Abstract
Background & Aims
The cyclophilin (Cyp) inhihibitors - cyclosporine A (CsA), NIM811, Debio 025 and SCY 635 - block HCV replication both in vitro and in vivo, and represent a novel class of potent anti-HCV agents. We and others showed that HCV relies on cyclophilin A (CypA) to replicate. We demonstrated that the hydrophobic pocket of CypA, where Cyp inhibitors bind, and which controls the isomerase activity of CypA, is critical for HCV replication. Recent studies showed that under Cyp inhibitor selection, mutations arose in the HCV nonstructural 5A (NS5A) protein. This led us to postulate that CypA assists HCV by acting on NS5A.
Methods
We tested this hypothesis by developing several interaction assays including GST pulldown assays, ELISA and two-hybrid mammalian binding assays.
Results
We demonstrated that full-length NS5A and CypA form a stable complex. Remarkably, CsA prevents the CypA-NS5A interaction in a dose-dependent manner. Importantly, the CypA-NS5A interaction is conserved among genotypes and is interrupted by CsA. Surprisingly, the NS5A mutant protein, which arose in CsA-resistant HCV variants, behaves similarly to wild-type NS5A in terms of both CypA-binding and CsA-mediated release from CypA. This latter finding suggests that HCV resistance to CsA does not correlate with a resistance of the CypA-NS5A interaction to Cyp inhibitors. Moreover, we found that CypA, devoid of its isomerase activity, fails to bind NS5A.
Conclusions
Altogether these data suggest that CypA, via its isomerase pocket, binds directly to NS5A, and most importantly, that disrupting this interaction stops HCV replication.
INTRODUCTION
HCV is the main contributing agent of acute and chronic liver diseases worldwide [1]. Primary infection is often asymptomatic or associated with mild symptoms. However, persistently infected individuals develop high risks for chronic liver diseases such as hepatocellular carcinoma and liver cirrhosis [1]. The combination of IFN alpha and ribavirin that serves as current therapy for chronically HCV-infected patients not only has a low success rate (about 50%) [2], but is often associated with serious side effects [2]. There is thus an urgent need for the development of novel anti-HCV treatments [2].
Cyclosporine A (CsA) was reported to be clinically effective against HCV [3]. Controlled trials showed that a combination of CsA with IFN alpha is more effective than IFN alpha alone, especially in patients with a high viral load [4–5]. Moreover, recent in vitro studies provided evidence that CsA prevents both HCV RNA replication and HCV protein production in an IFN alpha-independent manner [6–10]. CsA exerts this anti-HCV activity independently of its immunosuppressive activity because non-immunosuppressive CsA derivates - more recently termed Cyp inhibitors [11] - also block HCV RNA and protein production [9,12–15]. Recent clinical data demonstrated that these Cyp inhibitors profoundly decreased HCV viral load in HCV-infected patients [16–17]. More recently, the anti-HCV effect of Debio 025 in combination with peginterferon alpha 2a (peg-IFNα2a) was demonstrated in patients with chronic hepatitis C [17]. These findings are critical because they suggest that Cyp inhibitors represent a novel class of anti-HCV agents.
Although there was a growing body of evidence that Cyp inhibitors exert their antiviral effect by targeting Cyps, a disagreement existed on the respective roles of Cyp members in HCV replication. One study suggested that CypB, but not CypA, is critical for HCV replication [18], another suggested that CypA, but not CypB and CypC, was critical for HCV replication [19], and a third study suggested that three Cyps - CypA, B and C - are all required for HCV replication [9]. In order to attempt to clarify this apparent controversy, we recently re-analyzed the respective contribution of Cyp members to HCV replication by specifically and stably knocking down their expression by small RNA interference (sRNAi). We found that only the CypA knockdown drastically decreased HCV replication [20]. The re-expression of an exogenous CypA escape protein, which contains escape mutations at the sRNAi recognition site, restored HCV replication, demonstrating the specificity for the CypA requirement [23]. We also mutated residues, which reside in the hydrophobic pocket of CypA where proline-containing peptide substrates and CsA bind, and which are vital for the enzymatic or the hydrophobic pocket binding activity of CypA [20]. Remarkably, these CypA mutants fail to restore HCV replication, suggesting that HCV exploits the isomerase activity of CypA to replicate in hepatocytes and that CypA is the principal mediator of the Cyp inhibitor anti-HCV activity [20]. These results have now been confirmed by two independent studies from the Tang lab and from the Bartenschlager lab [21–22].
