Abstract
Background:
Alcohol dependence is a complex disease, and although linkage and candidate gene studies have identified several genes associated with the risk for alcoholism, these explain only a portion of the risk.
Methods:
We carried out a genome-wide association study (GWAS) on a case-control sample drawn from the families in the Collaborative Study on the Genetics of Alcoholism. The cases all met diagnostic criteria for alcohol dependence according to the Diagnostic and Statistical Manual of the American Psychiatric Association Fourth Edition (DSM-IV); controls all consumed alcohol but were not dependent on alcohol or illicit drugs. To prioritize among the strongest candidates, we genotyped most of the top 199 SNPs (p ≤ 2.1 × 10−4) in a sample of alcohol dependent families and performed pedigree-based association analysis. We also examined whether the genes harboring the top SNPs were expressed in human brain or were differentially expressed in the presence of ethanol in lymphoblastoid cells.
Results:
Although no single SNP met genome-wide criteria for significance, there were several clusters of SNPs that provided mutual support. Combining evidence from the case-control study, the followup in families, and gene expression provided strongest support for the association of a cluster of genes on chromosome 11 (SLC22A18, PHLDA2, NAP1L4, SNORA54, CARS, and OSBPL5) with alcohol dependence. Several SNPs nominated as candidates in earlier GWAS studies replicated in ours, including CPE, DNASE2B, SLC10A2,ARL6IP5, ID4, GATA4, SYNE1 and ADCY3.
Conclusions:
We have identified several promising associations that warrant further examination in independent samples.
Keywords: alcohol dependence, genome-wide association study, case-control study, family study, gene expression
INTRODUCTION
Alcohol dependence (alcoholism) is a major health and social issue affecting 4-5% of the United States population at any given time (Li et al., 2007), with a lifetime prevalence of 12.5% (Hasin et al., 2007). Alcohol dependence is characterized by serious problems in multiple domains. Family, twin and adoption studies have consistently demonstrated a substantial genetic contribution to disease etiology (Kendler et al., 1994; McGue, 1999; Nurnberger et al., 2004; Pickens et al., 1991). Alcohol dependence is a complex disease in which both genetic and environmental factors affect susceptibility.
Strategies for identifying genes in which variations contribute to the risk for alcohol dependence have employed linkage analysis or candidate gene approaches (Edenberg and Foroud, 2006). These methods have led to the identification of several genes associated with alcohol dependence, including GABRA2 (Covault et al., 2004; Drgon et al., 2006; Edenberg et al., 2004; Enoch et al., 2008; Enoch et al., 2006; Fehr et al., 2006; Lappalainen et al., 2005; Lind et al., 2008; Soyka et al., 2008), ADH4 (Edenberg and Foroud, 2006; Edenberg et al., 2006; Guindalini et al., 2005; Kuo et al., 2008; Luo et al., 2006; Luo et al., 2005b), GABRG3 (Dick et al., 2004), CHRM2 (Luo et al., 2005a; Wang et al., 2004), NFKB1 (Edenberg et al., 2008b), OPRK1 (Edenberg et al., 2008a; Xuei et al., 2006), PDYN (Xuei et al., 2006), NPY2R (Wetherill et al., 2008), ANKK1/DRD2 (Dick et al., 2007b); CHRNA5 (Wang et al., 2009); GRM8 (Chen et al., 2009), TACR3 (Foroud et al., 2008) and GABRR2 (Xuei et al., 2010). However, the effect of variation in each of these genes is small, and much of the genetic contribution to the risk for alcoholism remains to be discovered.
We completed a genome-wide association study (GWAS) to identify genes contributing to alcohol dependence, defined according to DSM-IV criteria (American Psychiatric Association, 1994). Both the alcohol dependent cases and the controls have completed a rigorous clinical evaluation, with controls being drinkers who are free from alcohol dependence or abuse by several diagnostic classification systems (DSM-IIIR, DSM-IV, ICD-10) (American Psychiatric Association, 1987; American Psychiatric Association, 1994; World Health Organization, 1993), and also free of dependence on illicit drugs. To prioritize among the most promising single nucleotide polymorphisms (SNPs) identified from the GWAS, we genotyped the top SNPs in a sample of alcohol dependent families and performed family based association analysis, and tested whether they fell within or near genes whose expression is affected by alcohol exposure, and were expressed in brain.
MATERIALS AND METHODS
Ascertainment of subjects
Alcohol dependent probands were ascertained through alcohol treatment programs and evaluated at seven centers in the United States: Indiana University, State University of New York Health Science Center Brooklyn, University of Connecticut, University of Iowa, University of California/San Diego, Washington University/St. Louis and Howard University as part of the Collaborative Study on the Genetics of Alcoholism (Begleiter et al., 1995; Edenberg, 2002; Foroud et al., 2000; Reich et al., 1998). The institutional review boards of all participating institutions approved the study. After providing informed consent, the probands and their relatives were administered a validated poly-diagnostic instrument, the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) (Bucholz et al., 1994; Hesselbrock et al., 1999), which allows assessment of alcohol dependence under several diagnostic systems. Details of the ascertainment and assessment have previously been published (Begleiter et al., 1995; Foroud et al., 2000; Reich et al., 1998) and are available at zork.wustl.edu/niaaa/coga_instruments/resources.html.
The same seven centers also recruited community probands through driver's license records, random mailings to employees and students at a university, and attendees at medical and dental clinics. After providing informed consent, the community probands and their relatives were administered the same SSAGA interview as the alcohol dependent probands and their families.
Genome-wide association study
Case-Control Sample Selection
For the GWAS, a sample of genetically unrelated cases and controls were selected from this pool of alcohol dependent and community ascertained families. Cases were all alcohol dependent by DSM-IV criteria at some time during their lives, and were selected from the families ascertained through alcohol dependent probands. The selected controls were required to have consumed alcohol but not to have a diagnosis of alcohol abuse, dependence or harmful use by any of the 4 diagnostic systems assessed in the SSAGA (Feighner, DSM-IIIR, DSM-IV, ICD-10) at any time in their lives. Because there might be shared genetic vulnerability with other substance use disorders, the selected controls also could not meet criteria for DSM-IIIR or DSM-IV diagnoses of abuse or dependence on cocaine, marijuana, opioids, sedatives, or stimulants. Controls were selected from both the community recruited families and the families recruited through an alcohol dependent proband, but could not share a known common ancestor with a case. Controls older than 25 years were preferentially selected, to increase the probability that they were beyond the peak age of risk for developing alcohol dependence. A subset of the COGA participants were assessed on multiple occasions; if a subject was assessed more than once, only one whose diagnosis or lack thereof was consistent across time was selected.
Microarray Genotyping, Quality Assessment and Statistical Analysis
Genotyping was performed by the Center for Inherited Disease Research (CIDR). DNA sources included blood (n=1453) and lymphoblastoid cell lines (LCL, n=492). Genotyping was performed using the Illumina Infinium II assay protocol (Gunderson et al., 2006) with hybridization to Illumina HumanHap1M BeadChips (Illumina, San Diego, CA, USA), which contain 1,199,187 markers with a mean spacing of 2.4 kb. Allele cluster definitions for each SNP were determined using Illumina BeadStudio Genotyping Module version 3.1.14 and the combined intensity data from 96% of study samples. The resulting cluster definition file was used on all study samples to determine genotype calls and quality scores. Genotype calls were made when a genotype yielded a quality score (Gencall value) of 0.15 or higher. The final raw dataset released by CIDR to the investigators and to dbGaP contained 1,041,465 SNPs for 1,945 unique DNA samples from case and control subjects. Twenty seven samples were removed due to poor sample quality before release of the dataset on dbGaP (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap; Accession number: phs000125.v1.p1). Blind duplicate reproducibility was 99.97%.
Starting with the dbGaP data, additional quality control measures were applied to both the samples and the SNPs. Samples having genotypes for at least 98% of the SNPs were considered for inclusion in analyses. These samples were rigorously checked for cryptic relatedness, population stratification, and related issues. Thirteen additional samples were removed from further analyses due to poor sample quality (n=4) or cryptic relatedness among subjects (n=9). A principal component-based analysis was performed in PLINK (Purcell et al., 2007) to cluster these samples along with HapMap reference samples (CEU, YRI, CHB, and JPT) to assign the study subjects to groups of predominantly European and African ancestry. The final European American (EA) sample included 847 alcohol dependent cases and 552 controls (n=1,399 individuals). The African American (AA) sample contained 345 cases and 140 controls (n=485 individuals). The remaining 21 individuals did not cluster with either of the two samples and were not analyzed.
SNPs with a call rate of 98% or greater in the EAs (n=1,015,550) were included and subjected to further quality control. From these, SNPs were removed if the minor allele frequency was less than 0.01 in the combined case and control dataset (n=161,048) or if among those with minor allele frequency ≥ 0.01 there was significant deviation from Hardy Weinberg equilibrium (p<10−4) in the controls (n=1,127). The final dataset for analysis of EAs consisted of 853,375 SNPs that passed all quality control measures. Similar quality control thresholds were applied to the 1,010,230 SNPs with a call rate of 98% or greater in the AAs. This resulted in removal of 68,600 SNPs with a minor allele frequency below 0.01 in the combined AA case and control sample, and 332 SNPs that demonstrated significant deviation from Hardy Weinberg equilibrium in AA controls. The final dataset for the AA sample consisted of 941,298 SNPs. Positions of all SNPs are from dbSNP build 130, genome build 36.3.
Given the limited number of non-EA subjects in the sample, and that self-defined ethnicity carries non-genetic as well as genetic differences, we focused our primary analyses on the larger EA subsample. Logistic regression covarying for gender was employed to test for the association of each SNP with alcohol dependence, using an additive model. Odds ratios and p-values were computed to assess the strength of the association. All analyses were performed using PLINK (Purcell et al., 2007).
