Abstract
The application of FRET-based molecular bio-sensors provided confirmation of the central model of Ran GTPase function and led to important new insights into its physiological role. In many fields of cell biology, methods employing FRET are a standard approach that is becoming increasingly accessible due to advances in instrumentation and available fluorophores. However, the optimal design of a FRET sensor remains to be the cornerstone of any successful FRET application. Utilizing the recent literature on FRET applications and our studies on Ran, we outline the basic considerations involved in designing molecular FRET sensors. We point to several broadly applicable principles that were used in many different FRET sensors that can detect a wide range of molecular events. Using the FRET sensors for Ran that we created as examples, we then focus on the practical aspects of FRET assays. We describe the preparation of a bipartite FRET sensor consisting of ECFP-Ran and EYFP-importin β and its validation as a reporter for FRET-based high throughput screening in small molecule libraries. Finally, we review the design and optimization of monomolecular FRET sensors that monitor the RanGTP-RanBP1 interaction, and of sensors detecting the RanGTP-regulated importin β cargo release.
Keywords: FRET, Förster, fluorescence, sensor, FLIM, GFP, Ran GTPase, importin β, IBB, RanBP1
1. Introduction
The GTPase Ran regulates many essential functions throughout the cell cycle in Eukaryotes, such as nucleo-cytoplasmic transport, mitotic spindle assembly and post-mitotic nuclear envelope and nuclear pore complex formation (1, 2). Central to the understanding of these Ran functions is the idea of an intracellular RanGTP concentration gradient that is driven by the localization of the regulators of Ran. Initially, such a model was strongly predicted by biochemical and genetic evidence (3, 4). The visualization with FRET molecular biosensors provided the definitive proof that the RanGTP gradient indeed exists and functions as the key regulator of nucleo-cytoplasmic transport and mitotic spindle assembly (1) (2–4). Moreover, the application of FRET revealed some unexpected features of Ran-regulated functions, for example, showing that only partial activation of spindle assembly factors (SAFs) around chromosomes by the RanGTP gradient is sufficient to drive mitotic spindle assembly (3).
More recently, evidence for important RanGTP gradient-independent roles of Ran in axon regeneration (5) and in cytoplasmic microtubule dynamics in megakaryocytes (6) was obtained. It is to be expected that the application of FRET in the future will lead to better understanding of such specialized functions of Ran in differentiated cells.
The full understanding of virtually any cell biological process requires its non-destructive observation in living cells. FRET techniques are uniquely suitable to fulfill this requirement and can also be used for studies of cellular mechanisms in intact live organisms (7). In addition, FRET biosensors are applicable in many in vitro formats, from single molecule assays, to ultra-high throughput screening. A number of technological advances during the past few years have made the design and application of FRET sensors more accessible. Most importantly, the dramatic expansion of the palette of fluorescent proteins (FPs) and the improvement of their properties (12–14) enabled the creation of many new genetically encoded FRET sensors. Fluorescent nanoparticles and systems combining the advantages of genetically encoded tags and organic fluorescent dyes have been added to the arsenal of fluorophores used in FRET techniques (8). Contemporary microscopes, flow cytometers and high throughput readers are often designed to be used for FRET-based applications. Finally, several broadly applicable principles of FRET sensor design can be identified in the large number of published studies and used as a guide in designing new sensors. The published molecular structures of proteins or molecular complexes can often be viewed as an opportunity to visualize their function in live cells using FRET. Using the review of published literature and our experience in designing FRET sensors for Ran (2, 3), here we present an introduction to FRET sensor design
2. FRET sensor design
2.1 FRET basics
An excited fluorophore can deactivate by emitting photons (fluorescence emission), by non-radiative dissipation (molecular collisions, heat), by transiting to a non-fluorescent triplet state (followed by phosphorescence) or by transferring the energy to a suitably oriented dipole of another nearby fluorophore. The non-radiative transfer of energy from the excited dipole of the donor fluorophore to the dipole of the acceptor fluorophore is described as resonance energy transfer (9). The acceptor excited by energy transfer can return to its ground state by the same mechanisms as the excited donor, including photon emission (sometimes called “sensitized” or indirectly excited acceptor fluorescence), non-radiative dissipation (fluorescence quenching, where no photons are emitted by the acceptor) or again by resonance energy transfer to another acceptor(10, 11).
In contrast to the potentially misleading term fluorescence resonance energy transfer, no light photons (or fluorescence) are transferred to the acceptor and no acceptor fluorescence is required for resonance energy transfer to occur. Many authors (for example, (10, 12–14)) prefer to name the process as Förster resonance energy transfer (FRET) after the pioneering German physicist Theodore Förster who accurately described the process decades before it became widely used (9).
The distance at which the probability of the energy transfer between donor and acceptor is 50% is called the Förster distance, R0. The R0 depends on the spectral overlap of the fluorophores (J; the integral of the donor emission and acceptor excitation spectrum overlap), on the orientation of the electric dipoles (κ2), on the refractive index of the media (n) and on the quantum yield of the donor QD (Eq.(1)) (11):
| (1) |
In standard conditions, the R0 is considered a characteristic property of each donor-acceptor (D-A) pair as a population of molecules. For FRET to occur, the acceptor emission spectrum must overlap with the donor excitation spectrum. In many applications, the donor emission is a shorter wavelength compared to the acceptor excitation spectrum. However, FRET also occurs between identical fluorophores (homo-FRET or energy migration FRET, EM-FRET). In EM-FRET, the donor quantum yield and spectral properties of both the donor and acceptor do not change but the process is detectable via fast anisotropy decay of the D dipole. A spatially resolved detection of EM-FRET in cells can be achieved by fluorescence anisotropy measurements using fluorescence lifetime microscopy (22–24).
The efficiency of FRET (E) increases with R0 and declines with the 6th power of the distance R between the D-A:
| (2) |
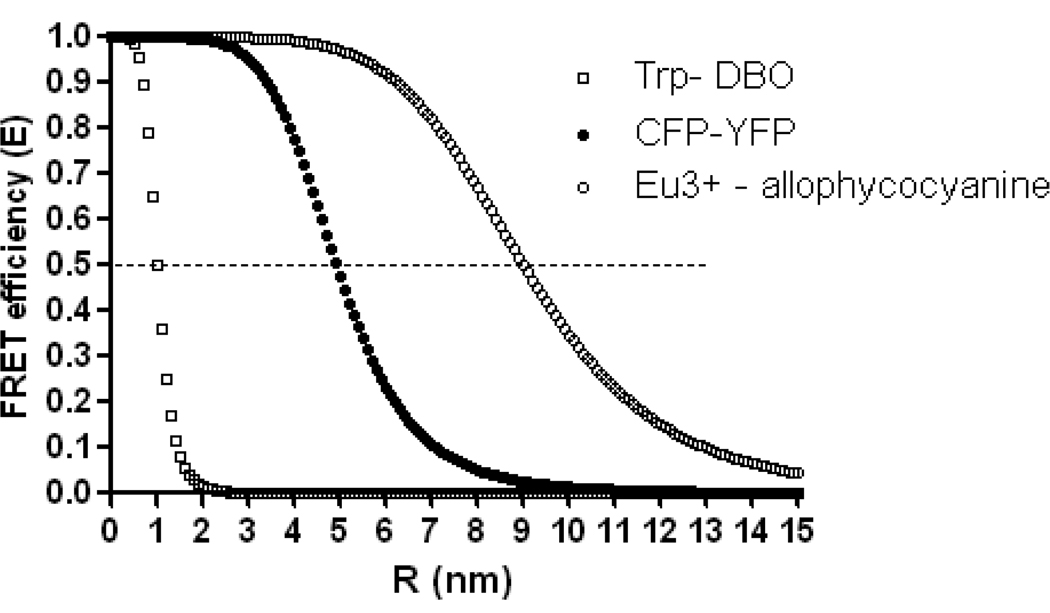
The FRET detection range ~1–10nm (Fig. 1) matches the size of many biological molecules and complexes, underlying FRET usefulness in biology. The distance range for sensitive FRET measurements lies between the inflexion points of the E vs. R curve, i.e. ~ 0.5–1.5 R0, corresponding to ~3–8 nm in the case of the CFP-YFP pair. Small molecule D-A pairs with very short R0 enable measurements even at distances as short as 1nm (15) (Fig. 1). Because the FRET signal is usually easily detectable at R< R0, the minimal requirement of a new sensor is to achieve smaller D-A separation than is its R0. Ideally, in FRET sensors whose function depends on the R changes, the R fluctuates around the R0 value.
Figure 1. FRET efficiency (E) dependence on the D-A distance (R) and on the Förster distance R0.
The dependence is plotted for ECFP-EYFP pair (filled circle, R0=4.9 (103)) and for the D-A pairs representing the extreme known values of R0: the Europium cryptate – crosslinked allophycocyanine (empty circle (104)}, R0 = 9nm) and tryptophan – 2,3-diazabicyclo[2.2.2]oct-2-ene-labeled asparagine (DBO), (empty square, R0=1nm (15)). Note that the useful distance detection range of the D-A pairs depends on the R0 and that the maximum sensitivity of the FRET sensor is achieved when the distance between the D and A changes around R0.
The orientation of the D-A dipoles has a strong effect on E: no FRET occurs when the dipoles are perpendicular (κ2 =0) and the energy transfer is maximal when the dipoles are aligned in parallel (κ2 =4). However, it is difficult to control and measure the κ2 effect on E experimentally (11) and the role of κ2 in the FRET sensors remains incompletely understood. For example, although it is expected that between freely moving fluorophores κ2=2/3 (11), computational simulations showed that κ2 converges to ~1 in FRET sensor where the D-A pair is presumed to be freely mobile (16). (See more discussion below).
2.2. Planning for a FRET-based assay
The basic considerations in planning an effective FRET assay fall into the following areas:
The objective of the assay
FRET detection method
FRET sensor design
Fluorophore choice
In cell biology, FRET sensors that can be applied both in vivo and in vitro are advantageous because the in vitro assays with defined components can validate the FRET sensor function in extremely complex in vivo conditions. In addition, the dose-response calibrations of the FRET sensor in vitro can be used to quantify the in vivo data (3, 17). However, designing such versatile FRET sensors can be difficult and is not always necessary. For example, a FRET sensor well suitable for in vitro assays may be toxic when expressed in cells and a validated cell-expressed sensor could be difficult to isolate for large scale in vitro assays. The objective of the assay therefore strongly influences how the FRET sensor is designed and detected.
2.3. FRET detection methods
Emission intensity-based FRET detection takes advantage of the FRET-induced decrease of the fluorescence emission from the donor and the increase of the acceptor emission (unless the acceptor acts as a quencher). With appropriate controls (see below), the ratio of the acceptor and donor emission intensities detected in samples exposed to donor excitation light is often used as a measure of FRET efficiency. Alternatively, the ratio of the donor emission over the directly excited acceptor emission (which is constant under moderate excitation) can help to evaluate potential sensor concentration-induced artifacts in FRET imaging (18). The spatial and temporal resolution of the intensity ratio- based methods is excellent particularly in low E conditions (10) and these methods are easy to perform on most fluorescence microscopes. In all variations of the intensity-based methods, controls for the spectral cross-bleeds between the donor and acceptor are critical in extracting the correct E estimate (19–21). This is best realized with single-construct FRET sensors where the D-A stoichiometry is fixed (provided the sensors are intact) and is possible with bipartite sensors whose concentration can be sufficiently controlled in the control samples.
Acceptor depletion techniques
If the A is destroyed as a fluorophore by strong illumination at its excitation wavelength, the D emission intensity recovers to the level observed in the absence of FRET. The ratio of the D emission after and before the A photodestruction is a measure of E which can be conveniently used for spatially resolved imaging in live samples (10). The disadvantages of this method include artifacts induced by sample movement during the bleach period and the only single time point measurements. This so called “acceptor bleach” technique is easily performed on confocal laser scanning microscopes (22) and serves as an excellent control for other FRET detection methods.