Since recent studies demonstrated that NS5A mutations arose when HCV were grown under CsA selection, we postulated for the existence of an interplay between CypA and NS5A. We thus tested this hypothesis and found that full-length NS5A and CypA directly associate. Remarkably, CsA prevents the CypA-NS5A interaction in a dose-dependent manner. The CypA-NS5A interaction is conserved among HCV genotypes and is prevented by CsA. Surprisingly, the interaction between CypA and the NS5A mutant protein identified in CsA-resistant HCV variants remains sensitive to CsA. Moreover, we found that CypA, devoid of its isomerase activity due to the introduction of a mutation in its enzymatic pocket, fails to bind to full-length NS5A. Altogether these data suggest that CypA, via its isomerase pocket, binds directly to NS5A, and most importantly, that disrupting this interaction stops HCV replication.
EXPERIMENTAL PROCEDURES
Production of Recombinant CypA and NS5A Proteins
Recombinant GST-CypA was produced and purified as we described previously [23], whereas full-length NS5A Con1 (pET-Ub-NS5A Con1-His) was expressed as described previously [24]. GST-CypA H126Q and NS5A D320E mutants were created by PCR mutagenesis. The NS5A genes from genotype 1a (H77), 1b (Con1), 2a (JFH-1) and 2b (MD2b-1) were cloned and expressed as described previously [24].
CypA-NS5A Pull-Down Studies
Glutathione beads were incubated for 2 h in dialysis buffer (50 mM Tris pH 7.4, 100 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.5% NP-40, 1 mM DTT) with 5 mg/ml BSA and washed twice at 4 °C in binding buffer (20 mM Tris pH 7.9, 0.5 M NaCl, 10% glycerol, 10 mM DTT and 1% NP-40). Meanwhile, 100 ng of GST-CypA or GST was mixed with 10 ng of NS5A-His in a total volume of 200 μl of binding buffer for 3 h at 4° C on wheel. Glutathione beads (25 μl) were added to the GST-CypA/NS5A mixture for 30 min at 4° C, washed 3 times with 400 μl of binding buffer. Beads were pelleted for 30 sec at 2000 g in a microfuge and bound material was eluted with 25 μl of 2× SDS sample buffer, heated for 5 min, and frozen at −20° C. Bound material was then analyzed by Western blotting using anti-GST, anti-CypA and anti-His antibodies as described previously [20].
CypA-NS5A ELISA
Nunc MaxiSorb 8-well strip plates were coated with GST, GST-CypA and GST-H126Q CypA for 16 h at 4° C and blocked as we described previously [26]. Recombinant NS5A-His (1 ng/ml) was added to wells in 50 μl of binding buffer (20 mM Tris pH 7.9, 0.5 M NaCl, 10% glycerol, 10 mM DTT and 1% NP-40) for 16 h at 4° C. Captured NS5A-His was subsequently detected using mouse anti-His antibodies (1 μg/ml) (anti-6xHis, Clontech) and rabbit anti-mouse-horseradish peroxidase phosphatase (HRP) antibodies (1:1000 dilution) as we described previously [23].
Scatchard Analyses
Recombinant NS5A proteins were labeled with [125I] (New England Nuclear, Boston) using Iodogen (Pierce Chemical Co., Rockford) to a specific radioactivity of 400 cpm/fmol. Nunc MaxiSorb 8-well strip plates were coated with GST or GST-CypA for 16 h at 4° C and blocked as we described previously [23]. For KD measurements, 125I-NS5A was added to GST or GST-CypA coated wells for 2 h at 4°C in the presence of increasing concentrations of unlabeled NS5A. Wells were then washed three times and bound 125I-NS5A was solubilized by incubating wells for 30 min at 50°C with 2% SDS, collected, and measured in a liquid scintillation counter. The amount of 125I-NS5A bound to GST-coated wells was used as nonspecific binding and was subtracted from all values. KD values were analyzed by the Scatchard plot procedure [25].
Mammalian Two-Hybrid System
Evaluation of intracellular CypA-NS5A interactions using the two-hybrid screening technology was accomplished using the Checkmate Mammalian Two-Hybrid System according to the manufacturer’s instructions (Promega). Briefly, pACT- and pBIND-based plasmids were co-transfected (using Genejuice) with the pG5luc reporter construct into Huh7 cells and incubated for 37° C. After 72 h, cell lysates were assessed for luciferase activity using the “Steady-Glo Luciferase Assay System” according to the manufacturer’s instructions (Promega).