Previously, stronger association results were often obtained when the alcohol-dependent cases were limited to those with greater correlates of disease severity, such as earlier age of onset, comorbid drug dependence or higher likelihood of conduct disorder symptoms (Agrawal et al., 2006; Dick et al., 2007a; Edenberg et al., 2008b; Foroud et al., 2008). We performed secondary analyses to identify potential associations with early onset alcohol dependence. A median split of our sample (Table 1) was performed and those alcohol dependent cases with an onset of 22 years or younger (n=454) were classified as having earlier onset of alcohol dependence. Logistic regression using the additive model with sex as a covariate was employed to test for association between the genotyped SNPs and the phenotype of early onset alcohol dependence.
Table 1.
Demographics of the GWAS sample
| Alcohol Dependent Cases | Controls | |||
|---|---|---|---|---|
| EA | AA | EA | AA | |
| Number | 847 | 345 | 552 | 140 |
| Gender (% female) | 30.3 | 35.9 | 71.6 | 71.4 |
| Alcohol dependent | 847 | 345 | 0 | 0 |
| Age at onset of dependence, Median (25%ile, 75%ile) |
22 (18, 27) |
25 (20, 31) |
NA* | NA* |
| Age at last interview, Median (25%ile, 75%ile) |
NA* | NA* | 47 (38, 55) |
39 (32, 49) |
| Marijuana dependent (%) | 34.7 | 33.9 | 0 | 0 |
| Cocaine dependent (%) | 35.1 | 56.2 | 0 | 0 |
| Dependent on other illicit substances (opioids, stimulants, sedatives) (%) |
29.5 | 13 | 0 | 0 |
EA = European American. AA = African American.
Not applicable
Family-based replication study
Family Sample
Previously, a set of 262 genetically informative, multiplex alcohol dependence pedigrees, each with at least three first degree relatives who met lifetime criteria for both DSM-IIIR alcohol dependence (American Psychiatric Association,1987) and Feighner definite alcoholism (Feighner et al., 1972) were selected from the larger COGA sample and used in linkage and family based association studies (Edenberg et al., 2004; Edenberg and Foroud, 2006; Foroud et al., 2000; Reich et al., 1998); 59 additional trios were also genotyped. After reviewing the results from the primary analysis of DSM-IV alcohol dependence, the 199 SNPs with the smallest p values for association (p ≤ 2.1 × 10−4) were selected for genotyping in this family-based sample. Genotypes were completed on 180 of these SNPs; the others did not design into assays or did not provide useable data.
Some members of these families, primarily probands, were included in the GWAS case control sample. There were 426 individuals included in the EA GWAS sample (326 cases; 100 controls) that were also a member of the families used for replication. The analytic methods applied to detect association in the case control and family-based tests of association are statistically independent (Abecasis et al., 2000; Vogel et al., 2009; Zuo et al., 2009).
Genotyping, Quality Assessment and Statistical Analysis
Genotyping assays were designed for the Sequenom MassArray system (Sequenom, San Diego, CA) using MassArray Assay Design Software. Genotyping used iPLEX Gold assays (Sequenom, San Diego, CA), with alleles discriminated by mass spectrometry. Assays were tested on two independent groups, 40 unrelated EA individuals and 40 unrelated African-American (AA) individuals obtained from Coriell Cell Repositories. SNPs that were not in Hardy Weinberg equilibrium in both groups were not genotyped in the COGA families. All SNPs were tested for Mendelian inheritance using the program PEDCHECK (O'Connell and Weeks, 1998).
Family based association analyses were performed using the Pedigree Disequilibrium test (PDT) (Martin et al., 2000) as implemented in the program UNPHASED (version 2.404) (Dudbridge, 2003). Affected individuals were defined as those meeting criteria for DSM-IV alcohol dependence. The PDT utilizes data from all available trios in a family, as well as discordant sibships. We report results using the PDTaverage option, which weights each family equally in computing the overall test statistic. Analysis was also performed classifying as affected only those DSM-IV alcohol dependence cases who also met criteria for early onset, defined as with the case control sample, as onset by age 22 or younger. After examining results in the family sample, five additional SNPs (cSNPs or SNPs in 3′ UTRs) from regions with evidence for replication were selected for genotyping in the family sample.
Gene expression analyses
In a separate study, we are analyzing the effects of ethanol exposure on gene expression in immortalized lymphoblastoid cell lines (LCLs). Details of the gene expression studies will be reported separately; here we note whether there was an effect of alcohol exposure on the expression of any of our top candidate genes. Briefly, 42 LCLs from subjects in the COGA study were either treated for 24 h with 75 mM ethanol or left untreated. The RNA was extracted and purified, and aliquots were analyzed on Affymetrix GeneChip® Human Genome U133 Plus 2.0 microarrays following standard procedures. Results were analyzed by ANOVA with factors of treatment, phenotype (cells from alcoholics or controls) and sex (Edenberg et al., in preparation). False discovery rates were calculated using the Storey qvalue package (Storey and Tibshirani, 2003).
Gene expression was assayed using standard Affymetrix procedures on samples from 9 different brain regions (prefrontal cortex, cerebral cortex, thalamus, visual cortex, hippocampus, amygdala, caudate, putamen and cerebellum) of 4 individuals (1 each male alcoholic, female alcoholic, male control, female control), obtained from the NIAAA-supported brain bank at the Tissue Resource Center located in the Neuropathology Unit of the Department of Pathology, University of Sydney, Australia. Hybridization was to GeneChip® Human Gene 1.0 ST Arrays. Genes were presumed expressed in brain if the average expression level of the 4 arrays from any region was higher than the average expression level of the negative probe sets on the arrays (Edenberg et al., in preparation). Genes from the expression studies were matched to genes from the SNP microarrays by matching gene symbols based on Affymetrix annotations.
RESULTS
Association analysis in the GWAS case control sample
The final EA case-control analytic sample included 1399 individuals. Cases all met lifetime DSM-IV criteria for alcohol dependence; many were also dependent upon other drugs (Table 1). Controls all had consumed alcohol, but were not affected by alcohol or drug abuse or dependence; most were well past the primary age of risk for alcohol or illicit drug disorders (Table 1). Thus the controls provide a strong phenotypic contrast to the cases.
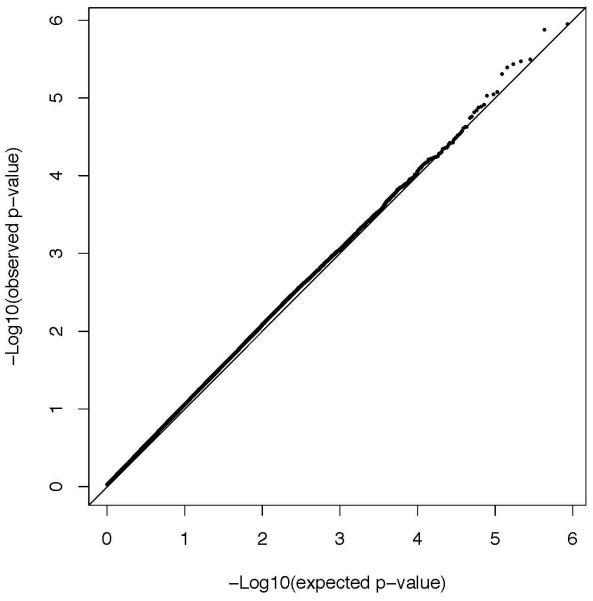
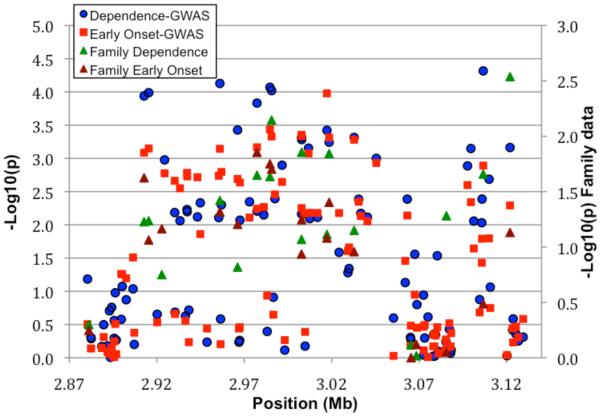
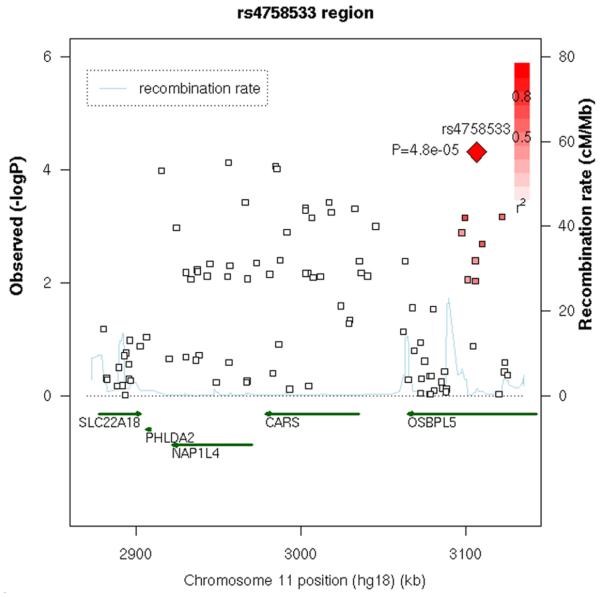
Association of the SNPs with alcohol dependence was carried out employing an additive model, with sex as a covariate. The λ value (inflation factor) after these adjustments was 1.049; the Q-Q plot for this analysis is shown in Figure 1. None of the SNPs analyzed met conventional criteria for genome-wide significance. However, eleven SNPs had p-values <10−5, and a total of 93 SNPs had p-values ≤10−4 (Supplemental Table 1 shows data from all SNPs with p≤10−3). There were many genes or regions in which surrounding SNPs provided additional support for association (Supplemental Table 1). A region of chromosome 11 (Figure 2A) had many SNPs among those with lowest p values. This was not due to a single SNP in high LD with other SNPs; including the most significant SNP (rs4758533) as a covariate in the logistic regression model decreased the evidence of association with several SNPs near the right end but did not substantially alter the evidence of association with most of the SNPs in the region (see also Figure 2B, which shows that most of the significant SNPs in the chromosome 11 region are not in high LD with rs4758533).
Figure 1. Q-Q plot for association analysis of the alcohol dependence phenotype.