FRET detection by fluorescence lifetime imaging microscopy (FLIM)
Fluorescence is a stochastic process that can be described by the rates of radiative (kr) and non-radiative (knr) processes deactivating the fluorophore. The decay of fluorescence over time is characterized by the exponential constant τ, the fluorescence lifetime. In standard conditions, τ is thought to be a characteristic molecular property of each fluorophore which corresponds to the average time the fluorophores spend in the excited state. τ is the inverse of the sum of the radiative (kr) and non-radiative (knr) deactivation rates:
| (3) |
The presence of FRET increases the non-radiative deactivation rate (knr), resulting in the shortening of the donor lifetime in the presence of the acceptor (τDA) as well as the decrease of the donor emission intensity. The FRET efficiency E is straightforwardly related to the changes of the τ due to FRET:
| (4) |
In Eq. 4, the τD (τ of the isolated donor) can be considered as a constant which can be measured separately. The τDA measurements by the fluorescence lifetime imaging microscopy (FLIM) are therefore sufficient for the quantitative detection of the E. Importantly, FLIM is not affected by the FRET sensor concentration both in theory and within a wide range of concentrations also in practice (7, 33, 34). This is a decisive advantage of FLIM in quantitative measurements with bipartite FRET sensors (4, 23). A related advantage of FLIM- FRET detection is the reduced (although not absent) concern for the D- to -A spectral cross-bleed.
Because the fluorescence lifetime is sensitive to the refractive index n (24), controls should be employed in samples where n can locally vary, such as live cells. FLIM can be actually used to visualize the refractive index in live cells (25).
FLIM is performed either in the time- or the frequency domain (26). The widely used time-domain FLIM method is the time-correlated single photon counting (TCSPC) where τ is measured directly by detecting the time of individual photons arrival to a detector correlated with a high frequency pulsed laser (27). In frequency-domain FLIM, τ is calculated from the shift in phase and the decrease of amplitude of the D emission from samples exposed to modulated excitation light (26).
These three widely used FRET detection methods are only a subset of theoretically possible options (17, 40) that are awaiting experimental demonstration or wider use. For example, the large size of FPs enables the detection of FRET between FP-tagged proteins by loss of emission polarization (28). Another technique called acceptor saturation (satFRET) was recently demonstrated as a live cell FRET imaging method feasible on conventional confocal microscopes. The satFRET is related to the “acceptor bleach” method and provides successive FRET measurements (29).
2.4. Widely applicable principles of FRET sensor design
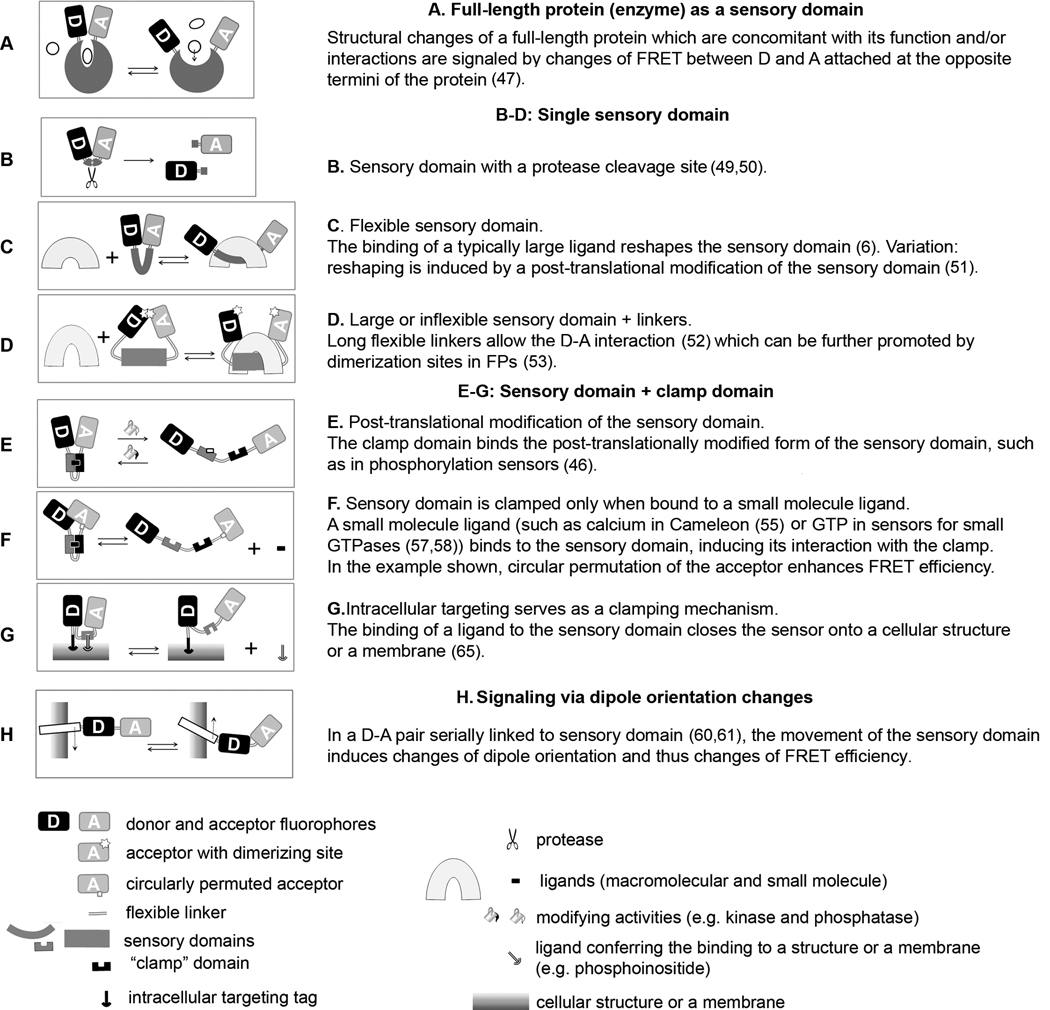
The donor and acceptor FPs can be located either within one molecule or on two or more separate molecules.
Bipartite FRET sensors are created by separately attaching the donor and acceptor to two molecules whose interaction is to be measured by FRET. No detectable FRET is expected in the absence of the interaction. Biochemical or structural evidence can be used to ensure that the donor and acceptor are positioned at a distance < R0 during the interaction. Choosing between the N- and C- terminal position of the FPs or deciding for the appropriate chemical labeling strategy is often sufficient in average-size proteins. Either of the interacting proteins (or domains) can also be incorporated within the structure of the FPs (30, 31).
In monomolecular FRET sensors, the donor and acceptor are both physically linked, in most cases flanking the sensory domain. The structural rearrangement of the sensory domain and/or its occupancy with a ligand induces change of the distance of the D-A pair, resulting in a change in E. The switch from the “closed” (the smallest R and maximum E) to the “open” (the maximum R, low E) sensor conformation can be driven by the absence or presence of the molecular event to be sensed. Both scenarios are equally useful. In principle, the function of a stable multimolecular complex containing subunits labeled with the donor and acceptor (for example, Arp 2/3 complex with two labeled subunits (32)) is similar to the monomolecular FRET sensor. Several broadly applicable design principles were used to create FP-based FRET sensors for many different purposes.
Some monomolecular FRET constructs, including FRET sensors for several kinases (33), contain full-length protein molecules as the sensory domain. In the Ran field, the CFP-RCC1-YFP FRET sensor was used to monitor the dynamic structural changes of RCC1 (RanGEF) involved in its binding to chromatin and in its interaction with Ran (34).
However, not many full length signaling molecules can be effectively used as FRET-sensors with the currently available labeling and detection techniques. A sensor for Ran function was created by labeling Ran with a C-terminally attached GFP (D) and Alexa546-maleimide covalently attached to an internal cysteine (A) (35). Although this sensor displayed the expected behaviors in vitro, also as expected, its “open” conformation was induced by its interactions with RanBP1 as well as with importin β, limiting the use of the sensor in live cells (35).
Because protein domains are smaller and often specialized in one function which can be preserved in new molecular contexts, it is easier to create protein domain-based FRET sensors.
In the simplest scenario, a flexible sensory domain is flanked by the D-A pair and undergoes significant structural change in the absence or presence of the signaling event. In sensors detecting large ligands such as interacting proteins, the ligand binding to the sensory domain pushes the flanking FPs away from each other, decreasing the FRET (2, 3). In the sensors for proteases, the donor and acceptor are joined by a peptide with specific substrate sequence whose cleavage results in irreversible D-A separation (49) (36). In another of the many variations, the flexible sensory domain contains cysteine pairs whose reversible connection through disulphide bridges signals redox potential (37).
To minimize the R, up to 5 N-terminal and 11 C-terminal amino acids can be removed from the CFP and YFP, respectively (38). If the sensory domain is still too large, it could be joined to the donor and acceptor with long flexible linkers (glycine- and serine-containing repeats) to allow the FP interaction in the ligand-free sensor. The D-A interaction in such sensors could be further promoted by the use of moderately dimerizing FPs such as CyPet-YPet (39, 40).
The function of many monomolecular FRET sensors is based on the conditional interaction of domains from two different proteins. That is the case for already the first highly successful FP-based FRET sensor “Cameleon” (41). In Cameleon, the binding of calcium to calmodulin (CaM) induces the CaM- binding domain to clamp onto the calcium - CaM complex, driving the molecule to the closed conformation where the flanking D-A pair can interact. The clamp (the M13 domain in most Cameleons) is either joined to CaM with a flexible linker or even placed internally as a hinge joining two lobes of the CaM structure (in YC6.1 (42)). Variations on the sensory domain linked to a “clamp” are used in numerous FRET sensors, particularly for detecting small ligands and wherever the sensory domain does not undergo sufficient structural change. In kinase sensors, a domain containing a kinase-specific substrate sequence is clamped by a neighboring phosphopeptide-binding domain once phosphorylation occurs (33). In sensors for small GTP-ases, the GTPase is linked to a protein specifically binding to its GTP-bound state (43, 44). Alternatively, the clamp can drive the sensor to the closed conformation in the absence of the signal. In a FRET sensor for phosphoinositides, a negatively charged clamp peptide competes with phospoinositide binding to a PH sensory domain (45).
FRET sensors using dipole orientation changes
Rapidly responding FRET sensors for voltage-gated channels were created where the FRET signal presumably results from changes of κ2 in serially linked FPs attached to a rotationally moving channel subunit (46, 47) (Fig. 2). A promising lead in controlling κ2 is the application of circularly permuted FPs (cpFP), where the wt N-and C-termini (located on the same side of the GFP β-barrel) are fused and new termini are created at various locations of the FP (48). By testing combinations of the wt and differently permuted FPs as D-A pairs, the optimal κ2 can be found. The cpYFP173 used in Cameleon 3.60 indeed resulted in a ~5x larger E amplitude compared to Cameleons with wt YFP (62). However, only cpYFP173 increased the performance of several different FRET sensors, although their structural diversity would predict the requirement for different permutations (49). This raises the question of whether the benefit obtained with cpYFP173 is indeed solely due to the optimal κ2.
Figure 2. Monomolecular FP-based FRET sensors.
The schematic drawings and brief summaries illustrate functional principles of genetically encoded FRET sensors. In most cases the same functional principle was used in a number of different FRET sensors and only examples of references are included. Some of the design principles can be combined between categories, e.g. circularly permuted FPs (F) with any other category. Most sensors exploit the changes of R and κ2 between the D-A pair. FRET sensors with circularly permuted FPs (F) presumably benefit from the optimal κ2 and the changes of κ2 are thought to be the major cause of FRET signal changes in H (46).
Intracellular targeting and tuning of FRET sensor
The great advantage of genetically encoded FRET sensors is their possible delivery to specific sites within cells by the appended intracellular targeting sequences. However, it is important to realize that the FRET measurements with targeted rather than freely diffusible sensors can significantly differ, due to considerations such as limiting ligand concentration. The art of developing intracellularly targeted FRET sensors is therefore linked to the proper “tuning” of their dynamic range. This question was extensively explored in the history of calcium measurements with Cameleons (50). Anchoring the sensor to membranes (51) or perhaps to other structures can be used as a part of the clamp mechanism in a FRET sensor reporting on regulated binding (Fig. 2).
2.5. The choice of fluorophores
In addition to the intrinsically fluorescent natural amino acids for in vitro studies, the most commonly used D-A pairs in biology are synthetic organic dyes and FPs (52, 53). During the past few years, these options were expanded by FRET sensors based on semiconductor nanoparticles (Quantum dots) (54)), non-natural autofluorescent aminoacids (55), several systems combining genetically encoded protein tags targeted by synthetic dyes (31, 53, 56, 57) and by the introduction of bioluminescent donors (58) (59)
The major advantages of synthetic organic fluorophores include small size (<1KD) and in some cases favorable spectral and photochemical properties compared to FPs. Due to their high quantum yield and photostability, several dyes (Cy3, Cy5, Cy3.5, Alexa dyes) are the preferred choice for single molecule FRET studies (60). Many organic dyes and quenchers are available commercially as conjugates with reactive moieties for protein labeling.