RESULTS
Specific and Direct Interaction Between CypA and Full-Length NS5A
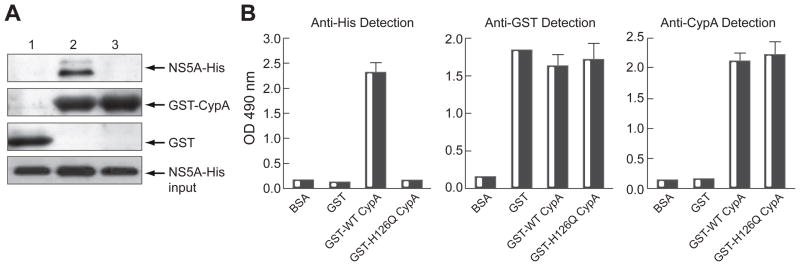
Since recent studies demonstrated that NS5A mutations arose when HCV was grown under CsA selection, we postulated for the existence of an interplay between CypA and NS5A. To test this hypothesis, we asked whether CypA possesses the capacity to interact with HCV NS5A. Specifically, we conducted binding studies between recombinant CypA and full-length wild-type NS5A. Importantly, we found that NS5A efficiently binds to GST-CypA (Fig. 1A, lane 2), but not GST (Fig. 1A, lane 1). This demonstrates that full-length NS5A possesses the ability to bind CypA directly. To further demonstrate a direct contact between NS5A and CypA, we developed an ELISA using CypA as solid phase to capture NS5A. Specifically, plates were coated with BSA, GST or GST-CypA and incubated with NS5A Con1-His. CypA-captured NS5A proteins were detected using anti-His antibodies. Importantly, NS5A binds to wells coated with GST-CypA, but not to those coated with GST or BSA (Fig. 1B). The ELISA data are perfectly in accordance with those of the GST-CypA pull down assay (Fig. 1A). Thus, full-length NS5A binds directly and specifically to CypA.
Fig. 1. Specific and Direct Interaction Between CypA and Full-Length NS5A.
(A) GST (lane 1), GST-CypA (lane 2) or GST-H126Q CypA (lane 3) (100 ng) was mixed with NS5A Con1-His (10 ng) for 3 h at 4° C. Glutathione beads were added to the GST-CypA/NS5A mixture for 30 min at 4° C and washed. Bound material was eluted and analyzed by Western blotting using anti-His and anti-GST antibodies. (B) Plates were coated with BSA, GST, GST-CypA and GST-H126Q CypA (10 μg/ml) for 16 h at 4° C. Recombinant NS5A-His (1 ng/ml) was added to wells for 16 h at 4° C. Captured NS5A-His was detected using mouse anti-His IgG (1 μg/ml) followed by anti-mouse-HRP IgG (1:1000 dilution). Adsorded levels of GST, GST-CypA and GST-H126Q CypA were monitored using mouse anti-GST IgG (1 μg/ml) and rabbit anti-CypA IgG (1μg/ml) followed by anti-mouse and -rabbit-HRP IgG (1:1000 dilution).
The Isomerase Hydrophobic Pocket of CypA Contains the NS5A-Binding Site
We and others demonstrated that the introduction of mutations in the active enzymatic pocket of CypA, where proline-containing peptide substrates bind, blocks HCV replication [20–22]. This led us to postulate that HCV requires the isomerase activity of CypA to replicate in human hepatocytes [20]. In this study, we asked whether CypA binds to NS5A via its isomerase hydrophobic active pocket. To address this issue, we created in the context of our bacterial expression GST-CypA construct, a CypA mutant deprived of its isomerase activity. Specifically, we replaced the histidine located at position 126 (H126) in the hydrophobic pocket of CypA by a glutamine, creating the H126Q CypA mutant. This mutation diminishes CypA isomerase activity by more than 99% compared to wild-type CypA [26]. Moreover, in contrast to wild-type CypA, the H126Q CypA mutant fails to support HCV replication [20–22]. In contrast to GST-wild-type CypA, GST-H126Q CypA fails to bind NS5A (Fig. 1A, lane 3). Similar levels of GST, GST-CypA and GST-H126Q CypA were used as demonstrated by anti-GST and anti-CypA Western blotting (Fig. 1A, lanes 2 and 3). We obtained similar data using our NS5A ELISA (Fig. 1B). We verified that similar levels of GST, GST-CypA and GST-H126Q CypA proteins were adsorded using anti-GST antibodies (Fig. 1B). Altogether these findings demonstrate that the isomerase active site of CypA is essential for both HCV replication [20–22] and NS5A binding (Fig. 1).