Observed log p-values (black dots) and expected log p-values (line) are plotted versus the expected log pvalue (x-axis).
Figure 2. Chromosome 11 region with supporting evidence for association.
A. GWAS data from primary analysis and analysis of early onset, and data from family analysis. All SNPs are plotted as −log10(p-value) vs. position along the chromosome; circles = GWAS alcohol dependence, squares are GWAS early onset; for GWAS data the scale is on the left. Family data are also presented for those SNPs tested; green triangles are alcohol dependence, brown triangles are early onset; for the family data, the scale is on the right. B. LD between the SNP with lowest p value and other SNPs in the region, depicted in a SNAP plot (Johnson et al., 2008). Along the X axis is the physical position in the region (in kb) with known genes shown in their orientation. The left Y-axis denotes the association test result as −log(p-value) corresponding to squares in the figure. The red diamond identifies the primary SNP result labeled with an rs# and p-value. The darkness of the squares indicate the pairwise linkage disequilibrium with the primary SNP; the scale for this is shown in the top right corner. r2 values were obtained from this study using Haploview (Barrett et al. 2005). The right Y-axis indicates the recombination rate, obtained from the available HapMap data in the CEPH Caucasians, and shown within the figure by the solid blue line.
Secondary analysis tested for association within the subset of cases that met criteria for early onset of alcohol dependence (≤22 years; n=454). No SNP met genome-wide significance. The SNPs with the smallest p values in the two analyses substantially overlap: 25 of the 100 SNPs with p ≤10−4 for alcohol dependence also had p ≤10−4 for early onset alcohol dependence, despite the smaller sample size (Supplemental Table 1). Notably, the odds ratios for the early onset phenotype and the dependence phenotype are very similar: of the top 199 SNPs for dependence, 166 had odds ratios for both phenotypes within ± 0.10 of each other.
Data from the AA case control sample provided nominal evidence of association for DSM-IV alcohol dependence for six SNPs among the top 985 from European Americans; these were in TMEM132C, EPHA7, OPA3, KCNMA1 (encoding a potassium large conductance calcium-activated channel important in neuronal excitability), DMRTA2 and SPTA1 (Supplementary Table 1). EPHA7 encodes ephrin receptor A7; ephrin receptors are tyrosine kinases implicated in the development of the nervous system. A secondary analysis of the smaller AA sample showed a different set of SNPs that provided evidence for association with alcohol dependence (Supplementary Table 2). Among the 790 SNPs at p≤ 10−3, 41 were nominally significant in the EA group; prominent among them were 7 SNPs (several, but not all, in high LD) in the region of SELL and SELE, two selectins (adhesion/homing receptors), 4 in LOC91431, encoding the protein prematurely terminated mRNA decay factor-like, and SNPs in PPARG (peroxisome proliferator-activated receptor gamma), CTNN2 (catenin, alpha 2). Also of note is a SNP between the leptin receptor (LEPR) and PDE4B, a cAMP-specific phosphodiesterase that has been implicated in schizophrenia and bipolar disorder.
Results of family-based association analysis
Ten of the 180 SNPs genotyped in the sample of 262 genetically informative, alcohol dependent families provided nominally significant (p ≤ 0.05) evidence of association with alcohol dependence (Table 2, Supplementary Table 1); 8 of these were also nominally significant for the early onset phenotype. Five of these SNPs lie in one region of chromosome 11 (Figure 2A), which contained CARS (all 3 tested SNPs replicated), OSBPL5, NAP1L4. The other SNPs were in BBX, SLC9A8, OPA3, TOX2, and near CD53. Several additional SNPs near other clusters of significant SNPs were also genotyped and also provided evidence for association (Supplementary Table 1), including additional SNPs in and near CARS, OSPBL5, SLC9A8, SLC2A14, SOX5, COL8A1 and ADCK1.
Table 2.
SNPs and genes with supporting evidence
| EUROPEAN AMERICAN CASE-CONTROL |
FAMILY SAMPLE (PDT) |
Gene expression | AFRICAN AMERICAN CASE-CONTROL |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CH R |
POSITION | Gene within 15 kb |
MAF | AD (p-value) |
AD OR |
Early Onset AD (p- value) |
Early Onset AD OR |
AD (p- value) |
Early onset AD (p- value) |
Expr in brain |
FOLD Et/C |
FDR | MAF | AD (p- value) |
AD OR |
| rs3010931 | 1 | 12,606,911 | DHRS3 | 0.04 | 3.9E-04 | 0.5 | 1.9E-03 | 0.5 | yes | 1.22 | 2.9E-03 | 0.25 | 3.7E-01 | 1.2 | ||
| rs343819 | 1 | 111,144,013 | 0.27 | 1.5E-04 | 0.7 | 8.0E-06 | 0.6 | 0.051 | 0.0017 | yes | 1.04 | 1.8E-03 | 0.25 | 9.4E-01 | 1.0 | |
| rs6540681 | 1 | 209,560,196 |
RCOR3 TRAF5 |
0.16 | 6.8E-04 | 0.7 | 1.2E-02 | 0.7 | yes | −1.28 | 5.6E-04 | 0.17 | 6.0E-01 | 0.9 | ||
| rs12471136 | 2 | 72,642,825 | SEC15L2 | 0.11 | 8.0E-04 | 1.6 | 4.3E-03 | 1.6 | yes | 1.34 | 7.7E-16 | 0.21 | 8.4E-01 | 1.0 | ||
| rs13410476 | 2 | 161,118,788 | 0.27 | 1.2E-04 | 0.7 | 3.1E-03 | 0.7 | 0.16 | 0.12 | no | 1.21 | 1.6E-07 | 0.40 | 4.0E-01 | 1.1 | |
| rs16849209 | 2 | 213,319,466 | 0.23 | 5.0E-04 | 1.4 | 9.3E-03 | 1.4 | yes | −1.20 | 3.9E-02 | 0.27 | 5.8E-01 | 0.9 | |||
| rs237899 | 3 | 8,783,515 | OXTR | 0.37 | 2.2E-04 | 1.4 | 1.1E-03 | 1.4 | 0.12 | 0.20 | yes | 1.36 | 1.8E-15 | 0.36 | 1.2E-01 | 0.8 |
| rs1735460 | 3 | 60,666,645 | FHIT | 0.03 | 3.7E-05 | 0.3 | 3.4E-05 | 0.2 | 0.51 | 0.91 | yes | 1.25 | 3.2E-07 | 0.08 | 5.5E-01 | 1.2 |
| rs2700667 | 3 | 100,568,496 | 0.27 | 4.0E-04 | 1.4 | 7.8E-03 | 1.3 | 0.013 | 0.0065 | no | 0.43 | 3.1E-01 | 1.2 | |||
| rs2700586 | 3 | 100,577,480 | 0.27 | 3.5E-04 | 1.4 | 7.0E-03 | 1.3 | 0.0095 | 0.0056 | no | 0.43 | 2.7E-01 | 1.2 | |||
| rs2200019 | 3 | 100,578,586 | 0.30 | 4.1E-04 | 1.4 | 8.3E-03 | 1.3 | 0.019 | 0.012 | no | 0.32 | 9.7E-01 | 1.0 | |||
| rs2700648 | 3 | 100,590,373 | 0.26 | 8.2E-04 | 1.4 | 1.2E-02 | 1.3 | 0.046 | 0.022 | no | 0.44 | 6.3E-01 | 0.9 | |||
| rs10511260 | 3 | 108,715,787 | BBX | 0.29 | 3.4E-06 | 0.6 | 4.7E-04 | 0.7 | 0.010 | 0.028 | yes | 1.13 | 8.5E-05 | 0.06 | 9.6E-01 | 1.0 |
| rs11918092 | 3 | 135,996,046 | EPHB1 | 0.13 | 9.7E-04 | 0.7 | 1.3E-03 | 0.6 | yes | 1.21 | 8.6E-18 | 0.45 | 3.2E-01 | 1.2 | ||
| rs4384980 | 3 | 183,941,763 | 0.42 | 3.2E-04 | 0.7 | 1.1E-03 | 0.7 | yes | 1.22 | 8.9E-11 | 0.46 | 6.3E-01 | 0.9 | |||
| rs10029539 | 4 | 31,401,050 | 0.49 | 3.1E-04 | 0.7 | 2.1E-02 | 0.8 | 0.55 | 0.052 | 0.21 | 6.6E-01 | 1.1 | ||||
| rs6881283 | 5 | 91,091,413 | 0.24 | 5.6E-05 | 1.5 | 1.1E-04 | 1.6 | 0.69 | 0.83 | yes | 1.52 | 1.1E-16 | 0.07 | 8.7E-01 | 1.0 | |
| rs9327332 | 5 | 123,917,480 | 0.16 | 1.4E-04 | 1.6 | 2.1E-03 | 1.5 | 0.14 | 0.023 | yes | 0.14 | 6.1E-01 | 1.1 | |||
| rs4388209 | 5 | 125,786,272 | GRAMD3 | 0.29 | 4.7E-04 | 0.7 | 1.1E-03 | 0.7 | yes | 1.25 | 1.2E-14 | 0.25 | 6.0E-01 | 1.1 | ||
| rs1883339 | 6 | 132,642,677 | 0.05 | 2.7E-04 | 0.5 | 2.6E-03 | 0.5 | yes | 1.31 | 1.3E-14 | 0.42 | 8.7E-01 | 1.0 | |||
| rs1982389 | 7 | 37,693,476 | 0.05 | 2.3E-04 | 0.5 | 3.2E-03 | 0.5 | 0.16 | 0.021 | |||||||
| rs4728699 | 7 | 87,006,685 | ABCB1 | 0.04 | 7.9E-04 | 0.5 | 1.6E-02 | 0.6 | yes | 1.39 | 2.4E-07 | 0.01 | 1.9E-01 | 2.9 | ||
| rs6955063 | 7 | 139,723,897 | SLC37A3 | 0.10 | 4.0E-05 | 0.6 | 1.5E-03 | 0.6 | 0.50 | 0.049 | yes | 0.30 | 1.9E-01 | 1.2 | ||
| rs17637986 | 8 | 6,621,112 | 0.09 | 5.5E-05 | 0.6 | 5.2E-05 | 0.5 | 0.57 | 0.76 | yes | −1.31 | 4.9E-14 | 0.03 | 7.6E-01 | 1.2 | |
| rs2094372 | 9 | 77,186,279 | 0.25 | 1.1E-03 | 1.4 | 1.9E-03 | 1.4 | yes | −1.30 | 1.8E-05 | 0.15 | 4.6E-01 | 1.2 | |||
| rs17527069 | 10 | 10,317,351 | 0.21 | 5.1E-04 | 0.7 | 1.7E-03 | 0.7 | yes | −1.35 | 7.5E-11 | 0.03 | 4.8E-01 | 0.8 | |||
