Abstract
Over the past 40 years there have been giant steps forward in our understanding of cellular and molecular biology that have given us the framework by which to understand tissue organization and tissue function on a range of scales. However, although the progress has been great, the more we have discovered, the more we are aware of what we don’t yet know. In this article, I would like to flag up some issues of cartilage biology, function and pathology where we still have significant ignorance. As scientists we all provide contributions to add to the greater understanding of science and progress is on a broad front, but gaps are left where particular difficulty is encountered and in life sciences it is no different. Progress is fast where new knowledge and techniques pave the way, but where study is complex and relevant techniques poorly developed the gaps are left behind. In cartilage research and matrix biology, the gaps can particularly be seen at interfaces between disciplines and where technology development has lagged behind and in the particular challenges of understanding how molecular properties can explain tissue macro properties.
Keywords: cartilage, chondrocyte, epigenetics, extracellular matrix
Cartilage matrix
Cartilage provides a wonderful example of a tissue predominantly formed of extracellular matrix (ECM). Being avascular and aneural it lacks some of the complexity of other tissues, as no vessels or nerves permeate it and it is composed of an expanded and highly specialized ECM, in which is embedded a single cell type, the chondrocyte. In the past 40 years, the perception of cartilage has changed from being a connective tissue in which sparse cells are surrounded by an amorphous ground substance, to one in which our view of the amorphous ground substance has been transcended by new knowledge of exquisite detail of the ECM containing proteins and glycans in an array of intricate fibrillar and non-fibrillar assemblies (Figure 1) (Heinegard 2009). Whereas the amorphous ground substance was purely an image of a static physical structure, we now learn that almost every molecule can bind or interact with every other molecule and the whole organization is not static, but dynamic. However. although we know incredible molecular detail of isolated interactions, we know little of the priorities of the intermolecular interactions when all components are present in the concentrated tissue environments and thereby how they together contribute to the composite viscoelastic and biomechanical properties that provide the function of cartilage.
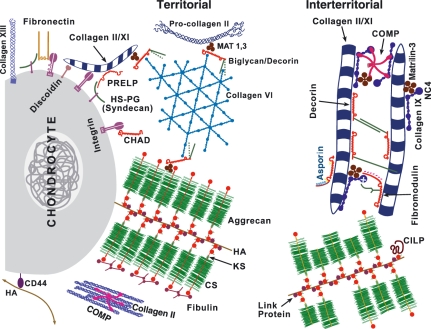
Figure 1.
Schematic to illustrate the range of molecular interactions in cartilage to matrix (from Heinegard 2009). Amongst interactions shown are territorial and interterritorial networks of type II/XI collagen fibres and type VI collagen networks; aggrecan aggregates with hyaluronan and link protein; possible retention of aggrecan G3 domain in territorial, but not interterritorial matrix and interaction of G3 with fibulins and tenascin.
An example that illustrates this complexity that arises from the detail we know can be found in aggrecan. It was established long ago that aggrecan forms supramolecular aggregates by binding to hyaluronan through its G1 domain and this interaction is stabilized by link protein (see Figure 1) (Hardingham & Muir 1972; Hardingham 1979). This provides a mechanism to immobilize aggrecan within the collagen network in cartilage and thereby create a large Donnan osmotic pressure drawing water into cartilage, placing the collagen under tension and generating compressive resilience. The aggrecan G2 domain was shown to have no function in the formation of aggregates (Fosang & Hardingham 1989), but the G3 domain which includes a C-type lectin fold was shown to bind to several matrix molecules, including tenascins and fibulins (Day et al. 2004). These interaction could thus potentially link aggrecan aggregates into a more complex network (see territorial matrix in Figure 1), however, when this G3 function is active and with which ligands remains unclear. The role of aggrecan G3 is also compounded by the proteolytic cleavage of the G3 domain during biosynthesis and secretion in cartilage, which results in much of the aggrecan in mature cartilage lacking this domain (see interterritorial matrix Figure 1). The conclusion is that G3 interactions are not crucial to most of aggrecan function in mature cartilage. However, there is evidence that during cartilage development the G3 domain remains more intact and the formation of an enhanced aggrecan network may then have a function in the nascent ECM during chondrogenesis (Day et al. 2004). This role during cartilage formation may have relevance to strategies for cartilage repair and regeneration, as conditions that favour G3 retention and expression of the G3 ligands could enhance matrix formation and the recovery of tissue properties. This example is just one of many molecular interactions in cartilage matrix, where we know what is possible, but not what is probable and we know little of the dynamic nature of the assemblies formed and what processes they affect.