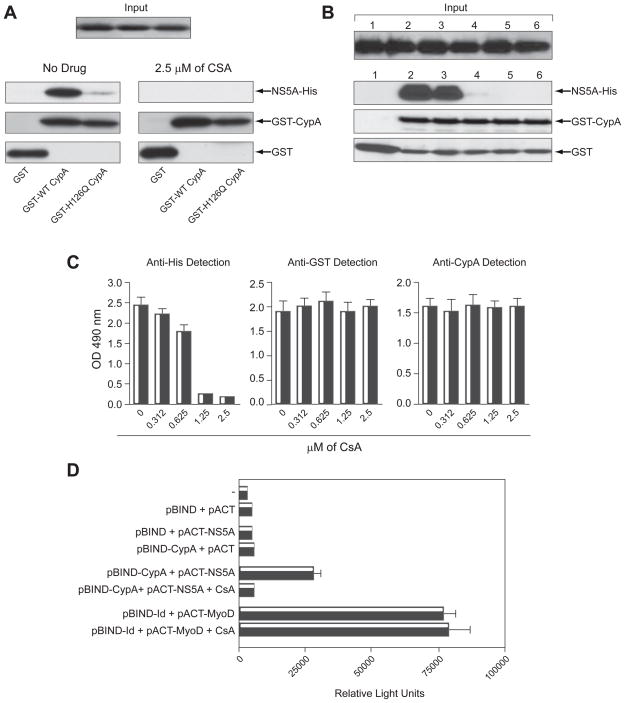
CsA Disrupts the CypA-NS5A Interaction in a Dose-Dependent Manner
We previously demonstrated that HCV highly relies on host CypA to replicate in hepatocytes and that the enzymatic active pocket of CypA is absolutely required for HCV replication [20]. In this study, we present evidence that CypA binds directly to full-length NS5A and that the enzymatic active pocket of CypA contains the NS5A-binding site (Fig. 1). Together these findings suggest that there is a direct correlation between CypA assistance to HCV replication and CypA binding to NS5A. If this model is correct, one could postulate that Cyp inhibitors, which block HCV replication, would interfere with the contact between CypA and NS5A. We thus tested this hypothesis, by examining the effect of the Cyp inhibitor CsA on CypA-NS5A interaction. As above, NS5A binds efficiently to GST-CypA in the absence of CsA (Fig. 2A). In sharp contrast, NS5A fails to bind to GST-CypA in the presence of the drug (Fig. 2A). This suggests that CsA, by binding to the hydrophobic active pocket of CypA, prevents the contact between viral NS5A and host CypA. Remarkably, CsA inhibits the CypA-NS5A interaction in a dose-dependent manner (Fig. 2B). Similar to the GST-CypA pull-down assay, we found by ELISA that CsA inhibits in a dose-dependent manner the binding of NS5A to GST-CypA (Fig. 2C). Altogether these findings demonstrate for the first time that there is a direct correlation between the CsA-mediated inhibition of HCV replication and the CsA-mediated inhibition of the CypA-NS5A interaction. Our demonstration that CsA blocks the CypA-NS5A interaction in a dose-dependent manner, further suggests that the interaction is specific. Our observation that CsA blocks the CypA-NS5A interaction (Fig. 2) is perfectly in accordance with our finding above that the enzymatic hydrophobic pocket of CypA, where CsA binds, is vital for NS5A binding (Fig. 1).
Fig. 2. CsA Disrupts the CypA-NS5A Interaction in a Dose-Dependent Manner.
(A) Same as Fig. 4A, except that CsA (2.5 μM) was added to GST, GST-CypA and GST-H126Q CypA 15 min prior to the addition of NS5A Con1-His. (B) Same as A, except that increasing concentrations of CsA were used: 0 (lane 2), 0.3125 (lane 3), 0.625 (lane 4), 1.25 (lane 5) and 2.5 μM (lane 6). (C) Same as 1B. (D) Huh7 cells were co-transfected with pBIND, pact and pG5luc. Three days post-transfection, luciferase activity was measured in cell lysates. Data (triplicates) are representative of four independent experiments.
We then examined whether this interaction also occurs in a cell. To address this issue, we took advantage of the mammalian two-hybrid system, which permits the analysis of protein-protein interaction in a cell. In this system, a pBIND vector contains the yeast GAL4 DNA-binding domain upstream of a multiple cloning region. A pACT vector contains the herpes simplex virus VP16 activation domain upstream of a multiple cloning region. Two genes encoding two potentially interactive proteins of interest (CypA and NS5A) are cloned into the pBIND and pACT vectors to generate fusion proteins with the DNA-binding domain of GAL4 and the activation domain of VP16, respectively. The pG5luc Vector contains five GAL4 binding sites upstream of a minimal TATA box, which in turn is upstream of the firefly luciferase gene. More specifically, we cloned CypA into the pBIND vector and NS5A into the pACT vector, co-transfected Huh7 cells and measured luciferase activity in cell lysates three days post-transfection. As expected, co-transfection of (i) the empty pBIND and pACT vectors; (ii) pBIND-CypA and empty pact; and (iii) empty pBIND and pACT-NS5A produce minimal luciferase activity (Fig. 2D). In contrast, co-transfection of pBIND-CypA and pACT-NS5A plasmids produces significant amounts of luciferase activity (Fig. 2D), suggesting that CypA and NS5A interact in a hepatocyte. Importantly, this production of enzymatic activity is abolished when Huh7 cells were pre-treated with CsA (Fig. 2D), suggesting that the CypA-NS5A interaction is specific. More importantly, CsA has no effect on the interaction between the two control proteins (MyoD and Id) included in the mammalian two-hybrid kit (Fig. 2D), demonstrating the specificity of the rupture of the NS5A-CypA interaction in hepatocytes by CsA. Together the mammalian two-hybrid system data demonstrate that a specific interaction between CypA and NS5A also occurs in a cell.