| rs7078551 | 10 | 25,214,732 | PRTFDC1 | 0.32 | 6.1E-04 | 1.4 | 2.7E-03 | 1.4 | yes | −1.21 | 1.3E-07 | 0.25 | 3.1E-01 | 0.8 | ||
| rs12219105 | 10 | 78,843,546 | KCNMA1 | 0.14 | 1.5E-04 | 0.6 | 9.7E-05 | 0.6 | 0.12 | 0.036 | yes | 0.17 | 3.9E-02 | 0.7 | ||
| rs196335 | 10 | 121,401,851 |
RPS8P4 BAG3 |
0.10 | 1.3E-04 | 1.7 | 1.9E-02 | 1.5 | 0.52 | 0.42 | yes | 1.39 | 1.1E-06 | 0.13 | 5.3E-01 | 0.9 |
| rs2583442 | 11 | 2,912,742 |
SLC22A18 PHLDA2 |
0.30 | 1.2E-04 | 0.7 | 8.2E-04 | 0.7 | 0.059 | 0.024 | yes | 1.14 | 4.2E-02 | 0.09 | 3.3E-01 | 0.8 |
| rs8505 | 11 | 2,922,869 |
NAP1L4-
3′UTR |
0.18 | 0.07 | yes | ||||||||||
| rs3814964 | 11 | 2,956,083 |
NAP1L4 SNORA54 |
0.31 | 7.4E-05 | 0.7 | 7.2E-04 | 0.7 | 0.038 | 0.048 | yes | −1.08 | 9.2E-06 | 0.15 | 1.4E-01 | 0.7 |
| rs4758621 | 11 | 2,966,216 |
NAP1L4 CARS |
0.32 | 3.7E-04 | 0.7 | 2.0E-03 | 0.7 | 0.15 | 0.06 | yes | 0.09 | 3.0E-01 | 0.8 | ||
| rs12805661 | 11 | 2,977,316 |
NAP1L4 CARS |
0.29 | 1.5E-04 | 0.7 | 6.8E-04 | 0.7 | 0.022 | 0.014 | yes | 0.09 | 3.0E-01 | 0.8 | ||
| rs729662 | 11 | 2,984,716 |
NAP1L4 CARS |
0.29 | 8.5E-05 | 0.7 | 3.7E-04 | 0.7 | 0.023 | 0.018 | yes | 1.03 | 5.0E-02 | 0.09 | 3.2E-01 | 0.8 |
| rs7481584 | 11 | 2,985,665 | CARS | 0.29 | 9.5E-05 | 0.7 | 4.7E-04 | 0.7 | 0.007 | 0.020 | yes | 0.13 | 1.2E-01 | 0.7 | ||
| rs377765 | 11 | 3,002,874 | CARS | 0.41 | 5.2E-04 | 0.7 | 4.6E-04 | 0.7 | 0.01 | 0.06 | yes | 0.12 | 4.6E-01 | 0.8 | ||
| rs369461 | 11 | 3,018,568 | CARS | 0.40 | 5.7E-04 | 0.7 | 4.8E-04 | 0.7 | 0.014 | 0.039 | yes | 0.10 | 2.8E-01 | 0.8 | ||
| rs2277301 | 11 | 3,068,442 |
OSBPL5-
cSNP |
0.96 | 0.76 | |||||||||||
| rs3741350 | 11 | 3,085,603 |
OSBPL5-
cSNP |
0.05 | 0.89 | |||||||||||
| rs4758533 | 11 | 3,106,662 | OSBPL5 | 0.21 | 4.8E-05 | 0.7 | 1.3E-03 | 0.7 | 0.02 | 0.33 | 0.14 | 1.4E-01 | 0.7 | |||
| rs4468331 | 11 | 3,121,988 | OSBPL5 | 0.20 | 6.9E-04 | 0.7 | 5.1E-03 | 0.7 | 0.00 | 0.07 | 0.16 | 2.3E-01 | 1.3 | |||
| rs11032702 | 11 | 34,424,699 | CAT | 0.05 | 5.2E-04 | 0.5 | 2.9E-03 | 0.5 | 0.03 | 0.16 | 1.08 | 7.3E-05 | 0.31 | 7.8E-01 | 1.0 | |
| rs12366192 | 11 | 42,261,639 | 0.12 | 1.3E-04 | 0.6 | 7.5E-03 | 0.7 | 0.18 | 0.011 | 0.16 | 9.6E-02 | 0.7 | ||||
| rs10502172 | 11 | 112,704,356 | TTC12 | 0.48 | 7.0E-04 | 0.7 | 2.1E-04 | 0.7 | yes | −1.25 | 3.9E-03 | 0.10 | 5.3E-01 | 0.9 | ||
| rs7963376 | 12 | 796,509 |
WNK1 LOC647502 |
0.48 | 5.5E-04 | 0.7 | 8.7E-03 | 0.8 | yes | 1.30 | 5.0E-05 | 0.42 | 4.4E-01 | 0.9 | ||
| rs7965203 | 12 | 7,891,603 | SLC2A14 | 0.49 | 4.5E-04 | 1.4 | 3.7E-04 | 1.4 | 0.035 | 0.022 | −1.10 | 5.6E-04 | 0.46 | 1.8E-01 | 0.8 | |
| rs11612319 | 12 | 7,908,192 |
SLC2A14 NANOGP 1 |
0.32 | 6.6E-05 | 0.7 | 4.4E-05 | 0.6 | 0.55 | 0.033 | yes | −1.10 | 5.6E-04 | 0.13 | 6.0E-01 | 1.1 |
| rs550338 | 12 | 24,403,304 | SOX5 | 0.24 | 2.2E-04 | 0.7 | 8.9E-05 | 0.6 | 0.04 | 0.13 | yes | 0.29 | 9.7E-01 | 1.0 | ||
| rs7311374 | 12 | 27,851,172 | KLHDC5 | 0.14 | 3.9E-04 | 0.7 | 5.4E-03 | 0.7 | 0.52 | 0.83 | yes | 1.25 | 1.2E-14 | 0.04 | 9.3E-01 | 1.0 |
| rs1144962 | 12 | 67,621,252 | CPM | 0.35 | 1.6E-04 | 0.7 | 9.0E-04 | 0.7 | yes | 1.33 | 1.7E-03 | 0.42 | 7.3E-01 | 1.1 | ||
| rs1195812 | 12 | 67,627,292 | CPM | 0.34 | 5.3E-04 | 0.7 | 2.5E-03 | 0.7 | yes | 1.28 | 2.0E-02 | 0.29 | 6.5E-02 | 0.7 | ||
| rs10847023 | 12 | 125,010,473 |
RPL22P1 9 |
0.16 | 2.3E-04 | 1.6 | 8.4E-05 | 1.7 | 0.05 | 0.20 | 0.12 | 9.4E-01 | 1.0 | |||
| rs10483889 | 14 | 77,627,258 | 0.19 | 3.4E-04 | 0.7 | 8.3E-04 | 0.6 | 0.04 | 0.46 | 0.03 | 7.6E-01 | 1.1 | ||||
| rs7164726 | 15 | 57,502,958 | FAM81A | 0.12 | 6.0E-04 | 0.6 | 5.3E-04 | 0.6 | yes | −1.22 | 3.8E-04 | 0.36 | 6.6E-01 | 0.9 | ||
| rs7175167 | 15 | 68,312,713 | 0.16 | 3.1E-04 | 0.7 | 3.8E-03 | 0.7 | yes | 1.22 | 1.8E-09 | 0.27 | 9.0E-01 | 1.0 | |||
| rs1909766 | 16 | 59,618,891 | 0.15 | 9.9E-05 | 1.6 | 1.4E-03 | 1.6 | 0.15 | 0.031 | yes | 0.41 | 1.2E-01 | 0.8 | |||
| rs1037260 | 17 | 68,668,860 |
LOC390811 SSTR2 |
0.06 | 6.5E-05 | 2.3 | 4.3E-03 | 1.9 | 0.38 | 0.48 | yes | 1.37 | 1.1E-09 | 0.29 | 6.7E-01 | 1.1 |
| rs1568447 | 17 | 70,348,607 |
TMEM104 GRIN2C |
0.38 | 7.1E-04 | 0.7 | 2.6E-03 | 0.7 | 0.38 | 0.0087 | 0.36 | 7.5E-01 | 1.0 | |||
| rs11652088 | 17 | 70,351,427 |
TMEM104 GRIN2C |
0.38 | 4.4E-04 | 0.7 | 1.3E-03 | 0.7 | 0.33 | 0.023 | 0.31 | 2.2E-01 | 1.2 | |||
| rs532241 | 18 | 10,130,629 | 0.45 | 7.2E-04 | 0.7 | 2.9E-03 | 0.7 | yes | −1.47 | 8.1E-10 | 0.42 | 5.3E-01 | 0.9 | |||
| rs7245090 | 18 | 54,786,303 | ZNF532 | 0.02 | 1.3E-04 | 0.3 | 2.6E-03 | 0.4 | 0.46 | 0.70 | yes | −1.21 | 8.2E-03 | 0.22 | 7.3E-01 | 1.1 |
| rs7242186 | 18 | 66,132,841 | SOCS6 | 0.25 | 8.8E-04 | 1.4 | 1.4E-03 | 1.5 | yes | 1.25 | 9.7E-05 | 0.33 | 6.7E-01 | 0.9 | ||
| rs2353203 | 19 | 16,960,980 | CPAMD8 | 0.44 | 3.8E-04 | 1.4 | 1.6E-03 | 1.4 | yes | −1.37 | 4.1E-03 | 0.30 | 6.5E-02 | 0.7 | ||
| rs8111589 | 19 | 50,726,398 |
VASP OPA3 |
0.44 | 1.5E-04 | 1.4 | 8.3E-03 | 1.3 | 0.028 | 0.018 | yes | 0.48 | 1.6E-02 | 0.7 | ||
| rs11696193 | 20 | 41,958,904 | 0.26 | 6.2E-05 | 0.7 | 4.4E-04 | 0.7 | 0.030 | 0.010 | 0.29 | 3.0E-01 | 0.8 | ||||
| rs8118160 | 20 | 47,868,043 | SLC9A8 | 0.50 | 2.0E-04 | 0.7 | 1.7E-03 | 0.7 | 0.01 | 0.09 | yes | 0.24 | 2.8E-01 | 1.2 | ||
| rs4809760 | 20 | 47,888,078 | SLC9A8 | 0.43 | 2.5E-04 | 1.4 | 1.4E-03 | 1.4 | 0.02 | 0.18 | yes | 0.43 | 6.4E-01 | 1.1 | ||
| rs2281223 | 20 | 47,917,648 | SLC9A8 | 0.42 | 2.2E-04 | 1.4 | 1.4E-03 | 1.4 | 0.02 | 0.18 | yes | −1.15 | 1.3E-04 | 0.45 | 6.1E-01 | 1.1 |
| rs1996017 | 20 | 51,356,045 | TSHZ2 | 0.36 | 4.6E-04 | 1.4 | 4.5E-03 | 1.3 | 0.16 | 0.049 | 0.17 | 2.0E-01 | 1.3 | |||
| rs11704468 | 22 | 46,037,678 | 0.07 | 6.8E-04 | 0.6 | 4.1E-03 | 0.6 | yes | −1.37 | 1.3E-10 | 0.02 | 7.6E-02 | 0.4 | |||
SNPs with evidence for alcohol dependence in the GWAS that either had support from the family-based analyses (p ≤ 0.05) or evidence for differential expression in the presence of ethanol (≥1.2-fold change, with FDR ≤ 0.05) are listed by position along the genome based upon dbSNP build 130 and genome build 36.3. Genes within 15 kb of the SNP are shown (blank means no gene is within 15 kb). MAF = minor allele frequency. AD p value is the p value for association with alcohol dependence in the GWAS EA sample; AD OR is the odds ratio for this association. Early onset AD p value is the p value for association with alcohol dependence in the GWAS with affected status defined as having an onset of alcohol dependence at or below the median (22 years); Early Onset AD OR is the odds ratio for this association. Family Sample (PDT) AD and Early Onset AD use these same phenotypic definitions but are analyzed by PDT in the families. Expr in brain: yes = reliably detected in at least one of 9 brain regions, no = not reliably detected, blank = no data. FOLD ET/C = ratio of gene expression in immortalized lymphoblastoid cell lines treated with ethanol for 24 h compared to untreated cells; FDR = false discovery rate for this ratio, calculated according to Storey and Tibshirani (Storey and Tibshirani, 2003). MAF and p-value for alcohol dependence in the smaller African-American sample of the GWAS are also presented for comparison.
Fifteen of the SNPs with the smallest p values for alcohol dependence (p ≤ 2.1 × 10−4) provided nominal evidence of association in the family sample with the early onset alcohol dependence phenotype. In addition to the SNPs noted above, these included SLC37A3, KCNMA1, CDH8, ZNF608 and API5. A few candidate genes in which a SNP had p≤2.1 × 10−4 in the GWAS or cluster of SNPs with p≤10−3 were also tested in families, and significant support was found for CAT, and GRIN2C (Supplementary Table 1).
Expression of the genes containing the more significant SNPs
