heterozygous or homozygous for the reference allele (5).
We observed an important decrease of MPA exposure in patients receiving MMF combined with cyclosporine as compared to tacrolimus or sirolimus. This difference is well-known and is thought to result from the inhibitory effect of cyclosporine on MPAG biliary excretion. The mechanisms presumably involved in this drug-drug interaction and in the pharmacogenetic association we observed are very close. It is even tempting to hypothesise that the inhibitory effect of cyclosporine might not only concern MRP2 (as generally admitted) but also OATP1B3. Indeed, in-vitro studies showed that cyclosporine is a potent inhibitor of OATP1B1 and OATP1B3 (13). This inhibitory effect was associated with drug-drug interactions of clinical relevance, in particular with statins (14, 15).
Most of the pharmacogenetic studies performed regarding MMF focused on UGT enzymes or MRP2 but weak associations were mainly reported (see supplementary Table 1). In the present study, we tested the effect of SNPs previously associated with MPA pharmacokinetics using stringent criteria for statistical analysis (i.e., Bonferroni correction): none of the SNPs evaluated significantly influenced MPA pharmacokinetics, whatever the comedication. It is possible that this absence of effect, in particular for UGT1A9 SNPs previously associated in-vitro with a decreased enzyme activity or expression (16, 17) is related to differences in the post-transplantation periods at which patients were studied. Association between the UGT1A9 C-2152T, T98C or UGT1A8 C518G SNPs were indeed observed in patients studied at early post-transplantation periods (<30 days) (18–20), when MPA apparent clearance (CL/F) is known to be higher than at later time-points (21), presumably owing to several time-dependent factors such as the renal function, albumin and haemoglobin levels, decreasing cyclosporine predose levels (22) and inductive effect of corticosteroids on UGT (23). However, this cannot explain the discrepancy with the significant influence of UGT1A9 C-440T/T-331C on MPA dn-AUC0-12h found by Baldelli et al. in stable renal transplants receiving a steroid-free regimen (9).
Our study certainly has some limitations: in addition to those mentioned above, potential covariates such as serum albumin, previously associated with MPAG disposition in renal transplants (9), could not be taken in account in multivariate analyses to ponder the associations (or the absence of associations) found here, contrary to what van Shaik et al. recently did (20), because they were not available in our databases.
Apart from the association of SLCO1B3 genotype with MPA pharmacokinetics (of which the relevance for MMF efficacy or tolerance remains to be investigated in a larger population), our study demonstrates for the first time that OATP proteins are involved in the pharmacokinetics of MPA. It gives new hints regarding the mechanism of drug-drug interactions, in particular with cyclosporine whose known effects on MPA and MPAG plasma levels are similar to those of the mutant SLCO1B3 allele herein.
In conclusion, the pharmacokinetics of MPA in stable renal transplant patients receiving a cyclosporine-free regimen is influenced by genetic polymorphisms of the hepatic uptake transporter OATP1B3, of which MPAG is a substrate. Cyclosporine might inhibit the transporter function and thus attenuate the effect of its polymorphism.
Methods
Cellular assays
SLCO1B3-pcDNA5/FRT plasmids containing the reference sequence (NM_019844) or a sequence with two nucleotide exchanges corresponding to the SLCO1B3 T334G (rs4149117) and SLCO1B3 G699A (rs7311358) polymorphisms were constructed as described in supplementary data. SLCO1A2-PSG5 and SLCO757-PCMV6-XL4 plasmids were a generous gift from Theo J. Visser, Department of Internal Medicine, Erasmus University Medical Center.
Human embryonic kidney (HEK) 293 T/17 cell lines (American Type Culture Collection, ATCC) were routinely cultured at 37°C with 5% CO2 and 95% humidity in low-glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, 100U/ml penicillin, and 100μg/ml streptomycin. Cells were seeded in 10-cm dishes and transfected with 10μg of SLCOs cDNA (or empty vector), 20μl of lipofectamine LTX (Invitrogen, Carlsbad, CA) and 2ml of Optimem (Invitrogen). 24h after transfection, cells were trypsinized and seeded in 24-well poly-d-lysine-coated plates (BD Discovery Labware, Bedford, MA) at a density of 1.5×105 cells/well in order to reach confluence within 24h for uptake assays.