There also remains a major gap between molecular detail and macro tissue properties. Often our tissues, like tendons, ligaments, cartilage and bone, fail because of inadequacies in their macro properties and frequently we cannot pin this down to molecular explanations. We know that many of the properties of cartilage depend on its very dense collagen network. The organization of collagen into fibrils and fibres and the cross links that stabilize intermolecular interactions are well characterized (Kadler et al. 2007), but the length of the fibres and whether they exist as an entangled polymer network, or as a continuum is unknown. The link between molecular organization and cartilage tensile and compressive properties is therefore unclear and there is a great need for new experimental approaches to unravel this.
Chondrocytes and epigenetics
There has been a quiet revolution in cell and developmental biology over the past 10–15 years in which concepts of cell phenotype, lineage restriction, commitment and irreversible differentiation have been challenged. It has become accepted that plasticity in cell phenotype is easily demonstrated and although plasticity of phenotype in differentiated cells is probably uncommon in vivo, there is evidence for trans-differentiation and in vitro, nuclear reprogramming can be achieved with relatively simple procedures. We can conclude that the phenotype of the fully differentiated cell is not hard wired and permanent, but given appropriate physical and biochemical signals it is open to change.
The concept of the chondrocyte as a highly specialized cell remains unchallenged, but the stability of its phenotype and how it differs from other closely related cells is incompletely understood. The different patterns of gene expression in chondrocytes and in meniscus cells, tendon cells and ligament cells, may be underpinned by epigenetic differences i.e. differences in the organization of their nuclear DNA (Table 1). However, we have no easy tools by which to compare and contrast cell phenotypes and establish the relationships between different phenotypes and identify pathways by which to move from one to another. Progress is being made in assessing DNA methylation and histone acetylation and chromatin modifications, but these remain, for the moment, detailed areas of research rather than easily applied practical tools (Laird 2010). We know that tendon cells in vivo under compression express cartilage proteins and become more chondrocyte-like (Koob & Vogel 1987) and isolated chondrocytes can be induced to differentiate to other mesenchymal cell types (Barbero et al. 2003). To understand the processes involved in these changes there is a need to establish the principles of epigenetic control which define differentiation and the routes to and from differentiated states. This will provide the defining basis of concepts such as progenitor cell, committed cell, transit amplifying cell, fully differentiated cell and de-differentiated cell, which have all arisen by observation and functional tests, but without known mechanisms.
Table 1.
Questions on the biology and epigenetics of chondrocytes
| 1. What is the epigenetic relationship between chondrocytes and other differentiated cells and amongst chondrocytes from different cartilaginous tissues? |
| 2. To what degree does a chondrocyte have a range of responses to local physical, mechanical, chemical signals that determine its phenotype, or do changes in its phenotype involve epigenetic switches between different differentiation states? |
| 3. What are the epigenetic changes that accompany chondrocyte de-differentiation and are they reversible without gene transfer? |
| 4. How far is the differentiation pathway to articular chondrocytes and to epiphyseal chondrocytes, separate and epigenetically distinct, or similar and interconvertible? |
| 5. How similar and interconvertible are chondrocytes from hyaline cartilages (articular, nasal) and how less similar and less interconvertible are chondrocytes from meniscus cells, nucleus/annulus pulposus cells and from tendon and ligament cells? |
Chondrocytes: one or many?
Another area where epigenetic tools will have major impact is in resolving the debate on how chondrocytes from different zones of cartilage, or from different anatomical sources of cartilage and from growth plate, differ from each other (Table 1). A leading question yet to be resolved is: how far do different chondrocyte phenotypes result from different programming signals determined during development and how far from different signals in the tissue environment in which each differentiated cell functions. In terms of epigenetics this is asking if all chondrocytes are epigenetically the same, or do they differ and by how much and how easy is it to move from one to the other? The issue here is if we place a nasal chondrocyte in an articular environment does it become an articular chondrocyte? Can the articular environment reprogram the nasal chondrocyte and if so does this involve changes in its epigenetics? This is an interesting biological question, but does it matter? Well, there is also much discussion in cartilage repair that the delivery of chondrocytes from differentiated bone marrow stromal cells (BMSC) risks delivering epiphyseal rather than articular chondrocytes (Murdoch et al. 2007). The background to this issue is because in chondrogenic differentiation tests, BMSC strongly up-regulate type II collagen, but also up-regulate type X collagen, which is expressed in epiphyseal growth plate, but not in articular cartilage (eg Murdoch et al. 2007). As chondrocytes in growth plate become hypertrophic and subsequently die, to allow vascular invasion and bone development, this would be highly undesirable in any long term cartilage repair (Pelttari et al. 2006). However, we have no understanding, as yet, why the in vitro chondrogenic differentiation protocol leads to the upregulation of type X collagen, or whether other conditions (in vitro or in vivo) may direct chondrogenesis to an articular phenotype without type X collagen expression. So our knowledge of the relationship between cells is very important to our understanding of how we may direct tissue repair.