Probably the two most common methods for protein labeling are with N-hydroxysuccinimide-esters (NHS) conjugates (for labeling N-terminal α-amines and ε-amines in lysines) and dye-maleimide conjugates (for labeling the sulfhydryl groups of cysteines). Such dye-protein conjugates are particularly well suited for in vitro applications with characterized proteins available in purified form. Several more advanced strategies were developed to selectively label proteins in vitro and even in live cells on specific sites with organic dyes (31, 56, 57, 61).
Compared to dyes, FPs are significantly larger (~25–30kD) and some still posses sub-optimal photochemical and/or spectral characteristics. However, the genetic control of the FRET sensor clearly outweighs such relative disadvantages. In addition, ongoing research leads to continuing improvement of FPs (53, 62, 63). The performance of the existing FRET sensors then can be upgraded by using the best available new D-A pairs. What are the properties of the current and future FPs to be considered for FRET sensor design?
The high molecular brightness and photostability under continuous illumination are among the most important features determining the suitability of FPs for FRET. In some FPs, the high brightness is not correlated with photostability (64). In ECFP and possibly other FPs, the instability under illumination can be detected as shortening of the fluorescence lifetime with little effect on the emission intensity (65). In all methods using the emission from both the donor and the acceptor, their brightness should be similar to avoid problems with the possibly limited detection range of the instruments. For the same reasons, the concentrations of the donor and acceptor in the samples should not differ by more than 10-fold (19).
Longer excitation and emission spectra of the sensor increase light penetration into the sample and induce less photodamage. The cellular damage caused by ultraviolet excitation compromises the observations of normal function and often also triggers rise of the cellular autofluorescence. Several common sources of autofluorescence such as flavins and NADH which are present in mammalian tissue culture cells are excited and emit below ~500nm (66). On the other hand in plant cells, the presence of chlorophyll favors the excitation in the UV range which is possible with the TSapphire-mOrange pair (exc. 399nm; (67)).
The calculation of monoexponential fluorescence lifetime requires <200 photons (68) while 10–100 -times more photons are required to resolve multiexponential decays with similar accuracy. Because of its monoexponential fluorescence lifetime, brightness and photostability, EGFP is an excellent donor for the detection of FRET with FLIM. Thus, EGFP was used for high resolution FLIM-FRET imaging in neurons despite the incomplete maturation of the mCherry and mRFP acceptors in the bipartite sensor (23). An improved brighter variant of ECFP called Cerulean has mostly monoexponential fluorescence lifetime (69), although its second fluorescence lifetime component is detectable with high resolution detectors (23). In our hands, the use of Cerulean moderately improved the resolution of FLIM-FRET imaging in live cells (18). However, the Cerulean- EYFP pair displays low E (39) and its replacement with the CyPet-YPet produced 2–3 fold larger amplitude of FLIM-FRET signal in vivo, despite the clearly 2-exponential τ of CyPet (Kalab, data not shown). Although the monoexponential τ is preferrable in FLIM, it is therefore not an overriding factor. The current promising alternative to CFP is the bright coral-derived mTFP1 with monoexponential τ (70).
Fast and complete maturation of both donor and acceptor FPs are desirable. The presence of immature non-fluorescent acceptor decreases the FRET signal detected by either FLIM or fluorescence intensity-based methods. The immature non-fluorescent donor introduces artifacts to all FRET detection schemes involving donor emission intensity. Although in FLIM the non-fluorescent donor is not recorded, the “invisible” donor population complicates the quantitative analysis of the FLIM results.
The environmental sensitivity of FPs is one of their potential weaknesses. The pH sensitivity of FPs is decreased by mutations resulting in overall lower pKa (63). The FPs can also negatively affect their molecular environment by producing reactive oxygen radicals during excitation (70). An extreme example is the “killer Red” coral fluorescent protein which can be used for light-induced local inactivation in cells (71).
An R0 of at least ~5nm is preferable because it maximizes the E amplitude of the FP-based FRET sensor. The minimal distance between the chromophores of two interacting FPs is ~2.5–3nm (in GFP, the chromophore location is fixed within the 4.2 × 2.4nm FP β- barrel (72)), corresponding to the first inflexion point of the E vs. R curve (Fig. 1) at R0 ~5nm. At this R0, even small separation of the FPs is detected with high sensitivity. The large R0 in some D-A pairs comes with significant spectral overlap on which the R0 depends and therefore with increased concern for spectral cross-bleed. Whether the large R0 is beneficial therefore often depends on the detection technique. For example, although the R0 of CFP-mRFP is smaller than that of GFP-mRFP, the cross-bleed subtracted FRET signal of CFP-mRFP in cells was greater (73). Because the spectral overlap factor J (Eq. (1)) increases with the wavelength, large R0 is displayed by some of the orange-red emitting D-A pairs (74). Some of the more advanced of today’s microscopes are equipped for spectrally-resolved detection of fluorescence which then can be used to un-mix the overlapping spectra and subtract the cross-bleeds of even GFP (emission 510nm) and YFP (emission 525nm) (75). Another approach to harnessing the large R0 is the use of acceptors converted by mutagenesis into fluorescence quenchers, i.e. lacking fluorescence emission. The first such usable “dark” acceptor for GFP called sReach (76, 77) was derived from YFP and has R0= 5.5–5.9nm with GFP (77). The FLIM imaging in neurons with GFP-sReach pair displayed a significantly improved signal to noise ratio compared to GFP-mRFP (77).
The wild type GFP is a dimer (Kd= ~100µM (78)) and many naturally occurring FPs are obligatory tetramers. In cell biological applications, strong oligomerization of FPs is unwelcome because it alters the interactions and localization of FP-tagged proteins and is toxic in cells. Only monomerized versions of red FPs could be used to produce transgenic animals(79). Mutagenesis of the dimerization sites is therefore commonly used to produce FPs for research. On the other hand, moderate dimerization decreases the average R, increasing the E. The CyPet-YPet D-A pair developed by directed evolution (39) displayed ~7 fold increase of E compared to original ECFP-EYFP mostly due to the re-introduction of dimerizing S208F mutation in YPet (80). As a result, the CyPet-YPet dimerization skewed the measurements of low affinity peptide interactions by FRET (80). On the other hand our results indicate that the detection of high affinity interactions (Kd =2nM for Snp1 IBB- Importin β interaction (18)) with CyPet-YPet FRET sensors in live cytoplasm is not appreciably altered (Fig. 4). Such a conclusion was also reached in a study which evaluated the effect of the A206K monomerizing mutations in CFP and YFP on the behavior of the tagged estrogen receptors in cells (81). Although the higher dimerization of the FP tags slightly improved FRET detection, the interactions of the tagged estrogen receptors were not detectably affected (81).
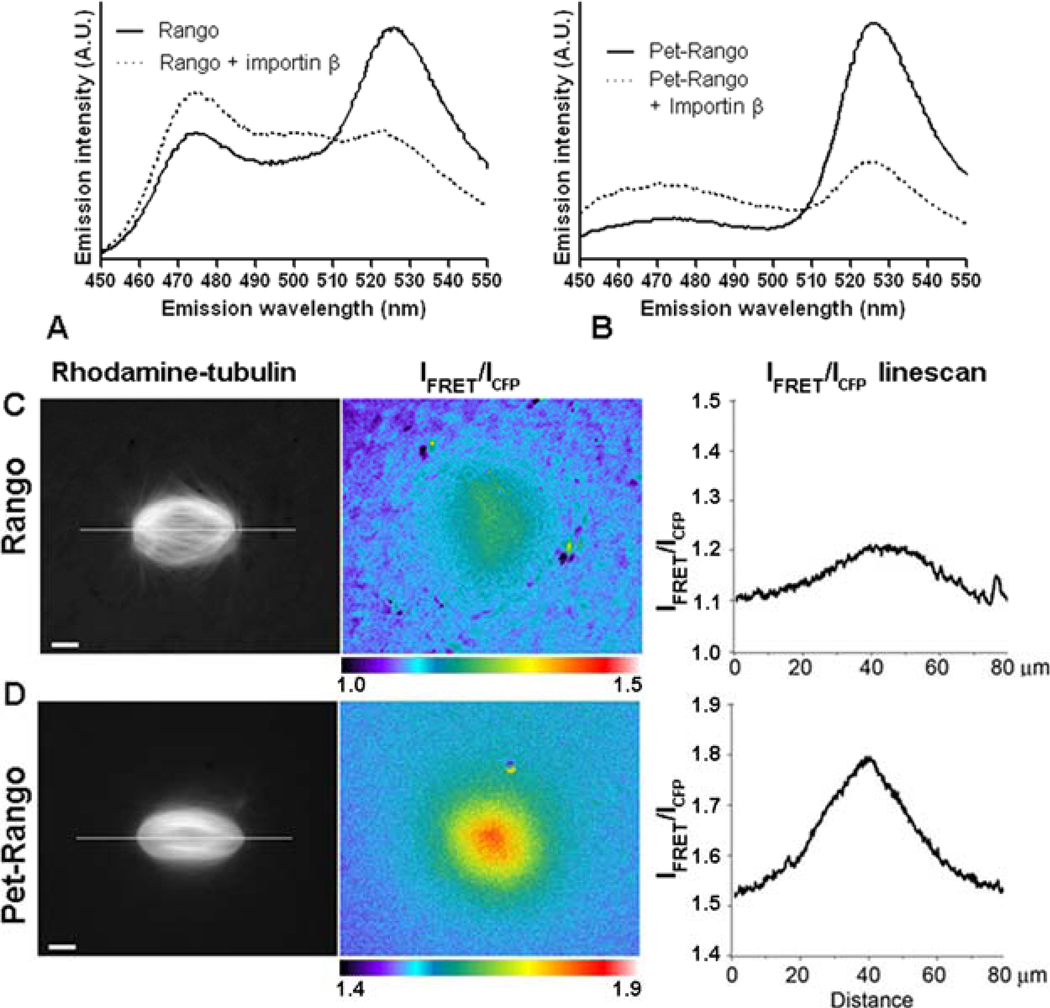
Fig. 4.
The CyPet-YPet FRET pair increases the dynamic range of the Rango sensor in the mitotic cytoplasm. In A and B, samples containing Rango sensor containing either ECFP-EYFP pair (left) or CyPet-YPet donor-acceptor pair (right) were analyzed in parallel. The samples with 0.5 µM Rango sensor alone (red lines) or in the presence of 1 µM importin β (blue lines) were excited at 435 nm and the emission was measured between 460-550nm. The IFRET/ICFP ratios are summarized beneath the panels. C and D show representative images of the mitotic Ran-regulated gradients detected by FRET imaging in Xenopus laevis egg extracts. The mitotic spindle assembly was induced by the addition of sperm nuclei to the extract containing Rhodamine-labeled tubulin to visualize microtubules (left panels in C and D) and 2 µM Rango. The IFRET/ICFP ratios are displayed as pseudocolored images (center) and as linescans (right panels in C and D) through the regions of the mitotic spindles indicated by a line shown in the left panels. An identical span of the IFRET/ICFP range was used in ratio images and linescans.
Photo-convertible FPs respond to illumination at a specific wavelength by reversible switching to alternate excitation and emission spectrum or by switching between fluorescent and dark states. Donor-induced photoswitching of the acceptor by FRET was used to create an inducible tag suitable for the measurements of fast intracellular diffusion (82).
Several combinations of the D-A pairs and FRET detection techniques were developed to detect two FRET pairs simultaneously in a single cell (93–96).
3 FRET sensors for Ran
3.1 Introduction to Ran GTPase
The GTP- vs. GDP-bound state of Ran is controlled by two Ran-specific regulators: the guanine nucleotide exchange factor (GEF) RCC1 and the RanGTP-ase activating protein RanGAP. Both RCC1 and RanGAP accelerate their respective intrinsic Ran activities ~105-fold. RCC1 is highly enriched around chromosomes and several mechanisms control this localization. First, RCC1 is imported to nuclei and then it binds to DNA and histones on the chromatin in a regulated fashion (83). In contrast, RanGAP is mostly cytoplasmic, although a fraction of RanGAP binds to kinetochores in mitotic cells (84) and RanGAP is present also in the nuclei in some organisms (85). In combination with the high mobility of Ran, the localized generation of RanGTP by RCC1 near chromosomes and the acceleration of GTP hydrolysis by Ran induced by Ran-GAP in the cytoplasm, produces the intracellular RanGTP concentration gradient (86).