Transport activities were analyzed through measuring intra-cellular accumulation of MPA and MPAG. (20μM). Uptake was initiated by adding the compounds in Hank’s Buffer Salt Solution (HBSS, Invitrogen) after cells had been washed twice and preincubated with HBSS at 37°C for 15 min. After 5 to 10 minutes, the cells were washed twice with ice-cold uptake buffer, lysed in 250μl of NaCl 0.2N (1 hour at 4°C) and lysates neutralized by adding 25μl of 2N HCl. MPA and MPAG were determined using a previously described validated liquid chromatography-tandem mass spectrometry (LC-MS/MS) assay (3). Results were normalized to per-well protein content as measured using the bicinchoninic acid protein assay (Pierce, Rockford, IL).
OATP1B3-mediated net uptake kinetics was studied after 10 min of incubation with MPAG, after subtracting the uptake in empty vector-transfected cells. Km and Vmax were obtained by fitting the Michaelis–Menten equation (GraphPad Software, San Diego, CA).
Clinical pharmacogenetic analysis
A first group of 70 kidney transplant patients enrolled in one of three phase 4 clinical trials gave their informed consent for providing blood samples for pharmacogenetic investigations related to the pharmacokinetics of MMF. 42 patients received MMF in combination with sirolimus and 28 with tacrolimus. MMF was administered at an initial fixed dose of 1g twice daily, which was reduced if necessary based on clinical criteria. Tacrolimus was administered twice daily and sirolimus once daily, targeting a whole blood trough concentration between 0.05 to 0.015 mg/l and 0.010 to 0.012 mg/l, respectively. For MMF pharmacokinetics, blood samples were collected in each patient between 3 and 6 months post-grafting at 0, 20, 40, 60, 90, 120, 180, 240, 360 and 540 min after the morning dose. MPA and MPAG plasma concentrations were determined by use of a validated LC-MS/MS method (24). Area under the concentration-time curve (AUC0-12h) were calculated using validated Bayesian models. In the absence of an adequate model for MPAG, AUC0-9h were estimated using the linear trapezoidal rule. The plasma metabolic ratio of MPAG to MPA was estimated as the AUC0-9h ratio. There was no difference in renal function between patients receiving tacrolimus or sirolimus as assessed by comparing serum creatinine levels at the time of blood sampling (p=0.7659).
A second group of 115 kidney transplants out of 137 patients enrolled in the multicenter, open-label, randomized trial Apomygre (Clinical Trial Registry No. NCT0019967) provided written informed consent for pharmacogenetic investigations related to MMF pharmacokinetics. These patients received the drug in combination with cyclosporine. Study aim and design, patients population and primary results were recently published (25). Blood samples were collected at the same times as in the first group at 3 or 6 months post-transplantation. MPA AUC0-12h were estimated using concentration at three sampling time (20, 60 and 180min) using previously published, validated Bayesian models (21, 26).
All these trials were approved by ethic committees.
There was no statistical difference in corticosteroid dose between patients co-treated with cyclosporine and tacrolimus or sirolimus at the time of blood sampling (p>0.05).
Patients’ DNA was isolated from EDTA-treated blood as previously described in details (27) and was characterized for SNPs in SLCO1B3, SLCO1B1, UGT1A8, UGT1A9 and ABCC2 genes using validated TaqMan allelic discrimination assays on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Courtaboeuf, France).
Statistical analysis
Pairwise linkage disequilibrium between polymorphisms was evaluated by means of D′ and r2 (calculated using Thesias® software (28)) and patients’ most probable haplotypes inferred using the PHASE program (29). Categorical variables were compared using the Fisher’s exact test and continuous variables using the Kruskal–Wallis or Mann-Whitney tests, or ANOVA when studying the interaction between variables. A Bonferroni correction was applied leading to a p threshold for significance of 0.005 (analysis of 10 SNPs). All statistical comparisons were performed using the Stat View program (SAS Institute, Cary, NC, version 5.0).
Supplementary Material
Acknowledgments
This work was supported by Roche Pharma and the Region Limousin.
Footnotes
Conflict of Interest/Disclosure: The authors declared no conflict of interest.