CypA-NS5A Interaction Conserved Among HCV Genotypes
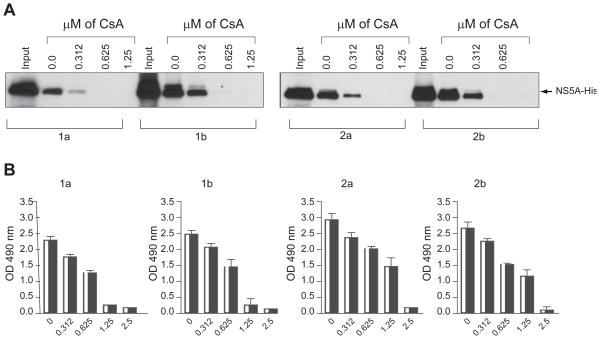
After demonstrating that HCV NS5A Con1 binds efficiently and specifically to human CypA, we asked whether it is also true for NS5A proteins derived from other genotypes. To address this issue, we cloned NS5A from genotype 1a (H77), 2a (JFH-1) and 2b (MD2b-1) into the bacterial expression pET-Ub vector. The four recombinant NS5A proteins were expressed, purified and tested for GST-CypA binding as described above. All four 1a, 1b, 2a and 2b NS5A proteins efficiently bind CypA (Fig. 3A), suggesting that the CypA-NS5A interaction is conserved among various HCV genotypes. No significant difference in CypA-binding was observed between NS5A genotypes. Importantly, CsA prevents all CypA-NS5A interactions in a dose-dependent manner (Fig. 3A). We confirmed the pulldown data by ELISA (Fig. 3B). We included the CsA IC50 in Table 1 (Table 1, right column). The IC50 for genotypes 2a and 2b were slightly superior to those for genotypes 1a and 1b (Table 1).
Fig. 3. CypA-NS5A Interaction Conserved Among HCV Genotypes.
(A) GST-CypA (100 ng) preincubated for 15 min with increasing concentrations of CsA (from 0.3125 to 1.25 μM) was mixed with recombinant NS5A-His (10 ng) derived from genotypes 1a, 1b, 2a and 2b for 3 h at 4° C. Glutathione beads were added to the GST-CypA/NS5A mixture for 30 min at 4° C and washed. Bound material was eluted and analyzed by Western blotting using anti-His antibodies. (B) Same as 2C.
Table 1. KD Values and CsA IC50 for NS5A-CypA Interactions.
Wells were coated with GST or GST-CypA for 16 h at 4° C and blocked. 125I-NS5A was added to wells for 2 h at 4°C in the presence of increasing concentrations of unlabeled NS5A. Wells were then washed and bound 125I-NS5A was measured in a liquid scintillation counter. The amount of 125I-NS5A bound to GST-coated wells was used as nonspecific binding and was subtracted from all values. KD values were analyzed by the Scatchard plot procedure. For the CsA IC50 values, unlabeled NS5A-His (1 ng/mL) was added to wells in the presence on increasing concentrations of CsA for 16 h at 4° C. Captured NS5A-His was subsequently detected using mouse anti-His and rabbit anti-mouse-HRP antibodies. Amounts of CsA necessary to inhibit 50% of NS5A binding in the absence of drug were calculated (IC50). Results (duplicates) are representatives of two independent experiments.
| Genotype | Strain | Sequence | Length | KD μM | CsA μM IC50 |
|---|---|---|---|---|---|
| 1a | H77 | Wild-type | Full | 131 | 0.72 |
| 1b | Con1 | Wild-type | Full | 126 | 0.91 |
| 2a | JFH-1 | Wild-type | Full | 108 | 1.40 |
| 2b | MD2b-1 | Wild-type | Full | 112 | 1.23 |
| 2a | JFH-1 | Wild-type | Domain 2 | 69 | 1.81 |
| 1b | Con1 | D320E | Full | 124 | 0.74 |
By Scatchard analysis, we measured the dissociation constants for each NS5A protein. We calculated KD values of 131 μM, 126 μM, 108 μM and 112 μM for wild-type H77, Con1, JFH-1 and MD2b-1 NS5A, respectively (Table 1). This low μM range is in accordance with the KD value of 64 μM that Hanoulle et al. recently calculated for the dissociation constant between CypA and the domain 2 of JFH-1 NS5A [27]. These relatively comparable KD values between NS5A proteins derived from four genotypes (Table 1) are in accordance with the relatively comparable concentrations (IC50) of CsA necessary to disrupt CypA-NS5A interactions between the four genotypes (Fig. 3B and Table 1). In order to be able to better compare our KD values with those of Hanoulle et al. [27], we also calculated the KD value for the domain 2 of JFH-1 NS5A. We obtained a KD value of 69 μM, very comparable to the KD value of 64 μM obtained by Hanoulle et al. (Table 1). This finding not only suggests that our dissociation constant measurements are similar to those previously reported [27], but it also suggests that the affinity of domain 2 JFH-1 NS5A (69 μM) is slightly superior to that of full-length JFH-1 NS5A (KD = 108 μM). This is also in accordance with our finding that the CsA IC50 for domain 2 NS5A-CypA interactions is superior to that of full-length NS5A-CypA interactions (see Table 1).