The functions of Ran are mediated by the interactions of RanGTP with nuclear transport receptors (NTRs) which control the binding of NTRs to their molecular cargos. The NTRs are usually grouped within a superfamily of importin β-like proteins, although they are comprised of several structurally diverse groups of proteins(87). Common to NTRs is their capacity to transit the molecular gate of the nuclear pore complex (NPC) channel either alone, while carrying their molecular cargos or while carrying both cargo and Ran (88). Depending on where they deliver their molecular cargos with respect to the nucleus, the NTRs are called either importins or exportins. The binding of RanGTP to the importins causes the dissociation of nuclear localization signal (NLS)-containing cargos from the importins. Because RanGTP is normally scarce in the cytoplasm and abundant in the nucleus, the RanGTP gradient across the nuclear envelope (NE) directs the nuclear import of NLS cargos. Nuclear export signal (NES) containing cargos are loaded on exportins in a complex that includes RanGTP. This hetero-trimeric export complex dissociates in the cytoplasm due to RanGAP-catalyzed GTP hydrolysis of Ran-GTP (reviewed in (83)).
The dynamic RanGTP-regulated binding of NTRs to their cargos continues after the NE is disassembled in mitotic cells of higher Eukaryotes. Instead of directing the nuclear-cytoplasmic transport, the RanGTP gradient surrounding the mitotic chromosomes directs the localized activation and/or targeting of spindle assembly factors (SAFs). The network of the RanGTP gradient- and NTR-regulated reactions can be thought of as a genome positioning system (GPS) that spatially directs many cellular functions with respect to the position of the chromosomes (83). For our studies of Ran with FRET sensors we used 3 different reactions involved in the formation and intracellular functions of the Ran-regulated cellular GPS.
3.2 Bipartite FRET sensors for the RanGTP-importin β interaction
Because Ran has many functions throughout the cell cycle, rapidly acting small molecule inhibitors would be useful to analyze Ran functions for example during the different phases of mitosis. Therefore, we were interested in designing FRET sensors that would be suitable as reporters in high throughput screens (HTS) for inhibitors of the RanGTP-importin β interaction in small molecule libraries. We compared chemically dye-labeled and FP-tagged D-A bipartite FRET sensors.
The RanGTP binding site on importin β (89) is a large N-terminal region that wraps around RanGTP. The single surface-exposed cysteine 112 in Ran is located 2–5 nm from multiple exposed lysine residues in importin β. Labeling Ran with a maleimide-conjugated dye and importin β with an NHS-dye conjugate therefore appeared to be a good starting strategy. We also noticed that the N-terminal aminoacids in Ran and importin β that are recorded in the structure of the bound proteins are only about 2 nm apart (89), indicating that labeling both proteins separately at N-termini with ECFP and EYFP (R0 5.17nm) should produce a functional FRET sensor.
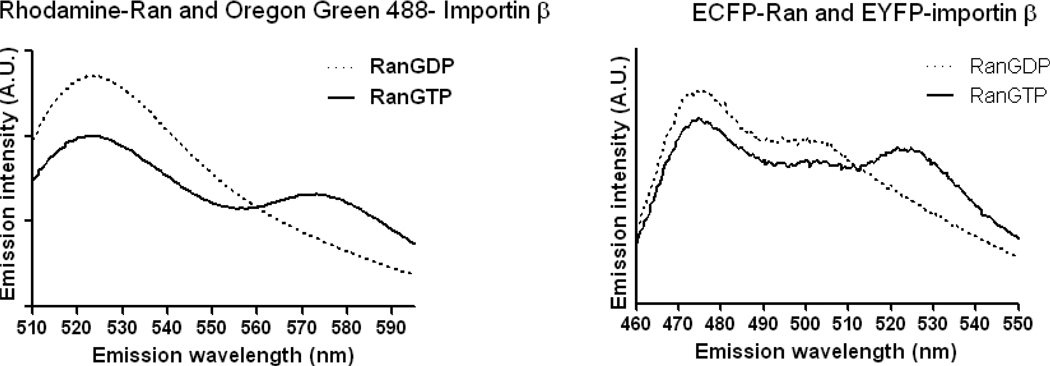
We prepared an Oregon Green 488- importin β and Rhodamine X- Ran pair and also cloned, expressed and purified an ECFP-Ran and EYFP-importin β FRET pair (see section 4). First, we tested the functionality of both FRET sensors by spectrometry in reactions where the nucleotide charge of Ran was controlled by the addition of either GTP or GDP in the presence of unlabeled RCC1. We expected that in both sensors, the amount of FRET should increase in the presence of GTP, resulting in an increase in acceptor fluorescence and a concomitant decrease in donor emission. In solutions containing GDP, FRET should decrease resulting in the opposite effects on the fluorescence emissions The sensors indeed displayed the expected behavior (Fig. 3).
Figure 3. Chemically labeled and FP-tagged bipartite FRET sensors for RanGTP-importin β interaction.
A. The D- A pair prepared by chemical labeling of purified proteins with activated fluorescent dyes. B. The D-A pair consisting of FP-tagged proteins. In both A and B, the emission spectra of the excited by the donor excitation wavelength (488nm in A and 435 in B) proteins were analyzed in spectrofluorimeter. The reactions were carried out in the presence of recombinant unlabelled RCC1 and either GDP or GTP. In both reactions, the interaction of RanGTP with importin β was signaled by the increase of acceptor emission and decrease of donor emission, as expected.
Next, we performed control experiments to determine whether the labeled proteins remained functional. For example, pull-down experiments with recombinant proteins confirmed the ECFP-RanGTP interaction with EYFP-importin β. This interaction did not occur in reactions containing ECFP-RanGDP. The addition of RanGAP and RanBP1 to RanGTP-containing reactions produced emission spectra resembling those seen in incubations with RanGDP, indicating that the interactions of the labeled proteins remain dynamic (data not shown).
The dye-conjugated FRET pairs can be prepared relatively easily starting with purified unlabeled proteins (2–3 days), compared to FP-based sensors requiring cloning. Different dyes could be used in the HTS assays to escape the fluorescence background present in small molecule libraries. However, sub-stoichiometric concentrations of the importin β relative to Ran were needed to achieve optimal detection of FRET by the intensity ratio method available for HTS (0.4 molar ratio, Fig. 3). Likely, this occurred because multiple Lysines were labeled with dye in importin β while only a single cysteine was dye-labeled in Ran. In addition, the multiple steps and reagents involved in the labeling inevitably result in batch-to-batch variability which complicates the analysis of the HTS.
We therefore decided to perform the HTS with the FP-tagged FRET sensor. In preliminary assays, we set up reactions in a 384-well format where CFP-Ran and YFP-importin β interactions were controlled by the nucleotide charge of Ran. For the detection of the FRET signal, we used the ratio of the acceptor over donor emission intensity in samples excited with the donor excitation. Potential “hits” in a small molecule screen would be expected to decreasethe FRET ratio, similar to control reactions containing CFP-Ran-GDP. To evaluate the robustness of the assay, we calculated the Z’-factor (90):
| (5) |
The μ in Eq.(5) are the averages of FRET efficiency values measured in sample wells containing EYFP-Importin β together with ECPF-RanGTP (maximum) or ECFP-RanGDP (minimum signal) and σ are the corresponding standard deviations. Because assays with Z’ between 0.5–1.0 are considered acceptable (90), the Z’=0.71 we detected encouraged us to proceed. Indeed, the HTS of ~130 000 compounds identified a promising inhibitor of the RanGTP-importin β interaction which is currently being analyzed in secondary assays.
3.2 YRC, the RanGTP-binding monomolecular FRET sensor
Initially we were interested to design FRET sensors that would allow us to directly verify whether the RanGTP gradient exist in cells and to visualize it by FRET microscopy. We reasoned that even if we succeeded in creating a sensor that would directly visualize the complex of Ran with GTP (as compared to RanGDP), it would not allow distinguishing the free RanGTP generated around chromatin from the many different types of RanGTP-NTR complexes.
For this reason we considered designing sensors that would detect RanGTP by specifically interacting with it or through a RanGTP-regulated event. Because we planned to detect such sensors using ratiometric FRET imaging, a monomolecular construct was clearly preferable. Given such considerations, we were looking in the published literature for highly flexible protein domains specifically responding to RanGTP, the size of which should not exceed about 5 nm (N- to C-terminus span). The guidance provided by the published molecular structures was sufficient to create two types of sensors described below: YRC that is interacting with RanGTP directly and YIC (and its version called Rango) that is released from the importin β by RanGTP.
The YRC sensor is based on the highly flexible Ran-binding domain (RBD) which functions as a co-factor of RanGAP in promoting the GTP hydrolysis on Ran. The RanGTP bound to NTRs is protected from RanGAP in the cytoplasm and accessory proteins carrying the RBD are required to release RanGTP from NTRs and present it to RanGAP (91, 92). In human cells, RBDs with this function are present in the large cytoplasmic nucleoporin RanBP2 (with 4 RBDs) and in the small, soluble cytoplasmic protein RanBP1.
In the structure of the RBD1 from RanBP2 (89) bound to Ran-GppNHp (GTP analogue resistant to hydrolysis), the N-terminus of the RBD extends ~2.2 nm and wraps around Ran, whose C-terminal domain is also extended. Because RBD1 is an internal component of RanBP2, we assumed that it would remain functional when flanked by FPs on both termini. However, we decided to create this sensor with the RBD derived from S. cerevisiae RanBP1 (Yrb1) which is highly homologous to RBD1 (93) and better characterized. We constructed E.coli expression plasmids coding for Yrb1-RBD flanked by ECFP and EYFP (YRC), and expressed and purified the protein from E.coli.
The unbound YRC protein should be capable of FRET and in fact it did display significant emission from EYFP when excited at the ECFP excitation (435nm) as compared to an equimolar mixture of RBD-EYFP and RBD-EYCFP proteins (6). In the presence of RanGTP, the YRC IFRET/ICFP ratio significantly decreased, consistent with the expected RanGTP-induced extension of the RBD N-terminus, while reactions with YRC and RanGDP were indistinguishable from YRC protein alone (2). We observed no importin β-induced decrease of the YRC FRET signal in reactions containing RanGDP (PK, unpublished data), indicating that unlike RanBP1 (35), the YRC does not form a trimeric complex with importin β and RanGDP.
The specificity of the YRC FRET signal was tested in a number of assays and the protein was used to detect the RCC1-generated RanGTP gradient in the Xenopus laevis egg extracts (2). The YRC contains nearly full length Yrb1, where the minimal RBD domain (94) is extended on both sides by several aminoacids. However, full truncation of the sensory domain in YRC to the minimal RBD domain did not produce functional FRET sensors (data not shown). Our currently favored ΔN YRC sensor contains a sensory domain shortened to the start of the minimal RBD and the same C-terminal extension of RBD as present in the original YRC. When equipped with the Cerulean-YPet D-A pair, this sensor allowed the detection of RanGTP gradient in the mitotic Xenopus laevis egg extracts using confocal FLIM microscopy (Fig. 5).
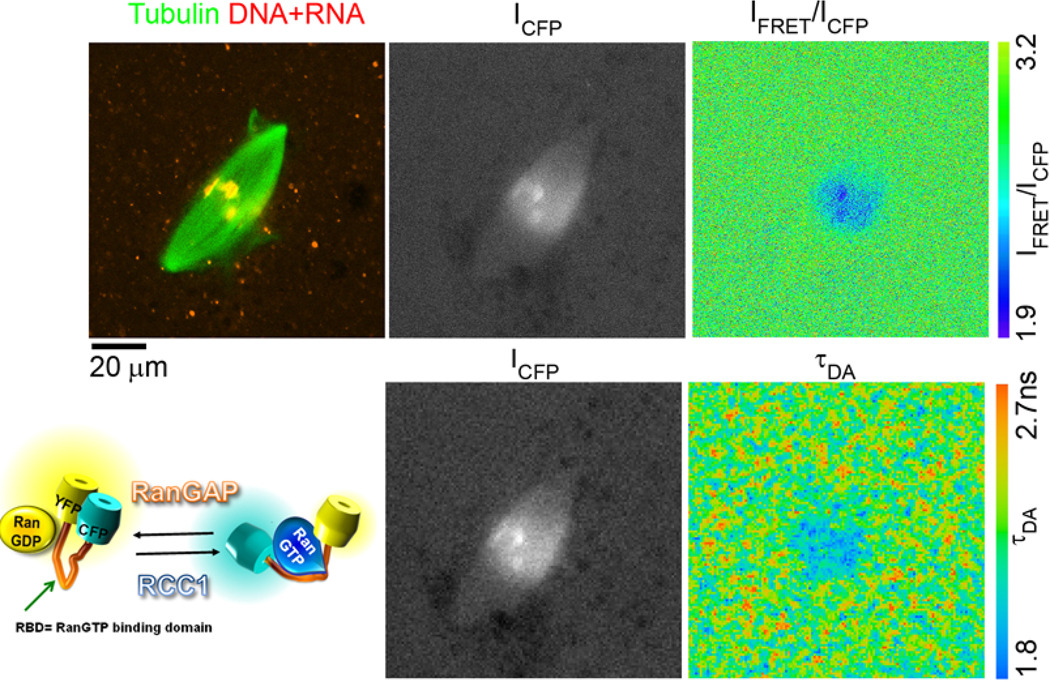
Figure 5.