We also tested whether the genes associated with alcohol dependence were differentially affected by ethanol exposure in LCLs. Forty of the SNPs listed in Table 2 were in genes expressed in brain and also significantly affected by ethanol in LCLs. One of the SNPs in CARS, rs729662 (synonymous, Pro706 in exon 19), was ranked 80 in the GWAS and was also significant in the family data (Supplemental Table 1).
DISCUSSION
Although the overall number of subjects was modest for a GWAS, the sample studied here had extensive phenotypic characterization and a strong contrast in substance dependence between affected and unaffected subjects. The affected subjects all met DSM-IV criteria for alcohol dependence, whereas the controls did not meet criteria for alcohol dependence or abuse or harmful use, nor were they dependent on other illicit drugs. Many of the affected subjects (56.5%) were also dependent upon an illicit drug; while it is possible that risk for a more general substance dependence contributed to the association signal, testing for this was not judged to be feasible, given the sample size and reduction in power that would result. None of the SNPs tested met conventional criteria for genome-wide significance, probably due to the modest size of the sample. Therefore, it was important to use other lines of evidence to prioritize potential candidates. Regions in which several SNPs provided evidence for association were found. Evidence of association, with similar estimates of effect size (odds ratios), was found for many SNPs when the sample of alcohol dependent individuals was dichotomized and only those with early onset alcohol dependence were defined as cases. Early onset generally marks a group of more severe cases, often with comorbid psychiatric problems. Analyses of this subgroup have yielded smaller p values, despite the reduced sample size, in some previous studies (Agrawal et al., 2006; Dick et al., 2007a; Edenberg et al., 2008b; Foroud et al., 2008). We assessed segregation within alcohol dependent families as a means to prioritize among our SNP results. We also used gene expression data as additional lines of support, testing whether a gene in or nearest the SNP was expressed in human brain samples. We tested whether the expression of the gene in LCLs was altered by ethanol exposure; 80% of the genes differentially expressed in LCLs after ethanol exposure are also expressed in at least one of the 9 brain regions we analyzed (Edenberg et al., in preparation).
Overall, the convergence of evidence supports a region on chromosome 11 as the strongest candidate. It contained many of the top SNPs from the case control sample, and several SNPs genotyped in the family sample support the association of this region with alcohol dependence and the early onset of dependence (Table 2; Figure 2A,B; Supplementary Table 1). This region contains 6 genes: SLC22A18 (solute carrier family 22, member 18, a poly-specific organic cation transporter), PHLDA2 (pleckstrin homology-like domain, family A, member 2, important in regulating placental growth), NAPIL4 (nucleosome assembly protein 1-like 4, a likely histone chaperone), CARS (cysteinyl-tRNA synthetase), OSBPL5 (oxysterol-binding protein-like protein 5, an intracellular lipid receptor that interacts with retinoic acid and estradiol), and SNORA54 (small nucleolar RNA, H/ACA box 54). The four genes that were assayed (PHLDA2, NAPIL4, CARS, OSBPL5) are expressed in the human brain. In LCLs, expression of SLC22A18 and PHLDA2 are increased 9-14% by ethanol exposure (FDR < 0.05), while expression of NAP1L4 is decreased 8% by ethanol (FDR <10−5; Edenberg et al., in preparation). These lines of evidence suggest that one or more of these genes is a good candidate for affecting the risk for alcoholism. Although not previously considered candidates for affecting the risk for alcoholism, several of these genes have functions that might relate to alcoholism, such as growth regulation, cation transport and lipid signaling. Careful dissection of a Quantitative Trait Locus (QTL) hotspot on mouse distal chromosome 1 (Mozhui et al., 2008) that includes QTLs affecting alcohol dependence (Buck et al., 2002), alcohol withdrawal (Crabbe, 1996), and alcohol induced locomotor activity (Downing et al., 2003) showed that genes located in the QTL had a trans effect on expression in nervous tissue of a set of aminoacyl tRNA synthetases (Mozhui et al., 2008). As noted by Mozhui et al. (2008), the nervous system depends heavily on finely-tuned protein metabolism in dendrites and axons (Chang et al., 2006; Malgaroli et al., 2006). Thus, CARS is a reasonable candidate.
One of the top-ranked SNPs was in the promoter region of BBX (Table 2); this SNP was also significant in the family sample, for both alcohol dependence and early onset (Table 2). Several additional SNPs just upstream of BBX were associated with both phenotypes in the GWAS (Supplementary Table 1). BBX is widely expressed in human tissues, and encodes the human homolog of Drosophila Bobbysox, an HMG-BOX transcription factor. The potential role of BBX in affecting risk for alcohol dependence is likely to be complex; BBX mRNA levels are themselves significantly increased by alcohol exposure (13% increase in lymphoblastoid cell lines, FDR = 9 × 10−5; Table 2). A SNP in intron 1 of KCNMA1 provided evidence in subjects of both European and African ancestry, and in the family sample (Table 2), KCNMA1 is expressed in brain, and encodes potassium large conductance calcium-activated channel, subfamily M, alpha member 1, a protein important in controlling neuronal excitability.
We examined results in our case control sample for genes that were previously associated with alcoholism in our family-based sample. Among the best-supported findings was the ANKK1/DRD2 region (Dick et al., 2007b), in which 24 SNPs were nominally associated with dependence and 22 with early onset; SNPs extending through TTC12 were also significant, supporting results from Yang et al. (Yang et al., 2007). Three GWAS SNPs in NFKB1 were nominally associated with early onset alcoholism, as were more extending upstream, consistent with our previous work (Edenberg et al., 2008). Thirty-two SNPs in GRM8 (Chen et al., 2009), encoding metabotropic glutamate receptor 8, were associated with early onset of dependence. There was also nominal support for PDYN (Xuei et al., 2006), CHRNA3 and CHRNA5 (Wang et al., 2009), and (among AA only) CHRM2 (Wang et al., 2004). In the ADH gene cluster (Edenberg et al., 2006), SNPs in ADH4 were not significant in this GWAS, although 15 SNPs (particularly in the region between ADH1B and ADH1A) were. Genes encoding GABAA receptors continued to provide evidence for association with alcoholism. A SNP in GABRR2 was among those with the smallest p value in this GWAS (6 × 10−5); that SNP was not itself significant in the earlier family study, although other SNPs in the gene were (Xuei et al., 2010). Although GABRA2 has been associated with alcohol dependence (Edenberg et al., 2004) in many samples (Bauer et al., 2007; Covault et al., 2004; Drgon et al., 2006; Enoch et al., 2008; Enoch et al., 2006; Fehr et al., 2006; Lappalainen et al., 2005; Soyka et al., 2008), there was no evidence in this case-control GWAS, although 5 SNPs in GABRG1 were nominally associated with alcoholism, and 11 with early onset (cf Covault et al., 2008). Six SNPs in GABRG3 (Dick et al., 2004) were associated with alcohol dependence in the EA sample, and 17 SNPs in the much smaller AA sample. In GABRA1 (Dick et al., 2006), we found 3 SNPs associated with alcohol dependence and 4 with early onset. Many SNPs in and near GABRG2 were also associated with dependence (and early onset). Thus, overall, there continues to be evidence that variations in GABAA receptors affect alcohol dependence.