References
- 1.Bullingham RE, Nicholls AJ, Kamm BR. Clinical pharmacokinetics of mycophenolate mofetil. Clin Pharmacokinet. 1998;34:429–455. doi: 10.2165/00003088-199834060-00002. [DOI] [PubMed] [Google Scholar]
- 2.Hesselink DA, et al. Cyclosporine interacts with mycophenolic acid by inhibiting the multidrug resistance-associated protein 2. Am J Transplant. 2005;5:987–994. doi: 10.1046/j.1600-6143.2005.00779.x. [DOI] [PubMed] [Google Scholar]
- 3.Picard N, Premaud A, Rousseau A, Le Meur Y, Marquet P. A comparison of the effect of ciclosporin and sirolimus on the pharmokinetics of mycophenolate in renal transplant patients. Br J Clin Pharmacol. 2006;62:477–484. doi: 10.1111/j.1365-2125.2006.02509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smak Gregoor PJ, van Gelder T, Hesse CJ, van der Mast BJ, van Besouw NM, Weimar W. Mycophenolic acid plasma concentrations in kidney allograft recipients with or without cyclosporin: a cross-sectional study. Nephrol Dial Transplant. 1999;14:706–708. doi: 10.1093/ndt/14.3.706. [DOI] [PubMed] [Google Scholar]
- 5.Miura M, et al. Influence of Drug Transporters and UGT Polymorphisms on Pharmacokinetics of Phenolic glucuronide Metabolite of Mycophenolic Acid in Japanese Renal Transplant Recipients. Ther Drug Monit. 2008;30 doi: 10.1097/FTD.0b013e3181838063. [DOI] [PubMed] [Google Scholar]
- 6.Miura M, et al. Influence of SLCO1B1, 1B3, 2B1 and ABCC2 genetic polymorphisms on mycophenolic acid pharmacokinetics in Japanese renal transplant recipients. Eur J Clin Pharmacol. 2007;63:1161–1169. doi: 10.1007/s00228-007-0380-7. [DOI] [PubMed] [Google Scholar]
- 7.Hamada A, et al. Effect of SLCO1B3 haplotype on testosterone transport and clinical outcome in Caucasian patients with androgen-independent prostatic cancer. Clin Cancer Res. 2008;14:3312–3318. doi: 10.1158/1078-0432.CCR-07-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naesens M, Kuypers DR, Verbeke K, Vanrenterghem Y. Multidrug resistance protein 2 genetic polymorphisms influence mycophenolic acid exposure in renal allograft recipients. Transplantations. 2006;82:1074–1084. doi: 10.1097/01.tp.0000235533.29300.e7. [DOI] [PubMed] [Google Scholar]
- 9.Baldelli S, et al. C-440T/T-331C polymorphisms in the UGT1A9 gene affect the pharmacokinetics of mycophenolic acid in kidney transplantation. Pharmacogenomics. 2007;8:1127–1141. doi: 10.2217/14622416.8.9.1127. [DOI] [PubMed] [Google Scholar]
- 10.Niemi M. Role of OATP transporters in the disposition of drugs. Pharmacogenomics. 2007;8:787–802. doi: 10.2217/14622416.8.7.787. [DOI] [PubMed] [Google Scholar]
- 11.Franke RM, Scherkenbach LA, Sparreboom A. Pharmacogenetics of the organic anion transporting polypeptide 1A2. Pharmacogenomics. 2009;10:339–344. doi: 10.2217/14622416.10.3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letschert K, Keppler D, Konig J. Mutations in the SLCO1B3 gene affecting the substrate specificity of the hepatocellular uptake transporter OATP1B3 (OATP8) Pharmacogenetics. 2004;14:441–452. doi: 10.1097/01.fpc.0000114744.08559.92. [DOI] [PubMed] [Google Scholar]
- 13.Treiber A, Schneiter R, Hausler S, Stieger B. Bosentan is a substrate of human OATP1B1 and OATP1B3: inhibition of hepatic uptake as the common mechanism of its interactions with cyclosporin A, rifampicin, and sildenafil. Drug Metab Dispos. 2007;35:1400–1407. doi: 10.1124/dmd.106.013615. [DOI] [PubMed] [Google Scholar]
- 14.Shitara Y, Itoh T, Sato H, Li AP, Sugiyama Y. Inhibition of transporter-mediated hepatic uptake as a mechanism for drug-drug interaction between cerivastatin and cyclosporin A. J Pharmacol Exp Ther. 2003;304:610–616. doi: 10.1124/jpet.102.041921. [DOI] [PubMed] [Google Scholar]