The detection of RanGTP gradient in Xenopus laevis egg extracts with FLIM using improved YRC FRET sensors. The ΔN YRC sensor containing Cerulean and YPet (pKW2388) was used to detect the RanGTP gradient during mitotic spindle assembly in Xenopus laevis egg extracts. The image of the mitotic spindle labeled with Rhodamine-tubulin and TO-PRO3 dye (staining DNA in chromosomes and RNA in the cytoplasm) was acquired (top row, left), followed by confocal scan with pulsed 435 nm laser excitation and acquisition with CFP (ICFP image, top row, middle) and YFP bandpass (IFRET image, not shown) filters. Finally, 30 sec scan with 435 nm pulsed laser excitation was used to acquire FLIM dataset using Becker-Hickl SPC-830 TCSPC board (lower row; ICFP image). The IFRET/ICFP pseudocolored ratio image (top, right) serves as a control for the FRET detection using FLIM (lower row, right; the calculated τDA pseudocolored image). The schema of the YRC-like sensors is shown in the bottom row left.
3.3. Monomolecular FRET sensors for RanGTP-regulated importin β cargos
In addition to detecting the RanGTP gradient, we were interested to visualize with FRET sensors the RanGTP-regulated biochemical reactions that were thought to have an essential role in the mitotic spindle assembly. The evidence suggested that the function of Ran in the mitotic spindle assembly requires the RanGTP-induced localized release of spindle assembly factors (SAFs) from their inhibitory complexes with importin β (95, 96). The YIC (Rango) FRET sensors described below allowed testing this model directly.
Importin β carries its cargos either directly or indirectly in complexes with adaptor proteins such as importins α and Snurportin 1. The binding of both direct and indirect cargos to importin β is negatively regulated by RanGTP. The importins α contain a flexible N-terminal importin β binding domain (IBB) and a C-terminal Armadillo repeat domain where the NLS cargos are loaded. The cargos are efficiently loaded only when the IBB is sequestered by binding to importin β, because the IBB binds to the Armadillo domain and competes with the cargos for binding (110, 111). In the complex of importin β with the IBB of importin α1, the IBB extends into an α-helical rod >5nm long (97), while in isolation, the IBB is apparently unstructured. Because the IBB is necessary and sufficient for the RanGTP-regulated importin α-importin β binding (98), we reasoned that an IBB-based FRET construct could serve as indirect RanGTP sensor and direct sensor of importin α- and importin β cargo liberation. To test this prediction, we created an EYFP-IBB-ECFP (YIC) construct with IBB taken from human importin α1 (1–61, KPNA2).
When examined in spectrometer in isolation, YIC displayed a 1.2–1.4 IFRET/ICFP ratio and the addition of an equimolar concentration of importin β decreased the IFRET/ICFP ratio to 0.8, indicative of the expected importin β-induced transition to the extended conformation. Importantly, the addition of RanGTP to the importin β-YIC sample induced full FRET signal recovery, signaling the formation of RanGTP-importin β complex and the release of free form of YIC (2). After its validation (2), we used YIC together with YRC to analyze the function of the RanGTP gradient in mitotic spindle assembly. In the mitotic Xenopus laevis egg extract, we obtained direct evidence that the RCC1 generated RanGTP gradient is responsible for the formation of an importin β cargo gradient and that both gradients are required for the spindle assembly (6).
Because we were interested in analyzing the RanGTP gradient function in somatic cells, we tested the performance of YRC and YIC in HeLa cells. The FRET signal of YRC was difficult to detect in live HeLa cells, at least in part due to the relatively small amplitude between the maximal and minimal signal (data not shown). YIC on the other hand displayed toxic effects in transfected or microinjected cells, particularly during mitosis. After testing several modifications of the importin α1 YIC and sensors based on IBBs from other importins α, we settled on YIC based on the IBB from Snurportin 1 (Snp1) which we renamed Rango (Ran-regulated importin β cargo) (18). Another change in Rango was the replacement of the ECFP with Cerulean which improved the resolution of FLIM assays. In a variety of in vitro tests, Rango behaved indistinguishably from the importin α1 YIC and was well tolerated by mitotic HeLa cells. Using 3 modes of Rango FRET signal detection (FLIM, acceptor bleach and intensity ratio) we then analyzed the mitotic RanGTP gradient in HeLa cells (3). More recently, a structural study confirmed the RanGTP-regulated binding of Snp1-IBB to importin β (99).
In an effort to further improve the YIC-like sensors, we tested a variety of D-A combinations. A particularly encouraging improvement of the Rango performance was achieved when the Clean-EYFP pair was replaced with the CyPet-YPet pair (39) (Fig. 4).
3. Materials and methods
4.1 The construction of plasmids for FRET sensors
To generate 6His-S tag- EYFP-importin β fusion (pKW1532), the EYFP was polymerase chain reaction (PCR)- amplified from the pEYFP plasmid (Clontech) and inserted via BspHI and NcoI sites into a pET30a- human importin β expression vector (pKW485) (100). Similarly, the 6His-ECFP-Ran fusion (pKW1534) was created by in-frame insertion of ECFP (Clontech) and wt human Ran into pET30a vector. For the functionality of the FRET sensor it was critical that the ECFP and Ran were joined by GGG linker and EYFP to importin β by a linker containing GGSGS (data not shown).
In all monomolecular FRET constructs, the individual components were initially PCR-amplified with Pfu or Pfx polymerase (Invitrogen), inserted into pRSET-A (Qiagen) and sequenced. Linkers were either synthesized and annealed or included in PCR oligos. Ligations were performed using the Quick ligation kit (New England Biolabs, (NEB)) with home-made buffers (which are the expensive component of the kit) and purchased T4 ligase (NEB). Subsequently, we used subcloning and PCR (where necessary) for combining the individual segments in frame, followed by restriction analysis and sequencing of the products.
To create 6His-tagged YRC (pKW966), the RBD from Saccharomyces cerevisiae Yrb1 (amino acids 37–201) was PCR-amplified and inserted between EYFP and ECFP (both Clontech). The ORFs for EYFP and EYCFP were PCR amplified with the appropriate restriction sites. Similarly, the IBB from human importin α 1 (1–65) was combined with EYFP and EYCFP to create 6His-YIC. To create 6His-Rango (pKW1648), the Snurportin 1 IBB (1–65) (101) was PCR-amplified and inserted between Cerulean (69) and EYFP. In Pet-Rango (pKW2162) the ECFP-EYFP pair in pKW1648 was replaced with the PCR- amplified YPet and CyPet (39) by 3 step subcloning (due to Kpn1 sites in both FPs and the pKW1648). The 6His-ΔN YRC with D-A pair (pKW2388) was assembled by insertion of Yrb1 (65–201) between Cerulean (69) and YPet (39).
For expression in tissue culture cells, the YRC and YIC ORFs were transferred to the pSG8 plasmid. The vector for the expression of Rango in cells (pKW1608) was created by transferring the pKW1648 ORF into pSG8 and removing the N-terminal 6His tag. The Pet-Rango for tissue cell expression (pKW2303) was created by replacing the pKW1608 ORF with pKW2162 ORF.
In silico cloning and clone records
The creation of the multi-component FRET sensor constructs required systematic electronic database and cloning records that were matched with the physical oligo and plasmid archives. Electronic records for plasmids and oligos were stored in home-built FileMaker database (FileMaker, Inc.). The consecutive numbers assigned by the FileMaker program were used to label the corresponding materials stored at −20°C.
We constructed the plasmids in silico with Gene Construction Kit 2.5 (GCK2.5, Textco, Inc.) or with Vector NTI Advantage (Invitrogen). The optimal sequence of PCR oligos was checked with Oligo 6 and the oligos (no purification, 200µM concentration in water) were ordered for overnight delivery from a local supplier (Elim Pharmaceuticals). The sequencing reactions were checked against the original in silico constructs in GCK2.5.
Other recombinant protein constructs
The construct for the E.coli expression of the 6His-protein A(ZZ) tag- RanQ69L (pKW1234) was created in pRSET A (18). The 6His-Stag-wt Ran for dye-labeling was expressed from pET30a expression plasmid (pKW1517). The wt RCC1 protein was expressed from pET3b (117).
4.2. Protein production in E.coli
All FRET sensor proteins were expressed in BL21 DE3 E.coli cells (Invitrogen) using a similar protocol. A few colonies of freshly transformed cells were collected from LB/Agar plates and used to inoculate ~25 ml pre-cultures in LB with antibiotics and incubated at 37°C on a shaker for 3–4 hours. The production cultures were started by inoculating 1l LB + antibiotics in 2.8l glass Fernbach flasks (flat bottom, no side baffles) with 6–25ml of the pre-culture. The flasks were kept at 37°C on a horizontal shaker (220 rpm) until the OD600nm reached 0.5–0.6. The cultures were transferred to a shaker kept at 22°C, and protein expression was induced with 0.3 mM IPTG overnight (16–18 h) on the shaker. The cells were harvested by centrifugation at 4000 rpm in a Sorvall 6×1l H-6000A (15–20 min, 4°C). The supernatant was decanted and the pellets were resuspended in ice-cold lysis buffer (PBS, 10 mM imidazole, pH 7.4, 100 mM PMSF, 10 µg/ml each of chymostatin, pepstatin and leupeptin) and collected by centrifugation into 50ml Falcon tubes. The cell pellets were either frozen in liquid Nitrogen and stored at −80°C, or resuspended in lysis buffer and immediately lysed using a French pressure cell. The lysates were clarified by centrifugation for 20 min. at 15 000 rpm in a Sorvall SS34 rotor at 4°C. The 6His tagged proteins were then batch-bound to Ni-agarose beads (Qiagen or Sigma) and the beads were washed on disposable plastic Polyprep columns (BioRad) with PBS, pH 7.4 plus 10 mM imidazole.
The Ni-agarose beads were extensively washed in the cold room (4°C) by gravity-driven perfusion with the ice-cold lysis buffer (>20 bead volumes). The 6His-tagged proteins were eluted from the Ni-agarose beads with PBS, 0.2M imidazole and then dialyzed in 3 exchanges of XB (50 mM sucrose, 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, pH 7.7, 4°C).
To reduce the concentration of partially translated or incompletely matured FRET sensors, the proteins were purified by FPLC gel filtration on Sepharose 6 or Superdex 200 columns (Pharmacia). Absorbance at 435nm and 512nm was measured during gel filtration to detect CFP and YFP and eliminate trailing fractions with partially inactivated fluorophores. Proteins were concentrated on Amicon 30 ultrafiltration columns (Millipore) to 100–200 µM, passed through a 0.45 µm filter, snap-frozen in single use aliquots in liquid Nitrogen and stored at −80°C. The protein concentration was measured with Bradford reagent (Pierce) using BSA as a standard and the purity of the proteins was assessed by SDS PAGE with Coomassie Blue R250 staining.
The ECFP-Ran and EYFP-importin β proteins for the HTS screen were expressed in the UC Berkeley fermentation facility, each in single 200 l batches and using similar conditions as described above. Similarly, the pellets with cells were stored in liquid Nitrogen before lysis in the French press and batch purification of the 6His-tagged proteins.
The expression of Ran and importin β for dye labeling followed a similar protocol as for the FP-tagged proteins. The wt Ran protein was further purified by Thrombin cleavage from the Ni-agarose beads using the biotinylated thrombin kit (Novagen) and the instructions of the manufacturer. The wt RCC1 protein was expressed and purified as described (102).