Several SNPs nominated as candidates in earlier GWAS studies replicated in ours. Johnson et al. (Johnson et al., 2006) reported 51 regions, containing 181 SNPs, that provided the strongest evidence for association in a GWAS of alcohol dependence using pooled samples of 120 cases and 160 controls drawn from the COGA study (we do not know the overlap with the present study). Among the 68 of those SNPs for which we had data, 4 provided evidence of replication (p<0.02; Table 3). CPE encodes carboxypeptidase E, present in the central nervous system (Lynch et al., 1990), which catalyzes an important step in the processing of peptide hormones and neurotransmitters (Hook et al., 2008). A pair of closely linked SNPs in DNASE2B, encoding deoxyribonuclease II beta, provided evidence for replication, and other SNPs in that region provided further support. SLC10A2, which encodes a sodium/bile acid cotransporter, also replicated; expression of SLC10A2 is regulated in part by retinol (Neimark et al., 2004). Three other SNPs replicated when the subgroup with early-onset alcoholism was analyzed: rs35164 just downstream of CDH11 (a type II classical cadherin), rs1927384, which lies between FGF14 (fibroblast growth factor 14) and TPP2 (tripeptidyl peptidase II), and rs6729553 in DNAH6 (axonemal dynein heavy chain 6, a microtubule-associated motor protein important in retrograde axonal organelles (Schnapp and Reese, 1989)).
Table 3.
Replication of SNPs detected in a pooling study
| SNP | Chr | Position | GENE | Alcohol Dependence (p-value) |
Alcohol Dependence Odds Ratio |
Early Onset Alcohol Dependence (p-value) |
Early Onset Alcohol Dependence Odds Ratio |
|
|---|---|---|---|---|---|---|---|---|
| rs1506700 | 1 | 84,649,319 | DNASE2B | 0.007 | 0.80 | 0.001 | 0.73 | DLAD DNase II-like acid DNase |
| rs3121147 | 1 | 84,649,571 | DNASE2B | 0.009 | 0.80 | 0.001 | 0.73 | DLAD DNase II-like acid DNase |
| rs6729553 | 2 | 84,763,346 | DNAH6 | 0.302 | 0.90 | 0.050 | 0.79 | dynein, axonemal, heavy chain 6 |
| rs1370687 | 4 | 166,609,805 | CPE | 0.001 | 1.36 | 0.027 | 1.27 | CPE carboxypeptidase E |
| rs1927384 | 13 | 101,943,751 | FGF14 | 0.188 | 0.86 | 0.037 | 0.75 | Between FGF14 and TPP2 |
| rs279929 | 13 | 102,538,720 | SLC10A2 | 0.016 | 1.26 | 0.015 | 1.31 | flank SLC10A2 solute carrier family 10 (sodium/bile acid cotransporter family), member 2 |
| rs35164 | 16 | 63,532,702 | CDH11 | 0.314 | 0.91 | 0.022 | 0.78 | flank CDH11 cadherin 11, type 2 |
SNPs from Johnson et al. (Johnson et al., 2006) that provided evidence for association (p≤0.05) in the present GWAS. Alcohol dependence is the p value for association in the GWAS EA sample. Early onset alcohol dependence is the p value for the GWAS with affected status defined as having an onset of alcohol dependence at or below the median (22 years). MAF = minor allele frequency.
Treutlein et al. (2009) recently reported results from a GWAS of German alcoholics. Their cases were male alcoholics who had been hospitalized for treatment or prevention of severe withdrawal. We had data for 114 of their top 140 SNPs (121 at p<10−4 in their study and 19 they nominated from rodent studies); 14 were significant in our primary analysis of alcohol dependence; 11 of these were also significant in our smaller subset of early-onset alcoholics (Table 4). Only one of these, rs13273672 in GATA4, was among the 15 SNPs for which Treutlein et al. reported confirmation in their follow-up (Treutlein et al., 2009). Among the SNPs for which we had replication, 6 have the same risk allele; these lie in or near ARL6IP5, ID4, GATA4, SYNE1, ADCY3 and PRKCA; three of these (GATA4, ADCY3, SYNE1) were among the top 6 with our early onset phenotype. These are all good candidate genes; two regulate transcription (ID4, GATA4), two (ADCY3, PRKCA) regulate important second messenger systems, one (ARL6IP5) inhibits the glutamate transporter EAAC1 and one (SYNE1) is associated with autosomal recessive spinocerebellar ataxia 8. PRKCA expression is lower in the nucleus accumbens of alcohol-preferring P rats after operant ethanol self-administration (Rodd et al., 2008), and is reduced by chronic alcohol in vertebrae of Sprague-Dawley rats after chronic binge exposure to ethanol (Himes et al., 2008). We have confirming evidence for three of these (PRKCA, ADCY3, ARL6IP5) in our African-American sample.
Table 4.
Replication of SNPs detected in a GWAS of German alcoholics
| Treutlein et al. | COGA RESULTS - EA |
COGA RESULTS - AA | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Location (bp) |
Gene | Risk allele |
P-value | Dependence (p-value) |
OR | Risk allele |
Early onset p-value |
OR | MAF | Dependence (p-value) |
OR | MAF | Gene name |
| rs420033 | 1 | 111,097,197 | KCNA3 | A | 2.7E-05 | 9.6E-03 | 0.8 | G | 7.7E-03 | 0.7 | 0.26 | 6.6E-01 | 1.2 | 0.06 | potassium voltage-gated channel, shaker-related subfamily, member 3 |
| rs343802 | 1 | 111,118,652 | CD53 | T | 9.0E-05 | 1.8E-03 | 0.7 | C | 1.4E-03 | 0.7 | 0.24 | 7.9E-01 | 1.1 | 0.06 | CD53 molecule |
| rs17799872 | 2 | 24,898,461 | ADCY3 | A | 2.1E-03 | 4.4E-02 | 1.4 | A | 1.9E-02 | 1.5 | 0.08 | 3.1E-03 | 3.1 | 0.07 | adenylate cyclase 3 |
| rs11706542 | 3 | 28,303,587 | CMC1 | T | 4.3E-05 | 2.0E-02 | 0.8 | C | 4.1E-02 | 0.8 | 0.28 | 6.8E-01 | 0.9 | 0.12 | COX assembly mitochondrial protein homolog |
| rs1606388 | 3 | 28,309,831 | CMC1 | T | 5.2E-05 | 1.5E-02 | 0.8 | G | 3.2E-02 | 0.8 | 0.28 | 2.0E-01 | 0.7 | 0.08 | COX assembly mitochondrial protein homolog |
| rs4680758 | 3 | 28,576,547 | ZCWPW2 | A | 4.7E-05 | 2.5E-02 | 0.8 | C | 6.0E-02 | 0.8 | 0.25 | 5.1E-01 | 0.9 | 0.10 | |
| rs6549184 | 3 | 69,232,967 | ARL6IP5 | A | 7.6E-05 | 1.6E-02 | 1.7 | A | 6.4E-02 | 1.6 | 0.04 | 1.1E-04 | 2.1 | 0.25 | ADP-ribosylation-like factor 6 interacting protein 5 |
| rs12527834 | 6 | 19,929,760 | ID4 | G | 3.7E-05 | 2.1E-02 | 1.4 | G | 7.7E-02 | 1.4 | 0.10 | 2.7E-01 | 1.5 | 0.04 | inhibitor of DNA binding 4, dominant negative helix- loop-helix protein |
| rs214959 | 6 | 152,744,514 | SYNE1 | A | 4.7E-05 | 2.8E-02 | 1.2 | A | 1.8E-02 | 1.3 | 0.50 | 9.3E-01 | 1.0 | 0.12 | spectrin repeat containing, nuclear envelope 1 |
| rs2188594 | 7 | 20,645,811 | ABCB5 | A | 9.3E-05 | 6.9E-03 | 0.7 | G | 2.2E-02 | 0.7 | 0.18 | 4.6E-01 | 0.9 | 0.49 | ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| rs6958596 | 7 | 20,661,260 | ABCB5 | T | 1.4E-03 | 4.6E-03 | 0.7 | C | 1.7E-02 | 0.7 | 0.16 | 2.6E-01 | 0.8 | 0.21 | ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
|
rs13273672 * |
8 | 11,649,790 | GATA4 | C | 2.2E-03 | 2.2E-02 | 1.2 | C | 1.6E-03 | 1.4 | 0.31 | 2.9E-01 | 0.8 | 0.35 | GATA binding protein 4 |
| rs753708 | 13 | 100,219,370 | TMTC4 | C | 7.9E-05 | 3.1E-02 | 0.7 | A | 2.5E-02 | 0.6 | 0.07 | 3.2E-01 | 1.3 | 0.12 | transmembrane and tetratricopeptide repeat containing 4 |
| rs12603061 | 17 | 62,242,660 | PRKCA | A | 7.8E-05 | 1.2E-01 | 1.1 | A | 4.7E-02 | 1.2 | 0.48 | 3.3E-02 | 0.7 | 0.46 | protein kinase C, alpha |
| rs8082983 | 18 | 55,306,698 | CCBE1 | G | 9.0E-05 | 1.7E-02 | 0.8 | A | 2.3E-02 | 0.8 | 0.35 | 2.1E-01 | 0.8 | 0.49 | collagen and calcium binding EGF domains 1 |
| rs11702690 | 21 | 42,280,737 | ZNF295 | G | 2.0E-05 | 7.8E-02 | 1.3 | G | 4.6E-02 | 1.4 | 0.08 | 8.6E-01 | 0.9 | 0.02 | zinc finger protein 295 |
SNPs from Treutlein et al. (Treutlein et al., 2009) that provided evidence for association (p≤0.05) in the present GWAS. Results from Treutlein are shown for comparison with the present GWAS in EA and AA. OR = odds ratio. MAF = minor allele frequency. Bold SNPs have the same risk allele.