- 15.Simonson SG, et al. Rosuvastatin pharmacokinetics in heart transplant recipients administered an antirejection regimen including cyclosporine. Clin Pharmacol Ther. 2004;76:167–177. doi: 10.1016/j.clpt.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Bernard O, Guillemette C. The main role of UGT1A9 in the hepatic metabolism of mycophenolic acid and the effects of naturally occurring variants. Drug Metab Dispos. 2004;32:775–778. doi: 10.1124/dmd.32.8.775. [DOI] [PubMed] [Google Scholar]
- 17.Girard H, et al. Identification of common polymorphisms in the promoter of the UGT1A9 gene: evidence that UGT1A9 protein and activity levels are strongly genetically controlled in the liver. Pharmacogenetics. 2004;14:501–515. doi: 10.1097/01.fpc.0000114754.08559.27. [DOI] [PubMed] [Google Scholar]
- 18.Kuypers DR, Naesens M, Vermeire S, Vanrenterghem Y. The impact of uridine diphosphate-glucuronosyltransferase 1A9 (UGT1A9) gene promoter region single-nucleotide polymorphisms T-275A and C-2152T on early mycophenolic acid dose-interval exposure in de novo renal allograft recipients. Clin Pharmacol Ther. 2005;78:351–361. doi: 10.1016/j.clpt.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Johnson LA, Oetting WS, Basu S, Prausa S, Matas A, Jacobson PA. Pharmacogenetic effect of the UGT polymorphisms on mycophenolate is modified by calcineurin inhibitors. Eur J Clin Pharmacol. 2008;64:1047–1056. doi: 10.1007/s00228-008-0501-y. [DOI] [PubMed] [Google Scholar]
- 20.van Schaik R, et al. UGT1A9 -275T>A/-2152C>T Polymorphisms Correlate With Low MPA Exposure and Acute Rejection in MMF/Tacrolimus-Treated Kidney Transplant Patients. Clin Pharmacol Ther. 2009;86:319–327. doi: 10.1038/clpt.2009.83. [DOI] [PubMed] [Google Scholar]
- 21.Premaud A, et al. Maximum a posteriori bayesian estimation of mycophenolic acid pharmacokinetics in renal transplant recipients at different postgrafting periods. Ther Drug Monit. 2005;27:354–361. doi: 10.1097/01.ftd.0000162231.90811.38. [DOI] [PubMed] [Google Scholar]
- 22.van Hest RM, Mathot RA, Pescovitz MD, Gordon R, Mamelok RD, van Gelder T. Explaining variability in mycophenolic acid exposure to optimize mycophenolate mofetil dosing: a population pharmacokinetic meta-analysis of mycophenolic acid in renal transplant recipients. J Am Soc Nephrol. 2006;17:871–880. doi: 10.1681/ASN.2005101070. [DOI] [PubMed] [Google Scholar]
- 23.Cattaneo D, Perico N, Gaspari F, Gotti E, Remuzzi G. Glucocorticoids interfere with mycophenolate mofetil bioavailability in kidney transplantation. Kidney Int. 2002;62:1060–1067. doi: 10.1046/j.1523-1755.2002.00531.x. [DOI] [PubMed] [Google Scholar]
- 24.Premaud A, Rousseau A, Le Meur Y, Lachatre G, Marquet P. Comparison of liquid chromatography-tandem mass spectrometry with a commercial enzyme-multiplied immunoassay for the determination of plasma MPA in renal transplant recipients and consequences for therapeutic drug monitoring. Ther Drug Monit. 2004;26:609–619. doi: 10.1097/00007691-200412000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Le Meur Y, et al. Individualized mycophenolate mofetil dosing based on drug exposure significantly improves patient outcomes after renal transplantation. Am J Transplant. 2007;7:2496–2503. doi: 10.1111/j.1600-6143.2007.01983.x. [DOI] [PubMed] [Google Scholar]
- 26.Premaud A, et al. A double absorption-phase model adequately describes mycophenolic acid plasma profiles in de novo renal transplant recipients given oral mycophenolate mofetil. Clin Pharmacokinet. 2005;44:837–847. doi: 10.2165/00003088-200544080-00005. [DOI] [PubMed] [Google Scholar]
- 27.Le Meur Y, et al. CYP3A5*3 influences sirolimus oral clearance in de novo and stable renal transplant recipients. Clin Pharmacol Ther. 2006;80:51–60. doi: 10.1016/j.clpt.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Tregouet DA, Tiret L. Cox proportional hazards survival regression in haplotype-based association analysis using the Stochastic-EM algorithm. Eur J Hum Genet. 2004;12:971–974. doi: 10.1038/sj.ejhg.5201238. [DOI] [PubMed] [Google Scholar]
- 29.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