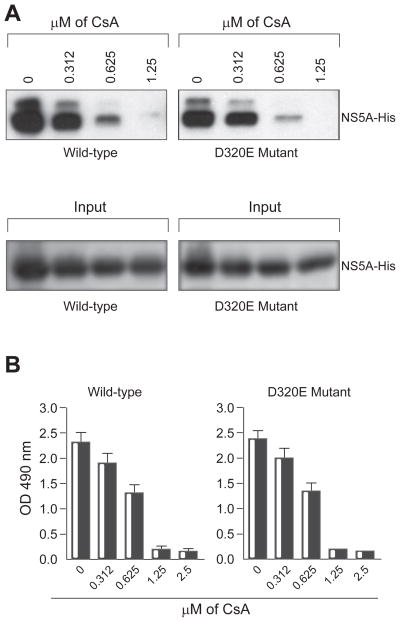
The D320E NS5A Mutation Does Not Render CypA-NS5A Interactions Resistant to CsA
Previous work showed that the emergence of HCV variants under CsA selection constantly correlates with the D320E NS5A mutation emergence [13,21,28–30]. Importantly recent studies demonstrated that the D320E NS5A mutation alone renders HCV resistant to Cyp inhibitors [28–30]. We thus asked whether this mutation has any effect on the binding of NS5A to CypA, or to the sensitivity of the CypA-NS5A interaction to CsA. We found that the D320E NS5A mutant binds to CypA similarly to wild-type NS5A (Fig. 4A). Importantly, the interactions between CypA and wild-type or D320E NS5A were similarly prevented by CsA (Fig. 4A). This was even true for a concentration of CsA that is lower than that used for the selection of CsA-resistant HCV variants [28–30]. We confirmed the pulldown data by ELISA (Fig. 4B). The CsA IC50 were included in Table 1. These findings indicate that the D320E NS5A mutation does not preserve the CypA-NS5A interaction from the CsA rupture.
Fig. 4. The D320E NS5A Mutation Does Not Render CypA-NS5A Interactions Resistant to Cs.
(A) GST-CypA (100 ng) preincubated for 15 min with increasing concentrations of CsA (from 0.3125 to 1.25 μM) was mixed with recombinant wild-type or D320E mutant NS5A-His (10 ng) for 3 h at 4 C. Glutathione beads were added to the GST-CypA/NS5A mixture for 30 min at 4° C and washed. Bound material was eluted and analyzed by Western blotting using anti-His antibodies. (B) Same as 2C.
DISCUSSION
A growing body of evidence suggests that mutations arose in NS5A when HCV is placed under various Cyp inhibitor selections [13,21,28–30]. Interestingly, two regions of NS5A undergo frequent mutations under Cyp inhibitor selection: domain 2 and the C-terminus of domain 3 of NS5A. Specifically, the D320E mutation [29–30], or the multiple E256G, V280A, F284S, L303Q, R356G and V444A mutations [28] were found in CsA-resistant Con1 variants. Importantly, two independent studies demonstrated that the D320E NS5A mutation alone governs CsA resistance [29–30]. The D253G, D294G and V445A mutations were found in a Debio 025-resistant JFH-1 variant [21].
This frequent emergence of mutations in NS5A under Cyp inhibitor selection suggests a link between NS5A and the antiviral action of Cyp inhibitors. Our demonstration that NS5A binds directly and specifically to CypA further supports this hypothesis. Specifically, we showed that CypA forms a stable complex with full-length NS5A derived from various HCV genotypes (1a, 1b, 2a and 2b), suggesting that the interaction is conserved among genotypes. Although this finding does not yet provide an explanation for the role of CypA in HCV replication, it strongly suggests that NS5A serves as the main viral locus for CypA action. Thus, the present study not only demonstrates for the first time that full-length NS5A forms a stable complex with CypA, it also demonstrates that this direct interaction is conserved among various genotypes.
We showed that CsA prevents the CypA-NS5A interaction in a dose-dependent manner. This was true for all NS5A proteins derived from various genotypes. We obtained similar inhibitory results with non-immunosuppressive CsA analogs (data not shown). This finding is in accordance with previous data, which showed that Cyp inhibitors block the replication of HCV derived from various genotypes in vitro [19–22] as well as in HCV-infected patients [16–17]. Evidently further work is required to determine how the CypA-NS5A interaction enhances HCV replication. In one scenario, the binding of CypA to NS5A facilitates or disrupts the contact between NS5A and NS5B [11], leading to activation of the NS5B polymerase complex. In another scenario, the binding of CypA to NS5A enhances HCV replication independently of NS5B, for example, by acting directly on the viral RNA.