A recently submitted manuscript (Bierut et al., in press) describes results from the Study of Addiction: Genetics and Environment consortium, which has performed a GWAS using the phenotype of alcohol dependence in a sample of cases and controls ascertained through three different studies (dbGaP accession phs000092.v1.p1). One of the three studies was COGA, with 612 EA alcohol dependent individuals and 413 EA controls included in both our analysis and the SAGE analyses. Two other studies also provided EA cases: one recruited cases with nicotine dependence through a population screening design (n=343 alcohol dependent EA cases) and the other recruited cases with cocaine dependence through treatment centers (n=278 alcohol dependent EA cases). While 52% of EA COGA subjects also reported another substance dependence, the inclusion of cases in the SAGE analysis recruited for different primary diagnoses will likely introduce a number of novel genes contributing to alcohol dependence and another comorbid condition, either nicotine dependence or cocaine dependence. For this reason, while we might hope for some commonality between the results from the COGA and SAGE studies due to the overlapping samples, it is not surprising that in practice results of GWAS from each study are quite different.
Overall, although we did not detect any SNP that met genome-wide significance, we have assembled several different lines of evidence to prioritize SNPs and genes from among the results for further study. In a multi-stage follow-up to the GWAS, we: a) analyzed a set of top SNPs by analyzing transmission in a family sample, which provided additional support for some of the SNPs; b) determined which among the top SNPs were expressed in human brain; and c) determined which top SNPs are affected by exposure to alcohol in human LCLs. The convergence of evidence from the GWAS, family-based association analyses and the response of genes to ethanol exposure provides a set of interesting candidate genes for further analyses in larger samples.
Supplementary Material
SNPs that were significant for alcohol dependence in the European-American GWAS study at p ≤ 10−3 are listed in order along the genome (build 36.3). Genes within 15 kb of the SNP are listed (blank means no gene is within 15 kb); gene name is based upon the gene symbol. MAF = minor allele frequency. Alcohol dependence is the p value for association in the GWAS. Early onset alcohol dependence is the p value for the GWAS with affected status defined as having an onset of alcohol dependence at or below the median (22 years). Analyses in the Family sample use these same phenotypic definitions but are analyzed by PDT; only some SNPs were tested (see text). The MAF, p-value and odds ratio for alcohol dependence in the smaller African-American sample of the GWAS are presented for comparison.
SNPs that were significant for alcohol dependence in the African-American GWAS study at p ≤ 10−3 are listed in order along the genome (build 36.3). Genes within 15 kb of the SNP, based upon the genome build (blank means no gene is within 15 kb). MAF = minor allele frequency. Alcohol dependence is the p value for association in the GWAS. Because of the small number of subjects, we did not analyze early onset. The MAF and p-values for alcohol dependence in the European-American sample of the GWAS are presented for comparison.
Acknowledgments
We thank Kim Doheny and Elizabeth Pugh from CIDR and Justin Paschall from the NCBI dbGaP staff for valuable assistance with genotyping and quality control in developing the dataset available at dbGaP.
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes ten different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice, K. Bucholz); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy), and Virginia Commonwealth University (D. Dick). A. Parsian and M. Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li (currently a consultant with COGA), P. Michael Conneally, and Raymond Crowe, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Funding support for GWAS genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the National Institute on Alcohol Abuse and Alcoholism, the NIH GEI (U01HG004438), and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). Family-based genotyping was performed using the facilities of the Center for Medical Genomics at Indiana University School of Medicine, which is supported in part by the Indiana Genomics Initiative of Indiana University (INGEN®); INGEN is supported in part by The Lilly Endowment, Inc.
Brain tissues were received from the New South Wales Tissue Resource Centre, which is supported by the National Health and Medical Research Council of Australia, The University of Sydney, Prince of Wales Medical Research Institute, Neuroscience Institute of Schizophrenia and Allied Disorders, National Institute of Alcohol Abuse and Alcoholism (Grant R01 AA12725) and NSW Department of Health.
References
- Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66(1):279–92. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Edenberg HJ, Foroud T, Bierut LJ, Dunne G, Hinrichs AL, Nurnberger JI, Crowe R, Kuperman S, Schuckit MA, Begleiter H, Porjesz B, Dick DM. Association of GABRA2 with drug dependence in the collaborative study of the genetics of alcoholism sample. Behav Genet. 2006;36(5):640–50. doi: 10.1007/s10519-006-9069-4. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. Third Edition. American Psychiatric Association Press; Washington, DC: 1987. Revised. [Google Scholar]
- American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. Fourth Edition American Psychiatric Association Press; Washington, DC: 1994. [Google Scholar]
- Bauer LO, Covault J, Harel O, Das S, Gelernter J, Anton R, Kranzler HR. Variation in GABRA2 predicts drinking behavior in project MATCH subjects. Alcohol Clin Exp Res. 2007;31(11):1780–7. doi: 10.1111/j.1530-0277.2007.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li T-K, Schuckit MA, Edenberg HJ, Rice AP. The Collaborative Study on the Genetics of Alcoholism. Alc.Health.Res.World. 1995;19:228–236. [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JIJ, Reich T, Schmidt I, Schuckit MA. A new semi-structured psychiatric interview for use in genetic linkage studies: A report of the reliability of the SSAGA. J.Stud.Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Rademacher BS, Metten P, Crabbe JC. Mapping murine loci for physical dependence on ethanol. Psychopharmacology (Berl) 2002;160(4):398–407. doi: 10.1007/s00213-001-0988-8. [DOI] [PubMed] [Google Scholar]
- Chang RC, Yu MS, Lai CS. Significance of molecular signaling for protein translation control in neurodegenerative diseases. Neurosignals. 2006;15(5):249–58. doi: 10.1159/000102599. [DOI] [PubMed] [Google Scholar]
- Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, Edenberg HJ, Hesselbrock V, Nurnberger J, Jr., Kuperman S, O'Connor SJ, Schuckit MA, Bauer LO, Tischfield J, Rice JP, Bierut L, Goate A, Porjesz B. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):359–68. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2004;129(1):104–9. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5′-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33(4):837–48. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC. Quantitative trait locus gene mapping: a new method for locating alcohol response genes. Addict Biol. 1996;1(3):229–35. doi: 10.1080/1355621961000124846. [DOI] [PubMed] [Google Scholar]
- Dick DM, Agrawal A, Wang JC, Hinrichs A, Bertelsen S, Bucholz KK, Schuckit M, Kramer J, Nurnberger J, Jr., Tischfield J, Edenberg HJ, Goate A, Bierut LJ. Alcohol dependence with comorbid drug dependence: genetic and phenotypic associations suggest a more severe form of the disorder with stronger genetic contribution to risk. Addiction. 2007a;102(7):1131–9. doi: 10.1111/j.1360-0443.2007.01871.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Kuperman S, Schuckit M, Crowe R, Smith TL, Porjesz B, Begleiter H, Foroud T. Association of GABRG3 with alcohol dependence. Alcohol Clin Exp Res. 2004;28(1):4–9. doi: 10.1097/01.ALC.0000108645.54345.98. [DOI] [PubMed] [Google Scholar]
- Dick DM, Plunkett J, Wetherill LF, Xuei X, Goate A, Hesselbrock V, Schuckit M, Crowe R, Edenberg HJ, Foroud T. Association between GABRA1 and drinking behaviors in the collaborative study on the genetics of alcoholism sample. Alcohol Clin Exp Res. 2006;30(7):1101–10. doi: 10.1111/j.1530-0277.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Wang JC, Plunkett J, Aliev F, Hinrichs A, Bertelsen S, Budde JP, Goldstein EL, Kaplan D, Edenberg HJ, Nurnberger J, Jr., Hesselbrock V, Schuckit M, Kuperman S, Tischfield J, Porjesz B, Begleiter H, Bierut LJ, Goate A. Family-Based Association Analyses of Alcohol Dependence Phenotypes Across DRD2 and Neighboring Gene ANKK1. Alcohol Clin Exp Res. 2007b;31(10):1645–53. doi: 10.1111/j.1530-0277.2007.00470.x. [DOI] [PubMed] [Google Scholar]
- Downing C, Rodd-Henricks KK, Flaherty L, Dudek BC. Genetic analysis of the psychomotor stimulant effect of ethanol. Genes Brain Behav. 2003;2(3):140–51. doi: 10.1034/j.1601-183x.2003.00022.x. [DOI] [PubMed] [Google Scholar]
- Drgon T, D'Addario C, Uhl GR. Linkage disequilibrium, haplotype and association studies of a chromosome 4 GABA receptor gene cluster: candidate gene variants for addictions. Am J Med Genet B Neuropsychiatr Genet. 2006;141(8):854–60. doi: 10.1002/ajmg.b.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25(2):115–21. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The collaborative study on the genetics of alcoholism: an update. Alcohol Res Health. 2002;26(3):214–8. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr., O'Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74(4):705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006;11(3-4):386–96. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Wang J, Tian H, Pochareddy S, Xuei X, Wetherill L, Goate A, Hinrichs T, Kuperman S, Nurnberger JI, Jr., Schuckit M, Tischfield JA, Foroud T. A regulatory variation in OPRK1, the gene encoding the kappa-opioid receptor, is associated with alcohol dependence. Hum Mol Genet. 2008a;17(12):1783–9. doi: 10.1093/hmg/ddn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15(9):1539–49. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Wetherill LF, Bierut L, Bucholz K, Dick DM, Hesselbrock V, Kuperman S, Porjesz B, Schuckit MA, Tischfield JA, Almasy LA, Nurnberger JI, Jr., Foroud T. Association of NFKB1, which encodes a subunit of the transcription factor NF-kappaB, with alcohol dependence. Hum Mol Genet. 2008b;17(7):963–70. doi: 10.1093/hmg/ddm368. [DOI] [PubMed] [Google Scholar]