4.3 Labeling of Importin β and Ran with dyes
The Oregon Green 488-NHS and Rhodamine-X-maleimide were purchased from Molecular Probes (Invitrogen; currently only Rhodamine-C2-maleimide is available). The labeling was carried out with minor modifications of the conditions recommended by the manufacturer (Invitrogen, Molecular Probes). For Rhodamine-X-maleimide labeling, the dye was dissolved in DMSO to 10mM final concentration just before use. The Ran concentration was adjusted to ~100 µM in XB and the protein solution was placed in a 1.5ml Eppendorf tube wrapped in aluminum foil on ice. The dye solution was added in small drops to achieve a 10-fold molar excess over the protein and the tube was quickly mixed by inversion and kept covered on ice for 2–4 hours. The reaction was stopped by a 10 mM final concentration DTT and the unincorporated dye first removed by filtration on NAP5 disposable gel filtration columns (Pharmacia, GE Life Sciences) and then by dialysis against XB. The dialyzed protein was concentrated on Amicon 10 columns (Millipore).
Similarly, Oregon Green 488-NHS was dissolved in DMSO to a 10 mM concentration and added drop-wise to ~100 µM importin β to achieve a final 10–20 molar excess of dye over the protein. Again, the protein solution was kept on ice in a covered 1.5ml Eppendorf tube for 2–4 hours. The reaction was quenched by the addition of 5% v/v Tris-glycine buffer (0.25M Tris, 1.92M glycine, pH 8.3) and the unincorporated dye removed by NAP5 gel filtration and dialysis in XB. The protein was concentrated on Amicon 30. Both proteins were snap-frozen in liquid Nitrogen in small aliquots and stored at −80°C.
4.4 FRET detection
Spectrophotometry
Spectra were analyzed with Fluorolog 2 spectrofluorimeter controlled by the Datamax 2.2 (Jobin Yvon Spex) and the Grams 3.04 II software package (Galactic Industries, Salem, NH). For purified protein solutions, the excitation wavelength was 435 +/− 2nm and the emission between 460–550 nm was acquired with 2nm slits. The emission was acquired at 0.5–2nm increments and the scanning speed was varied between 0.01 and 0.1s/wavelength, depending on the FP concentrations. The instrument and the samples were equilibrated at 25°C and all scans were performed with samples placed in quartz cuvettes. The fluorophore concentration was adjusted to 100–500 nM in most experiments. Background subtraction was not required for purified protein solutions above ~40nM.
To avoid autofluorescence in Xenopus egg extracts and HeLa lysates ((6, 28); data not shown), the excitation wavelength was lowered to 405nm and the emission slits were widened to 5–15nm. The fluorescence background was subtracted using control scans in identical samples diluted with buffer instead of the FPs.
For standard CFP-Ran/YFP-importin-β FRET assays, 50µl solutions were prepared containing 100 nM CFP-Ran, 20 nM RCC1 and 200 µM GDP or 200 µM GTP. This was followed by the addition of a 50 µl solution of 100 nM YFP-importin-β and analysis in the spectrofluorimeter (435 nm excitation and 460–550 nm emission).
FRET detection in HTS
The Analyst AD (Molecular Devices) automated high throughput fluorescence reader at the Small Molecule Discovery Center (SMDC, University of California San Francisco, Mission Bay campus) was used to measure FRET between ECFP-Ran and EYFP-importin β in the HTS for small molecule inhibitors. The reactions contained 62.5nM EYFP-importin β, 50nM ECFP-Ran, 20nM RCC1, 2mM MgCl2 and 200 µM GTP or GDP in PBS, pH 7.2.
Imaging of FRET in Xenopus egg extracts
The detection of FRET in the Xenopus egg extracts was performed essentially as described previously (2, 18). The freshly prepared Xenopus laevis CSF egg extracts were supplemented with Rhodamine tubulin (6), 2 µM FRET sensor, 500 demembranated Xenopus laevis sperm nuclei/ µl and incubated on water bath at 20°C. At selected time intervals, 2–4 µl aliquots were placed on microscope slides and mitotic spindles were found under wide field epifluorescence illumination. The 2–4 µl drop of the extract was placed on microscope slide, covered by 22/22mm coverslip and placed on the microscope (Olympus BX71). The mitotic spindles were found in the Rhodamine channel and 14 bit images in Rhodamine, CFP, YFP and CFP-to-YFP FRET channels were then acquired. The exposure time (0.5–2s) was constant for all 3 FP fluorescence images. The 8 bit ratio image of the IFRET/ICFP was produced using the Metamorph (v. 7 Molecular Dynamics, Inc) imaging software.
Detection of FRET with FLIM
The samples for the detection of FRET with FLIM in the Xenopus laevis egg extracts were prepared as above except that 5 µM TO-PRO 3 (Invitrogen) DNA and RNA dye was added to the sample. The detection conditions were essentially as described in (18) except that the SPC730 TCSPC board was replaced with SPC830. This allowed to increase the photon count rate to 3–4 MHz, increasing the image size to 128×128 pixels while reducing the acquisition time to 30–60 sec at 2 µM FRET sensor concentration in the sample. At selected time intervals, 2–4 µl aliquots were placed on microscope slides and mitotic spindles were found under wide field epifluorescence illumination for Rhodamine. The Rhodamine images were acquired with 543nm HeNe laser and TO-PRO-3, with the 633nm HeNe laser (both lasers Spectra Physics) for excitation. Next, the images for the control FRET intensity ratio file were acquired using a 435nm pulsed (80MHz) laser which was generated as the second harmonics of the 870 nm pulsed mode-locked MaiTai laser (Spectra Physics) excitation. The emission was acquired using 450 +/−30nm and 500 +/−20 nm bandpass filters. Finally, the 128×128 pixel FLIM images were acquired using the 435nm 1 photon pulsed laser excitation, 480nm emission bandpass filter, PMC-100 photomultiplier (Becker- Hickl) and SPC830 TCSPC board. The scan time was 30 seconds and the excitation intensity was set to limit the count rate to 3–4MHz. The average fluorescence lifetime of the Cerulean donor τDA was calculated from 2-exponential decay profiles using SPCI software package (Becker&Hickl). Binning was set to one and incomplete exponential decay and no fixed constants (shift and scatter) were assumed in the calculation.
Table 1.
Donor-acceptor FP pairs
| Category | Reference | Comment | |
|---|---|---|---|
| Blue -> green | |||
| mTagBFP –GFP | (1) | Bright photostable blue donor for GFP | |
| Cyan -> Yellow | |||
| Cerulean-EYFP | (2, 3) | Suitable for FLIM, EYFP obsolete; low E | |
| ECFP-cpYFP173 | (4) | Increased dynamic range due to circular permutation in EYFP; incompletely understood mechanism |
|
| SCFP1, SCFP3 and SYFP | (5) | Simultaneous detection of two spectrally identical sensors by FLIM |
|
| CyPet-YPet | (6) | Dimerizing D-A pair with high E; bright YPet does not fold well at 37°C |
|
| mTFP- GFP or orange FP | (7) | mTFP superior to other cyan FPs including Cerulean |
|
| Green -> orange or red | |||
| GFP-mCherry | (8) | Excellent for FLIM; incomplete mCherry maturation |
|
| Orange -> Red | |||
| mKO-Cherry | (9) | High FRET efficiency; R0=6.4nm | |
| Other | |||
| TSaphire- mOrange | (10) | Excitation of green fluorescence at 399nm. Particularly useful in plants |
|
| mOrange – TagRFP | (11) | Highly photostable monomeric D-A pair | |
| GFP-sReach | (12) | Dark acceptor for GFP, better than mCherry | |
| TagRFP-mPlum with ECFP-Venus |
(13) | Simultaneous detection of two FRET pairs | |
Acknowledgements
All the experimental work in this study was performed in the laboratories of Dr. Karsten Weis and Dr. Rebecca Heald at the University of California at Berkeley, Berkeley, California. We thank Dr. Valarie Barr for valuable suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mishin AS, Subach FV, Yampolsky IV, King W, Lukyanov KA, Verkhusha VV. Biochemistry. 2008;47:4666–4673. doi: 10.1021/bi702130s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizzo MA, Springer GH, Granada B, Piston DW. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 3.Kaláb P, Pralle A, Isacoff E, Heald R, Weis K. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 4.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kremers GJ, Goedhart J, van Munster EB, Gadella TW., Jr Biochemistry. 2005;45:6570–6580. doi: 10.1021/bi0516273. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen AW, Daugherty PS. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 7.Ai HW, Henderson JN, Remington SJ, Campbell RE. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasuda R, Harvey CD, Zhong H, Sobczyk A, van Aelst L, Svoboda K. Nat Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 9.Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW., Jr PLoS ONE. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayle V, Nussaume L, Bhat RA. Plant Physiol. 2008;148:51–60. doi: 10.1104/pp.108.117358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakoshi H, Lee SJ, Yasuda R. Brain Cell Biol. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant DM, Zhang W, McGhee EJ, Bunney TD, Talbot CB, Kumar S, Munro I, Dunsby C, Neil MA, Katan M, French PM. Biophys J. 2008;95:L69–L71. doi: 10.1529/biophysj.108.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimozono S, Hosoi H, Mizuno H, Fukano T, Tahara T, Miyawaki A. Biochemistry. 2006;45:6267–6271. doi: 10.1021/bi060093i. [DOI] [PubMed] [Google Scholar]
References
- 1.Kalab P, Heald R. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman B, Zheng Y. Biochem Soc Trans. 2006;34:716–721. doi: 10.1042/BST0340716. [DOI] [PubMed] [Google Scholar]
- 3.Clarke PR, Zhang C. Trends Cell Biol. 2001;11:366–371. doi: 10.1016/s0962-8924(01)02071-2. [DOI] [PubMed] [Google Scholar]
- 4.Kuersten S, Ohno M, Mattaj IW. Trends Cell Biol. 2001;11:497–503. doi: 10.1016/s0962-8924(01)02144-4. [DOI] [PubMed] [Google Scholar]
- 5.Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ, Verlhac MH. J Cell Biol. 2007;176:295–305. doi: 10.1083/jcb.200605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalab P, Weis K, Heald R. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 7.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 8.Caudron M, Bunt G, Bastiaens P, Karsenti E. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- 9.Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal-Ruder Y, Ben-Yaakov K, Hieda M, Yoneda Y, Twiss JL, Fainzilber M. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulze H, Dose M, Korpal M, Meyer I, Italiano JE, Jr, Shivdasani RA. J Biol Chem. 2008;283:14109–14119. doi: 10.1074/jbc.M709397200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasan MT, Friedrich RW, Euler T, Larkum ME, Giese G, Both M, Duebel J, Waters J, Bujard H, Griesbeck O, Tsien RY, Nagai T, Miyawaki A, Denk W. PLoS Biol. 2004;2:e163. doi: 10.1371/journal.pbio.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 13.Tsien RY. Keio J Med. 2006;55:127–140. doi: 10.2302/kjm.55.127. [DOI] [PubMed] [Google Scholar]
- 14.Shaner NC, Patterson GH, Davidson MW. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 15.Sapsford KE, Berti L, Medintz IL. Angew Chem Int Ed Engl. 2006;45:4562–4589. doi: 10.1002/anie.200503873. [DOI] [PubMed] [Google Scholar]
- 16.Förster T. Annalen der Physik. 1948;2:55–75. [Google Scholar]
- 17.Jares-Erijman EA, Jovin TM. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 18.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer Science; 2006. [Google Scholar]
- 19.Wallrabe H, Periasamy A. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, Aoki K, Matsuda M. Brain Cell Biol. 2008;36:19–30. doi: 10.1007/s11068-008-9028-5. [DOI] [PubMed] [Google Scholar]
- 21.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Gruenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lidke DS, Nagy P, Barisas BG, Heintzmann R, Post JN, Lidke KA, Clayton AH, Arndt-Jovin DJ, Jovin TM. Biochem Soc Trans. 2003;31:1020–1027. doi: 10.1042/bst0311020. [DOI] [PubMed] [Google Scholar]