We demonstrated in this study that CypA, devoid of its isomerase activity due to the introduction of a mutation in its enzymatic pocket, fails to bind NS5A. This suggests that CypA, via its isomerase pocket, binds directly to NS5A, and most importantly, that disrupting this interaction stops HCV replication. Further studies are required to determine how the peptidyl-prolyl isomerase CypA, by acting on NS5A, governs HCV replication. The hydrophobic pocket of CypA does not only contain the residues vital for the isomerase activity of CypA, it also contains the residues responsible for the binding of CypA to NS5A. Thus, the inability of the H126Q CypA mutant to support HCV replication [20–22] may arise from either its inability to isomerize peptidyl-prolyl bonds within NS5A or its inability to bind to NS5A.
Surprisingly, we found that the NS5A mutant protein (D320E), which arose in CsA-resistant HCV variants, behave similarly to wild-type NS5A in terms of both CypA-binding and CsA-mediated release from CypA. This latter finding strongly suggests that HCV resistance to CsA does not correlate with a resistance of the CypA-NS5A interaction to Cyp inhibitors. This critical finding also suggests that the D320E mutation bypasses the HCV need for CypA. How could we explain that the D320E NS5A mutation renders HCV resistant to CsA even when the CypA-NS5A interaction is disrupted? In one scenario, the main function of CypA is to decrease the affinity between NS5A and NS5B, freeing NS5B to mediate its polymerase activity. In this model, the D320E NS5A mutation would facilitate the dissociation of NS5A from NS5B, even when CypA is neutralized by Cyp inhibitors. In another scenario, the main function of CypA is to enhance the association between NS5A and NS5B, a precondition for the activation of the NS5B polymerase complex. In this model, the D320E NS5A mutation would preserve the contact between NS5A and NS5B, even in the absence of CypA-NS5A interactions. In another scenario, the D320E mutation preserves the NS5A-enhanced HCV replication independently of NS5B. Further work is required to determine which of these models is correct. Note that we only analyzed a single NS5A mutation and that possibly other mutations may act differently through alternative mechanisms.
In conclusion, this study shows for the first time that (i) full-length HCV NS5A binds directly to the isomerase pocket of CypA; (ii) the NS5A-CypA interaction also occurs in a cell; (iii) the NS5A-CypA interaction is conserved among various HCV genotypes; (iv) the contact between the host and the viral protein is disrupted by Cyp inhibitors in a dose-dependent manner; and (v) most importantly that HCV resistance to CsA does not correlate with a resistance of the CypA-NS5A interaction to Cyp inhibitors.
Acknowledgments
We thank J. Kuhns for secretarial assistance. We thank Gunter Fischer for careful reading of the manuscript. This is publication no. 20257-IMM from the Department of Immunology & Microbial Science, The Scripps Research Institute, La Jolla, CA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pawlotsky JM. Therapy of hepatitis C: from empiricism to eradication. Hepatology. 2006;43:S207–S220. doi: 10.1002/hep.21064. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi N, Takehara T. Antiviral therapy for chronic hepatitis C: past, present, and future. J Gastroenterol. 2006;41:17–27. doi: 10.1007/s00535-005-1740-7. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama H, Yoshinaga H, Tanaka T, Hiruma K, Tanikawa S, Sakamaki H, et al. Effects of cyclosporin A on hepatitis C virus infection in bone marrow transplant patients. Bone Marrow Transplantation Team Bone Marrow Transplant. 1997;20:993–995. doi: 10.1038/sj.bmt.1700996. [DOI] [PubMed] [Google Scholar]
- 4.Inoue K, Sekiyama K, Yamada M, Watanabe T, Yasuda H, Yoshiba M. Combined interferon alpha2b and cyclosporin A in the treatment of chronic hepatitis C: controlled trial. J Gastroenterol. 2003;38:567–572. doi: 10.1007/s00535-002-1104-5. [DOI] [PubMed] [Google Scholar]
- 5.Inoue K, Yoshiba M. Interferon combined with cyclosporine treatment as an effective countermeasure against hepatitis C virus recurrence in liver transplant patients with end-stage hepatitis C virus related disease. Transplant Proc. 2005;37:1233–1234. doi: 10.1016/j.transproceed.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 6.Goto K, Watashi K, Murata T, Hishiki T, Hijikata M, Shimotohno K. Evaluation of the anti-hepatitis C virus effects of cyclophilin inhibitors, cyclosporin A, and NIM811. Biochem Biophys Res Commun. 2006;343:879–884. doi: 10.1016/j.bbrc.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 7.Ishii N, Watashi K, Hishiki T, Goto K, Inoue D, Hijikata M. Diverse effects of cyclosporine on hepatitis C virus strain replication. J Virol. 2006;80:4510–4520. doi: 10.1128/JVI.80.9.4510-4520.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma S, Boerner JE, TiongYip C, Weidmann B, Ryder NS, Cooreman MP, et al. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob Agents Chemother. 2006;50:2976–2982. doi: 10.1128/AAC.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa M, Sakamoto N, Tanabe Y, Koyama T, Itsui Y, Takeda Y, et al. Suppression of hepatitis C virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology. 2005;129:1031–1041. doi: 10.1053/j.gastro.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 10.Watashi K, Hijikata M, Hosaka M, Yamaji M, Shimotohno K. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology. 2003;38:1282–1288. doi: 10.1053/jhep.2003.50449. [DOI] [PubMed] [Google Scholar]