- Enoch MA, Hodgkinson CA, Yuan Q, Albaugh B, Virkkunen M, Goldman D. GABRG1 and GABRA2 as Independent Predictors for Alcoholism in Two Populations. Neuropsychopharmacology. 2008 doi: 10.1038/npp.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Schwartz L, Albaugh B, Virkkunen M, Goldman D. Dimensional anxiety mediates linkage of GABRA2 haplotypes with alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141(6):599–607. doi: 10.1002/ajmg.b.30336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Dahmen N, Schmidt LG, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16(1):9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Feighner JP, Robins E, Guze SB, Woodruff RA, Winokur G, Munoz R. Diagnostic criteria for use in psychiatric research. Arch.Gen.Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24(7):933–45. [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Kramer J, Tischfield JA, Nurnberger JI, Jr., Schuckit MA, Xuei X, Edenberg HJ. The tachykinin receptor 3 is associated with alcohol and cocaine dependence. Alcohol Clin Exp Res. 2008;32(6):1023–30. doi: 10.1111/j.1530-0277.2008.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindalini C, Scivoletto S, Ferreira RG, Breen G, Zilberman M, Peluso MA, Zatz M. Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. Am J Psychiatry. 2005;162(5):1005–7. doi: 10.1176/appi.ajp.162.5.1005. [DOI] [PubMed] [Google Scholar]
- Gunderson KL, Steemers FJ, Ren H, Ng P, Zhou L, Tsan C, Chang W, Bullis D, Musmacker J, King C, Lebruska LL, Barker D, Oliphant A, Kuhn KM, Shen R. Whole-genome genotyping. Methods Enzymol. 2006;410:359–76. doi: 10.1016/S0076-6879(06)10017-8. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2007;64(7):830–42. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94(9):1361–70. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Himes R, Wezeman FH, Callaci JJ. Identification of novel bone-specific molecular targets of binge alcohol and ibandronate by transcriptome analysis. Alcohol Clin Exp Res. 2008;32(7):1167–80. doi: 10.1111/j.1530-0277.2008.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang SR. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, Foroud T, Uhl GR. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: Validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(8):844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am.J.Psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van den Oord EJ, Alexander J, Jiang C, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Association of ADH and ALDH genes with alcohol dependence in the Irish Affected Sib Pair Study of alcohol dependence (IASPSAD) sample. Alcohol Clin Exp Res. 2008;32(5):785–95. doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Somberg LK, Covault J, Kranzler HR, Krystal JH, Gelernter J. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29(4):493–8. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Li TK, Hewitt BG, Grant BF. The Alcohol Dependence Syndrome, 30 years later: a commentary. Addiction. 2007 doi: 10.1111/j.1360-0443.2007.01911.x. [DOI] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Montgomery GW, Heath AC, Martin NG, Whitfield JB. Effects of GABRA2 Variation on Physiological, Psychomotor and Subjective Responses in the Alcohol Challenge Twin Study. Twin Res Hum Genet. 2008;11(2):174–82. doi: 10.1375/twin.11.2.174. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology. 2006;31(5):1085–95. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Blumberg HP, Gelernter J. CHRM2 gene predisposes to alcohol dependence, drug dependence and affective disorders: results from an extended case-control structured association study. Hum Mol Genet. 2005a;14(16):2421–34. doi: 10.1093/hmg/ddi244. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: results from family controlled and population-structured association studies. Pharmacogenet Genomics. 2005b;15(11):755–768. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- Lynch DR, Braas KM, Hutton JC, Snyder SH. Carboxypeptidase E (CPE): immunocytochemical localization in the rat central nervous system and pituitary gland. J Neurosci. 1990;10(5):1592–9. doi: 10.1523/JNEUROSCI.10-05-01592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgaroli A, Vallar L, Zimarino V. Protein homeostasis in neurons and its pathological alterations. Curr Opin Neurobiol. 2006;16(3):270–4. doi: 10.1016/j.conb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67(1):146–54. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M. Phenotyping alcoholism. Alcohol.Clin.Exp.Res. 1999;23(5):757–758. doi: 10.1111/j.1530-0277.1999.tb04180.x. [DOI] [PubMed] [Google Scholar]
- Mozhui K, Ciobanu DC, Schikorski T, Wang X, Lu L, Williams RW. Dissection of a QTL hotspot on mouse distal chromosome 1 that modulates neurobehavioral phenotypes and gene expression. PLoS Genet. 2008;4(11):e1000260. doi: 10.1371/journal.pgen.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neimark E, Chen F, Li X, Shneider BL. Bile acid-induced negative feedback regulation of the human ileal bile acid transporter. Hepatology. 2004;40(1):149–56. doi: 10.1002/hep.20295. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr., Wiegand R, Bucholz K, O'Connor S, Meyer ET, Reich T, Rice J, Schuckit M, King L, Petti T, Bierut L, Hinrichs AL, Kuperman S, Hesselbrock V, Porjesz B. A family study of alcohol dependence: coaggregation of multiple disorders in relatives of alcohol-dependent probands. Arch Gen Psychiatry. 2004;61(12):1246–56. doi: 10.1001/archpsyc.61.12.1246. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Lykken DT, Heston LL, Clayton PJ. Heterogeneity in the inheritance of alcoholism: a study of male and female twins. Arch.Gen.Psychiatry. 1991;48:19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr., Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81(3):207–15. [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89(4):481–98. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp BJ, Reese TS. Dynein is the motor for retrograde axonal transport of organelles. Proc Natl Acad Sci U S A. 1989;86(5):1548–52. doi: 10.1073/pnas.86.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyka M, Preuss UW, Hesselbrock V, Zill P, Koller G, Bondy B. GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. J Psychiatr Res. 2008;42(3):184–191. doi: 10.1016/j.jpsychires.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66(7):773–84. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel CI, Scherag A, Bronner G, Nguyen TT, Wang HJ, Grallert H, Bornhorst A, Rosskopf D, Volzke H, Reinehr T, Rief W, Illig T, Wichmann HE, Schafer H, Hebebrand J, Hinney A. Gastric inhibitory polypeptide receptor: association analyses for obesity of several polymorphisms in large study groups. BMC Med Genet. 2009;10:19. doi: 10.1186/1471-2350-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP, Fox L, Goldstein E, Reyes O, Saccone N, Saccone S, Xuei X, Bucholz K, Kuperman S, Nurnberger J, Jr., Rice JP, Schuckit M, Tischfield J, Hesselbrock V, Porjesz B, Edenberg HJ, Bierut LJ, Goate AM. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14(5):501–10. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, Kwon JM, Wu W, Dick DM, Rice J, Jones K, Nurnberger JI, Jr., Tischfield J, Porjesz B, Edenberg HJ, Hesselbrock V, Crowe R, Schuckit M, Begleiter H, Reich T, Goate AM, Bierut LJ. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum Mol Genet. 2004;13(17):1903–11. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- Wetherill L, Schuckit MA, Hesselbrock V, Xuei X, Liang T, Dick DM, Kramer J, Nurnberger JI, Jr., Tischfield JA, Porjesz B, Edenberg HJ, Foroud T. Neuropeptide Y receptor genes are associated with alcohol dependence, alcohol withdrawal phenotypes, and cocaine dependence. Alcohol Clin Exp Res. 2008;32(12):2031–40. doi: 10.1111/j.1530-0277.2008.00790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . International Classification of Disease. Tenth Edition World Health Organization; Geneva: 1993. [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L, Goate A, Bucholz K, Schuckit M, Nurnberger J, Jr., Tischfield J, Kuperman S, Porjesz B, Begleiter H, Foroud T, Edenberg HJ. Association of the kappa-opioid system with alcohol dependence. Mol Psychiatry. 2006;11(11):1016–24. doi: 10.1038/sj.mp.4001882. [DOI] [PubMed] [Google Scholar]
- Xuei X, Flury-Wetherill L, Dick D, Goate A, Tischfield J, Nurnberger J, Jr., Schuckit M, Kramer J, Kuperman S, Hesselbrock V, Porjesz B, Foroud T, Edenberg HJ. GABRR1 and GABRR2, encoding the GABA-A receptor subunits rho1 and rho2, are associated with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.30995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Association of haplotypic variants in DRD2, ANKK1, TTC12 and NCAM1 to alcohol dependence in independent case control and family samples. Hum Mol Genet. 2007;16(23):2844–53. doi: 10.1093/hmg/ddm240. [DOI] [PubMed] [Google Scholar]
- Zuo L, Kranzler HR, Luo X, Yang BZ, Weiss R, Brady K, Poling J, Farrer L, Gelernter J. Interaction between two independent CNR1 variants increases risk for cocaine dependence in European Americans: a replication study in family-based sample and population-based sample. Neuropsychopharmacology. 2009;34(6):1504–13. doi: 10.1038/npp.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNPs that were significant for alcohol dependence in the European-American GWAS study at p ≤ 10−3 are listed in order along the genome (build 36.3). Genes within 15 kb of the SNP are listed (blank means no gene is within 15 kb); gene name is based upon the gene symbol. MAF = minor allele frequency. Alcohol dependence is the p value for association in the GWAS. Early onset alcohol dependence is the p value for the GWAS with affected status defined as having an onset of alcohol dependence at or below the median (22 years). Analyses in the Family sample use these same phenotypic definitions but are analyzed by PDT; only some SNPs were tested (see text). The MAF, p-value and odds ratio for alcohol dependence in the smaller African-American sample of the GWAS are presented for comparison.
SNPs that were significant for alcohol dependence in the African-American GWAS study at p ≤ 10−3 are listed in order along the genome (build 36.3). Genes within 15 kb of the SNP, based upon the genome build (blank means no gene is within 15 kb). MAF = minor allele frequency. Alcohol dependence is the p value for association in the GWAS. Because of the small number of subjects, we did not analyze early onset. The MAF and p-values for alcohol dependence in the European-American sample of the GWAS are presented for comparison.