- 23.Clayton AH, Hanley QS, Arndt-Jovin DJ, Subramaniam V, Jovin TM. Biophys J. 2002;83:1631–1649. doi: 10.1016/S0006-3495(02)73932-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altman D, Goswami D, Hasson T, Spudich JA, Mayor S. PLoS Biol. 2007;5:e210. doi: 10.1371/journal.pbio.0050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahoo H, Roccatano D, Zacharias M, Nau WM. J Am Chem Soc. 2006;128:8118–8119. doi: 10.1021/ja062293n. [DOI] [PubMed] [Google Scholar]
- 26.Pantano S. J Mol Graph Model. 2008;27:563–567. doi: 10.1016/j.jmgm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Kalab P, Pralle A. Methods in Enzymology. 2008;89:539–566. [Google Scholar]
- 28.Kaláb P, Pralle A, Isacoff E, Heald R, Weis K. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 29.Piston DW, Kremers GJ. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Puhl HL, 3rd, Koushik SV, Vogel SS, Ikeda SR. Biophys J. 2006;91:L39–L41. doi: 10.1529/biophysj.106.088773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karpova TS, Baumann CT, He L, Wu X, Grammer A, Lipsky P, Hager GL, McNally JG. J Microsc. 2003;209:56–70. doi: 10.1046/j.1365-2818.2003.01100.x. [DOI] [PubMed] [Google Scholar]
- 33.van Munster EB, Gadella TW. Adv Biochem Eng Biotechnol. 2005;95:143–175. doi: 10.1007/b102213. [DOI] [PubMed] [Google Scholar]
- 34.Festy F, Ameer-Beg SM, Ng T, Suhling K. Mol Biosyst. 2007;3:381–391. doi: 10.1039/b617204k. [DOI] [PubMed] [Google Scholar]
- 35.Yasuda R, Harvey CD, Zhong H, Sobczyk A, van Aelst L, Svoboda K. Nat Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 36.Tregidgo C, Levitt JA, Suhling K. J Biomed Opt. 2008;13:031218. doi: 10.1117/1.2937212. [DOI] [PubMed] [Google Scholar]
- 37.van Manen HJ, Verkuijlen P, Wittendorp P, Subramaniam V, van den Berg TK, Roos D, Otto C. Biophys J. 2008;94:L67–L69. doi: 10.1529/biophysj.107.127837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suhling K, French PM, Phillips D. Photochem Photobiol Sci. 2005;4:13–22. doi: 10.1039/b412924p. [DOI] [PubMed] [Google Scholar]
- 39.Becker W, Bergmann A, Hink MA, Konig K, Benndorf K, Biskup C. Microsc Res Tech. 2004;63:58–66. doi: 10.1002/jemt.10421. [DOI] [PubMed] [Google Scholar]
- 40.Jares-Erijman EA, Jovin TM. Curr Opin Chem Biol. 2006;10:409–416. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Piston DW, Rizzo MA. Methods Cell Biol. 2008;85:415–430. doi: 10.1016/S0091-679X(08)85018-2. [DOI] [PubMed] [Google Scholar]
- 42.Beutler M, Makrogianneli K, Vermeij RJ, Keppler M, Ng T, Jovin TM, Heintzmann R. Eur Biophys J. 2008;38:69–82. doi: 10.1007/s00249-008-0361-5. [DOI] [PubMed] [Google Scholar]
- 43.Miyawaki A, Nagai T, Mizuno H. Adv Biochem Eng Biotechnol. 2005;95:1–15. doi: 10.1007/b102208. [DOI] [PubMed] [Google Scholar]
- 44.Adams SR, Tsien RY. Nat Protoc. 2008;3:1527–1534. doi: 10.1038/nprot.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Allen MD. Mol Biosyst. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 47.Hao Y, Macara IG. J Cell Biol. 2008;182:827–836. doi: 10.1083/jcb.200803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plafker K, Macara IG. J Biol Chem. 2002;277:30121–30127. doi: 10.1074/jbc.M203006200. [DOI] [PubMed] [Google Scholar]
- 49.Ouyang M, Lu S, Li XY, Xu J, Seong J, Giepmans BN, Shyy JY, Weiss SJ, Wang Y. J Biol Chem. 2008;283:17740–17748. doi: 10.1074/jbc.M709872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Nat Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 51.Kolossov VL, Spring BQ, Sokolowski A, Conour JE, Clegg RM, Kenis PJ, Gaskins HR. Exp Biol Med (Maywood) 2008;233:238–248. doi: 10.3181/0707-RM-192. [DOI] [PubMed] [Google Scholar]
- 52.Shimozono S, Hosoi H, Mizuno H, Fukano T, Tahara T, Miyawaki A. Biochemistry. 2006;45:6267–6271. doi: 10.1021/bi060093i. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen AW, Daugherty PS. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 54.Shimozono S, Miyawaki A. Methods Cell Biol. 2008;85:381–393. doi: 10.1016/S0091-679X(08)85016-9. [DOI] [PubMed] [Google Scholar]
- 55.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 56.Truong K, Sawano A, Mizuno H, Hama H, Tong KI, Mal TK, Miyawaki A, Ikura M. Nat Struct Biol. 2001;8:1069–1073. doi: 10.1038/nsb728. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura T, Kurokawa K, Kiyokawa E, Matsuda M. Methods Enzymol. 2006;406:315–332. doi: 10.1016/S0076-6879(06)06023-X. [DOI] [PubMed] [Google Scholar]
- 58.Nakaya M, Kitano M, Matsuda M, Nagata S. Proc Natl Acad Sci U S A. 2008;105:9198–9203. doi: 10.1073/pnas.0803677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ananthanarayanan B, Ni Q, Zhang J. Methods Cell Biol. 2008;89:37–57. doi: 10.1016/S0091-679X(08)00602-X. [DOI] [PubMed] [Google Scholar]
- 60.Sakai R, Repunte-Canonigo V, Raj CD, Knopfel T. European Journal of Neuroscience. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- 61.Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Nat Methods. 2008;5:683–685. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- 62.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van der Krogt GN, Ogink J, Ponsioen B, Jalink K. PLoS ONE. 2008;3:e1916. doi: 10.1371/journal.pone.0001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Palmer AE, Tsien RY. Nat Protoc. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 65.Yoshizaki H, Aoki K, Nakamura T, Matsuda M. Biochem Soc Trans. 2006;34:851–854. doi: 10.1042/BST0340851. [DOI] [PubMed] [Google Scholar]
- 66.Medintz IL, Mattoussi H. Phys Chem Chem Phys. 2009;11:17–45. doi: 10.1039/b813919a. [DOI] [PubMed] [Google Scholar]
- 67.Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, Hohsaka T. Nat Methods. 2006;3:923–929. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- 68.Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 69.O’Hare HM, Johnsson K, Gautier A. Curr Opin Struct Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 70.Pfleger KD, Eidne KA. Nat Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 71.Ciruela F. Curr Opin Biotechnol. 2008;19:338–343. doi: 10.1016/j.copbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 72.Roy R, Hohng S, Ha T. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guerrero G, Siegel MS, Roska B, Loots E, Isacoff EY. Biophysical Journal. 2002;83:3607–3618. doi: 10.1016/S0006-3495(02)75361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shaner NC, Steinbach PA, Tsien RY. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 75.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tramier M, Zahid M, Mevel JC, Masse MJ, Coppey-Moisan M. Microsc Res Tech. 2006;69:933–939. doi: 10.1002/jemt.20370. [DOI] [PubMed] [Google Scholar]
- 77.Monici M. Biotechnol Annu Rev. 2005;11:227–256. doi: 10.1016/S1387-2656(05)11007-2. [DOI] [PubMed] [Google Scholar]
- 78.Bayle V, Nussaume L, Bhat RA. Plant Physiol. 2008;148:51–60. doi: 10.1104/pp.108.117358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kollner M, Wolfrum J. Chemical Physics Letters. 1992;200:199–204. [Google Scholar]
- 80.Rizzo MA, Springer GH, Granada B, Piston DW. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 81.Ai HW, Henderson JN, Remington SJ, Campbell RE. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bulina ME, Lukyanov KA, Britanova OV, Onichtchouk D, Lukyanov S, Chudakov DM. Nat Protoc. 2006;1:947–953. doi: 10.1038/nprot.2006.89. [DOI] [PubMed] [Google Scholar]
- 83.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 84.Galperin E, Sorkin A. Methods Enzymol. 2005;403:119–134. doi: 10.1016/S0076-6879(05)03011-9. [DOI] [PubMed] [Google Scholar]
- 85.Dinant C, van Royen ME, Vermeulen W, Houtsmuller AB. J Microsc. 2008;231:97–104. doi: 10.1111/j.1365-2818.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- 86.Ganesan S, Ameer-Beg SM, Ng TT, Vojnovic B, Wouters FS. Proc Natl Acad Sci U S A. 2006;103:4089–4094. doi: 10.1073/pnas.0509922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murakoshi H, Lee SJ, Yasuda R. Brain Cell Biol. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsien RY. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 89.Long JZ, Lackan CS, Hadjantonakis AK. BMC Biotechnol. 2005;5:20. doi: 10.1186/1472-6750-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. Protein Sci. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kofoed EM, Guerbadot M, Schaufele F. J Biomed Opt. 2008;13:031207. doi: 10.1117/1.2940366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matsuda T, Miyawaki A, Nagai T. Nat Methods. 2008;5:339–345. doi: 10.1038/nmeth.1193. [DOI] [PubMed] [Google Scholar]
- 93.Grant DM, Zhang W, McGhee EJ, Bunney TD, Talbot CB, Kumar S, Munro I, Dunsby C, Neil MA, Katan M, French PM. Biophys J. 2008;95:L69–L71. doi: 10.1529/biophysj.108.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Piljic A, Schultz C. ACS Chem Biol. 2008;3:156–160. doi: 10.1021/cb700247q. [DOI] [PubMed] [Google Scholar]
- 95.Peyker A, Rocks O, Bastiaens PIH. Chembiochem. 2005;6:78–85. doi: 10.1002/cbic.200400280. [DOI] [PubMed] [Google Scholar]
- 96.Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW., Jr PLoS ONE. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arnaoutov A, Dasso M. Cell Cycle. 2005;4:1161–1165. doi: 10.4161/cc.4.9.1979. [DOI] [PubMed] [Google Scholar]
- 98.Nishijima H, Nakayama J, Yoshioka T, Kusano A, Nishitani H, Shibahara K, Nishimoto T. Mol Biol Cell. 2006;17:2524–2536. doi: 10.1091/mbc.E05-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bastiaens P, Caudron M, Niethammer P, Karsenti E. Trends Cell Biol. 2006;16:125–134. doi: 10.1016/j.tcb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 100.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 101.Terry LJ, Shows EB, Wente SR. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 102.Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- 103.Zhang JH, Chung TD, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 104.Seewald MJ, Kraemer A, Farkasovsky M, Korner C, Wittinghofer A, Vetter IR. Mol Cell Biol. 2003;23:8124–8136. doi: 10.1128/MCB.23.22.8124-8136.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bischoff FR, Gorlich D. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 106.Novoa I, Rush MG, D’Eustachio P. Mol Biol Cell. 1999;10:2175–2190. doi: 10.1091/mbc.10.7.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lounsbury KM, Richards SA, Carey KL, Macara IG. J Biol Chem. 1996;271:32834–32841. doi: 10.1074/jbc.271.51.32834. [DOI] [PubMed] [Google Scholar]
- 108.Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 109.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 110.Cingolani G, Lashuel HA, Gerace L, Muller CW. FEBS Lett. 2000;484:291–298. doi: 10.1016/s0014-5793(00)02154-2. [DOI] [PubMed] [Google Scholar]
- 111.Harreman MT, Hodel MR, Fanara P, Hodel AE, Corbett AH. J Biol Chem. 2003;278:5854–5863. doi: 10.1074/jbc.M210951200. [DOI] [PubMed] [Google Scholar]
- 112.Cingolani G, Petosa C, Weis K, Muller CW. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 113.Weis K, Ryder U, Lamond AI. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 114.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. J Biol Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 115.Chi NC, Adam SA. Mol Biol Cell. 1997;8:945–956. doi: 10.1091/mbc.8.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huber J, Dickmanns A, Luhrmann R. J Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Azuma Y, Seino H, Seki T, Uzawa S, Klebe C, Ohba T, Wittinghofer A, Hayashi N, Nishimoto T. J Biochem. 1996;120:82–91. doi: 10.1093/oxfordjournals.jbchem.a021397. [DOI] [PubMed] [Google Scholar]
- 118.Patterson GH, Piston DW, Barisas BG. Anal Biochem. 2000;284:438–440. doi: 10.1006/abio.2000.4708. [DOI] [PubMed] [Google Scholar]
- 119.Bazin H, Preaudat M, Trinquet E, Mathis G. Spectrochim Acta A Mol Biomol Spectrosc. 2001;57:2197–2211. doi: 10.1016/s1386-1425(01)00493-0. [DOI] [PubMed] [Google Scholar]
References