- 11.Gallay PA. Cyclophilin inhibitors. Clin Liver Dis. 2009;13:403–417. doi: 10.1016/j.cld.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Coelmont L, Kaptein S, Paeshuyse J, Vliegen I, Dumont JM, Vuagniaux G, et al. Debio 025, a cyclophilin binding molecule, is highly efficient in clearing hepatitis C virus (HCV) replicon-containing cells when used alone or in combination with specifically targeted antiviral therapy for HCV (STAT-C) inhibitors. Antimicrob Agents Chemother. 2009;53:967–976. doi: 10.1128/AAC.00939-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopkins S, Scorneaux B, Huang Z, Murray MG, Wring S, Smitley C, Harris R, Erdmann F, Fisher G, Ribeill Y. SCY-635: A Novel Non-Immunosuppressive Analog of Cyclosporin A that Exhibits Potent Inhibition of Hepatitis C Virus RNA Replication in vitro. Antimicrob Agents Chemother. 2009 doi: 10.1128/AAC.00660-09. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathy JE, Ma S, Compton T, Lin K. Combinations of cyclophilin inhibitor NIM811 with hepatitis C Virus NS3–4A Protease or NS5B polymerase inhibitors enhance antiviral activity and suppress the emergence of resistance. Antimicrob Agents Chemother. 2008;52:3267–3275. doi: 10.1128/AAC.00498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paeshuyse J, Kaul A, De Clercq E, Rosenwirth B, Dumont JM, Scalfaro P, et al. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43:761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- 16.Flisiak R, Horban A, Gallay P, Bobardt M, Selvarajah S, Wiercinska-Drapalo A, et al. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology. 2008;47:817–826. doi: 10.1002/hep.22131. [DOI] [PubMed] [Google Scholar]
- 17.Flisiak R, Feinman SV, Jablkowski M, Horban A, Kryczka W, Pawlowska M, et al. The cyclophilin inhibitor Debio 025 combined with PEG IFNalpha2a significantly reduces viral load in treatment-naïve hepatitis C patients. Hepatology. 2009;49:1460–1468. doi: 10.1002/hep.22835. [DOI] [PubMed] [Google Scholar]
- 18.Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, et al. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Yang F, Robotham JM, Nelson HB, Irsigler A, Kenworthy R, Tang H. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J Virol. 2008;82:5269–5278. doi: 10.1128/JVI.02614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatterji U, Bobardt M, Selvarajah S, Yang F, Tang H, Sakamoto N, et al. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J Biol Chem. 2009;284:16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, et al. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Yang F, Robotham JM, Tang H. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J Virol. 2009;83:6554–6565. doi: 10.1128/JVI.02550-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saphire AC, Bobardt MD, Gallay PA. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 1999;18:6771–6785. doi: 10.1093/emboj/18.23.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Sineva EV, Hargittai MR, Sharma SD, Suthar M, Raney KD, et al. Purification and characterization of hepatitis C virus non-structural protein 5A expressed in Escherichia coli. Protein Expr Purif. 2004;37:144–153. doi: 10.1016/j.pep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Scatchard G. The attractions of proteins for small molecules and ions. Annu NY Acad Sci. 1949;51:660–672. [Google Scholar]
- 26.Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, et al. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanoulle X, Badillo A, Wieruszeski JM, Verdegem D, Landrieu I, Bartenschlager R, et al. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J Biol Chem. 2009;284:13589–13601. doi: 10.1074/jbc.M809244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandes F, Poole DS, Hoover S, Middleton R, Andrei AC, Gerstner J, et al. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology. 2007;46:1026–1033. doi: 10.1002/hep.21809. [DOI] [PubMed] [Google Scholar]
- 29.Goto K, Watashi K, Inoue D, Hijikata M, Shimotohno K. Identification of cellular and viral factors related to anti-hepatitis C virus activity of cyclophilin inhibitor. Cancer Sci. 2009;100:1943–1950. doi: 10.1111/j.1349-7006.2009.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abe KI, Ikeda M, Ariumi Y, Dansako H, Wakita T, Kato N. HCV genotype 1b chimeric replicon with NS5B of JFH-1 exhibited resistance to cyclosporine A. Arch Virol. 2009 doi: 10.1007/s00705-009-0502-x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]