- 1.Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ, Verlhac MH. J Cell Biol. 2007;176:295–305. doi: 10.1083/jcb.200605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalab P, Weis K, Heald R. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- 3.Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 4.Caudron M, Bunt G, Bastiaens P, Karsenti E. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- 5.Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, Vuppalanchi D, Segal-Ruder Y, Ben-Yaakov K, Hieda M, Yoneda Y, Twiss JL, Fainzilber M. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulze H, Dose M, Korpal M, Meyer I, Italiano JE, Jr, Shivdasani RA. J Biol Chem. 2008;283:14109–14119. doi: 10.1074/jbc.M709397200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasan MT, Friedrich RW, Euler T, Larkum ME, Giese G, Both M, Duebel J, Waters J, Bujard H, Griesbeck O, Tsien RY, Nagai T, Miyawaki A, Denk W. PLoS Biol. 2004;2:e163. doi: 10.1371/journal.pbio.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sapsford KE, Berti L, Medintz IL. Angew Chem Int Ed Engl. 2006;45:4562–4589. doi: 10.1002/anie.200503873. [DOI] [PubMed] [Google Scholar]
- 9.Förster T. Annalen der Physik. 1948;2:55–75. [Google Scholar]
- 10.Jares-Erijman EA, Jovin TM. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 11.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer Science; 2006. [Google Scholar]
- 12.Wallrabe H, Periasamy A. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura T, Aoki K, Matsuda M. Brain Cell Biol. 2008;36:19–30. doi: 10.1007/s11068-008-9028-5. [DOI] [PubMed] [Google Scholar]
- 14.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Gruenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahoo H, Roccatano D, Zacharias M, Nau WM. J Am Chem Soc. 2006;128:8118–8119. doi: 10.1021/ja062293n. [DOI] [PubMed] [Google Scholar]
- 16.Pantano S. J Mol Graph Model. 2008;27:563–567. doi: 10.1016/j.jmgm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Kalab P, Pralle A. Methods in Enzymology. 2008;89:539–566. [Google Scholar]
- 18.Kaláb P, Pralle A, Isacoff E, Heald R, Weis K. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 19.Piston DW, Kremers GJ. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Puhl HL, 3rd, Koushik SV, Vogel SS, Ikeda SR. Biophys J. 2006;91:L39–L41. doi: 10.1529/biophysj.106.088773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karpova TS, Baumann CT, He L, Wu X, Grammer A, Lipsky P, Hager GL, McNally JG. J Microsc. 2003;209:56–70. doi: 10.1046/j.1365-2818.2003.01100.x. [DOI] [PubMed] [Google Scholar]
- 23.Yasuda R, Harvey CD, Zhong H, Sobczyk A, van Aelst L, Svoboda K. Nat Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 24.Tregidgo C, Levitt JA, Suhling K. J Biomed Opt. 2008;13:031218. doi: 10.1117/1.2937212. [DOI] [PubMed] [Google Scholar]
- 25.van Manen HJ, Verkuijlen P, Wittendorp P, Subramaniam V, van den Berg TK, Roos D, Otto C. Biophys J. 2008;94:L67–L69. doi: 10.1529/biophysj.107.127837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suhling K, French PM, Phillips D. Photochem Photobiol Sci. 2005;4:13–22. doi: 10.1039/b412924p. [DOI] [PubMed] [Google Scholar]
- 27.Becker W, Bergmann A, Hink MA, Konig K, Benndorf K, Biskup C. Microsc Res Tech. 2004;63:58–66. doi: 10.1002/jemt.10421. [DOI] [PubMed] [Google Scholar]
- 28.Piston DW, Rizzo MA. Methods Cell Biol. 2008;85:415–430. doi: 10.1016/S0091-679X(08)85018-2. [DOI] [PubMed] [Google Scholar]
- 29.Beutler M, Makrogianneli K, Vermeij RJ, Keppler M, Ng T, Jovin TM, Heintzmann R. Eur Biophys J. 2008;38:69–82. doi: 10.1007/s00249-008-0361-5. [DOI] [PubMed] [Google Scholar]
- 30.Miyawaki A, Nagai T, Mizuno H. Adv Biochem Eng Biotechnol. 2005;95:1–15. doi: 10.1007/b102208. [DOI] [PubMed] [Google Scholar]
- 31.Adams SR, Tsien RY. Nat Protoc. 2008;3:1527–1534. doi: 10.1038/nprot.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Allen MD. Mol Biosyst. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 34.Hao Y, Macara IG. J Cell Biol. 2008;182:827–836. doi: 10.1083/jcb.200803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plafker K, Macara IG. J Biol Chem. 2002;277:30121–30127. doi: 10.1074/jbc.M203006200. [DOI] [PubMed] [Google Scholar]
- 36.Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Nat Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 37.Kolossov VL, Spring BQ, Sokolowski A, Conour JE, Clegg RM, Kenis PJ, Gaskins HR. Exp Biol Med (Maywood) 2008;233:238–248. doi: 10.3181/0707-RM-192. [DOI] [PubMed] [Google Scholar]
- 38.Shimozono S, Hosoi H, Mizuno H, Fukano T, Tahara T, Miyawaki A. Biochemistry. 2006;45:6267–6271. doi: 10.1021/bi060093i. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen AW, Daugherty PS. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 40.Shimozono S, Miyawaki A. Methods Cell Biol. 2008;85:381–393. doi: 10.1016/S0091-679X(08)85016-9. [DOI] [PubMed] [Google Scholar]
- 41.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 42.Truong K, Sawano A, Mizuno H, Hama H, Tong KI, Mal TK, Miyawaki A, Ikura M. Nat Struct Biol. 2001;8:1069–1073. doi: 10.1038/nsb728. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura T, Kurokawa K, Kiyokawa E, Matsuda M. Methods Enzymol. 2006;406:315–332. doi: 10.1016/S0076-6879(06)06023-X. [DOI] [PubMed] [Google Scholar]
- 44.Nakaya M, Kitano M, Matsuda M, Nagata S. Proc Natl Acad Sci U S A. 2008;105:9198–9203. doi: 10.1073/pnas.0803677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ananthanarayanan B, Ni Q, Zhang J. Methods Cell Biol. 2008;89:37–57. doi: 10.1016/S0091-679X(08)00602-X. [DOI] [PubMed] [Google Scholar]
- 46.Sakai R, Repunte-Canonigo V, Raj CD, Knopfel T. European Journal of Neuroscience. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- 47.Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Nat Methods. 2008;5:683–685. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- 48.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Krogt GN, Ogink J, Ponsioen B, Jalink K. PLoS ONE. 2008;3:e1916. doi: 10.1371/journal.pone.0001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer AE, Tsien RY. Nat Protoc. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 51.Yoshizaki H, Aoki K, Nakamura T, Matsuda M. Biochem Soc Trans. 2006;34:851–854. doi: 10.1042/BST0340851. [DOI] [PubMed] [Google Scholar]
- 52.Tsien RY. Keio J Med. 2006;55:127–140. doi: 10.2302/kjm.55.127. [DOI] [PubMed] [Google Scholar]
- 53.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 54.Medintz IL, Mattoussi H. Phys Chem Chem Phys. 2009;11:17–45. doi: 10.1039/b813919a. [DOI] [PubMed] [Google Scholar]
- 55.Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, Hohsaka T. Nat Methods. 2006;3:923–929. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- 56.Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 57.O'Hare HM, Johnsson K, Gautier A. Curr Opin Struct Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 58.Pfleger KD, Eidne KA. Nat Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 59.Ciruela F. Curr Opin Biotechnol. 2008;19:338–343. doi: 10.1016/j.copbio.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Roy R, Hohng S, Ha T. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guerrero G, Siegel MS, Roska B, Loots E, Isacoff EY. Biophysical Journal. 2002;83:3607–3618. doi: 10.1016/S0006-3495(02)75361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaner NC, Steinbach PA, Tsien RY. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 63.Shaner NC, Patterson GH, Davidson MW. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 64.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tramier M, Zahid M, Mevel JC, Masse MJ, Coppey-Moisan M. Microsc Res Tech. 2006;69:933–939. doi: 10.1002/jemt.20370. [DOI] [PubMed] [Google Scholar]
- 66.Monici M. Biotechnol Annu Rev. 2005;11:227–256. doi: 10.1016/S1387-2656(05)11007-2. [DOI] [PubMed] [Google Scholar]
- 67.Bayle V, Nussaume L, Bhat RA. Plant Physiol. 2008;148:51–60. doi: 10.1104/pp.108.117358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kollner M, Wolfrum J. Chemical Physics Letters. 1992;200:199–204. [Google Scholar]
- 69.Rizzo MA, Springer GH, Granada B, Piston DW. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 70.Ai HW, Henderson JN, Remington SJ, Campbell RE. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bulina ME, Lukyanov KA, Britanova OV, Onichtchouk D, Lukyanov S, Chudakov DM. Nat Protoc. 2006;1:947–953. doi: 10.1038/nprot.2006.89. [DOI] [PubMed] [Google Scholar]
- 72.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 73.Galperin E, Sorkin A. Methods Enzymol. 2005;403:119–134. doi: 10.1016/S0076-6879(05)03011-9. [DOI] [PubMed] [Google Scholar]
- 74.Goedhart J, Vermeer JE, Adjobo-Hermans MJ, van Weeren L, Gadella TW., Jr PLoS ONE. 2007;2:e1011. doi: 10.1371/journal.pone.0001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dinant C, van Royen ME, Vermeulen W, Houtsmuller AB. J Microsc. 2008;231:97–104. doi: 10.1111/j.1365-2818.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- 76.Ganesan S, Ameer-Beg SM, Ng TT, Vojnovic B, Wouters FS. Proc Natl Acad Sci U S A. 2006;103:4089–4094. doi: 10.1073/pnas.0509922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murakoshi H, Lee SJ, Yasuda R. Brain Cell Biol. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsien RY. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 79.Long JZ, Lackan CS, Hadjantonakis AK. BMC Biotechnol. 2005;5:20. doi: 10.1186/1472-6750-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. Protein Sci. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kofoed EM, Guerbadot M, Schaufele F. J Biomed Opt. 2008;13:031207. doi: 10.1117/1.2940366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matsuda T, Miyawaki A, Nagai T. Nat Methods. 2008;5:339–345. doi: 10.1038/nmeth.1193. [DOI] [PubMed] [Google Scholar]
- 83.Kalab P, Heald R. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arnaoutov A, Dasso M. Cell Cycle. 2005;4:1161–1165. doi: 10.4161/cc.4.9.1979. [DOI] [PubMed] [Google Scholar]
- 85.Nishijima H, Nakayama J, Yoshioka T, Kusano A, Nishitani H, Shibahara K, Nishimoto T. Mol Biol Cell. 2006;17:2524–2536. doi: 10.1091/mbc.E05-09-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bastiaens P, Caudron M, Niethammer P, Karsenti E. Trends Cell Biol. 2006;16:125–134. doi: 10.1016/j.tcb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 87.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 88.Terry LJ, Shows EB, Wente SR. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 89.Vetter IR, Arndt A, Kutay U, Gorlich D, Wittinghofer A. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- 90.Zhang JH, Chung TD, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 91.Seewald MJ, Kraemer A, Farkasovsky M, Korner C, Wittinghofer A, Vetter IR. Mol Cell Biol. 2003;23:8124–8136. doi: 10.1128/MCB.23.22.8124-8136.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bischoff FR, Gorlich D. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 93.Novoa I, Rush MG, D'Eustachio P. Mol Biol Cell. 1999;10:2175–2190. doi: 10.1091/mbc.10.7.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lounsbury KM, Richards SA, Carey KL, Macara IG. J Biol Chem. 1996;271:32834–32841. doi: 10.1074/jbc.271.51.32834. [DOI] [PubMed] [Google Scholar]
- 95.Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- 96.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 97.Cingolani G, Petosa C, Weis K, Muller CW. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 98.Weis K, Ryder U, Lamond AI. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 99.Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. J Biol Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- 100.Chi NC, Adam SA. Mol Biol Cell. 1997;8:945–956. doi: 10.1091/mbc.8.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huber J, Dickmanns A, Luhrmann R. J Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Azuma Y, Seino H, Seki T, Uzawa S, Klebe C, Ohba T, Wittinghofer A, Hayashi N, Nishimoto T. J Biochem. 1996;120:82–91. doi: 10.1093/oxfordjournals.jbchem.a021397. [DOI] [PubMed] [Google Scholar]
- 103.Patterson GH, Piston DW, Barisas BG. Anal Biochem. 2000;284:438–440. doi: 10.1006/abio.2000.4708. [DOI] [PubMed] [Google Scholar]
- 104.Bazin H, Preaudat M, Trinquet E, Mathis G. Spectrochim Acta A Mol Biomol Spectrosc. 2001;57:2197–2211. doi: 10.1016/s1386-1425(01)00493-0. [DOI] [PubMed] [Google Scholar]