Chondrocyte de-differentiation
Another aspect of chondrocyte biology where epigenetic insight would be helpful is in chondrocyte de-differentiation (Table 1.3). It was shown almost 30 years ago that when chondrocytes are isolated from cartilage they become fibroblastic in monolayer culture and progressively lose their characteristic gene expression pattern (Benya & Shaffer 1982). This was termed de-differentiation, as a description of this apparent loss of differentiated phenotype. When analysed there appear to be two distinct, but linked changes taking place; one is a fairly rapid decline in the gene expression of chondrogenic genes, which equates with the loss of matrix environment and signals that maintain a chondrogenic phenotype and the formation of actin stress fibres and focal adhesions in monolayer culture (Tew & Hardingham 2006); the second is a slow progressive loss of the ability for the cells to recover to be chondrogenic (Tew et al. 2008). Recovery is assessed by micromass, pellet, agarose, or alginate culture, which all result in new matrix formation by chondrocytes from healthy, or OA cartilage, if tested with early passage cells (P < 4); but is progressively lost with cells at higher passage (Tew et al. 2008; Katapodi et al. 2009). The loss of an ability to recover matrix formation suggests there are slow progressive changes, presumably epigenetic, in monolayer culture, which are not reversed by returning the chondrocytes to a cartilage-like matrix. Primary chondrocytes in monolayer culture are thus on a programme of progressive change and although frequently described as de-differentiated, it is unclear what has changed and how it might be reversed. As noted above, from our work with SOX9, we found that it was easy to recover a matrix forming phenotype in primary OA chondrocytes even at high passage, if SOX9 expression was increased following retroviral transduction (Tew et al. 2005). However, SOX9 transduction was unable to make a primary dermal fibroblast into a chondrocyte. So a passaged chondrocyte retains part of its epigenetic status as a chondrocyte and an ability to activate chondrocyte genes in response to increased SOX9 expression, but this is not a property shared with a differentiated fibroblast. The details of the epigenetic changes that accompany chondrocyte ‘de-differentiation’ in monolayer culture are as yet unknown.
Chondrocytes and osteoarthritis
The epigenetic status of chondrocytes is thus of much interest for generating cartilage repair, but it is also of great importance in understanding cartilage pathology (Table 2). Whilst there is a vast literature characterizing differences between chondrocytes in healthy and OA cartilage and also differences in an array of cell responses of isolated OA and normal chondrocytes, there is little clear evidence to conclude that chondrocytes from OA cartilage are permanently changed. This lack of clarity arises because a) of the difficulty in designing experiments in intact cartilage to identify permanent cellular changes and distinguish them from temporary changes in expression caused by the altered signals resulting from the pathology and b) with isolated OA chondrocytes, to distinguish the myriad of changes that arise in culture following their isolation, from changes that genuinely reflect a disease state. With the background evidence of phenotypic plasticity, as described above, even the concept of permanent change appears questionable and we certainly need more substantial experimental evidence to show if irreversible DNA changes occur in diseased cartilage. Based on our groups experience with chondrocytes isolated from OA cartilage and their ability to regenerate a cartilage matrix (Katapodi et al. 2009), this suggests that ‘permanent change’ is unlikely in most cells. This would be important for concepts of OA cartilage repair if existing chondrocytes could be guided back to a healthy phenotype and it is our knowledge of their epigenetic status that may enable us to design better ways of achieving this.
Table 2.
Questions on the pathogenesis of osteoarthritis
| 1. The joint is a biomechanical organ. How do genetic, biochemical and biomechanical factors contribute to the varied pathogenic processes that lead to OA? |
| 2. What different pathogenic processes contribute to the heterogeneity of OA and its progression and how do these processes result in different cartilage responses in different patients? |
| 3. What are the signals that drive similar changes in OA chondrocytes in both damaged (loaded) and intact (unloaded) cartilage on an OA joint? |
| 4. Do epigenetic changes occur in OA chondrocytes from loaded and unloaded sites and are they reversible? |
Osteoarthritis: one disease or many
Human OA is frequently discussed as one pathology with its effects on cartilage being driven by one mechanism, but there is an increasing appreciation that it is not just one pathology, but a complex and heterogeneous condition (see Hardingham 2008; Abramson & Attur 2009; Bay-Jensen et al. 2010). We recently published a simple study of gene expression in chondrocytes in normal and OA cartilage and showed changes that were common and changes that were heterogeneous (Brew et al. 2010). In cartilage from 25 late OA patients compared to age matched controls there was consistent down regulation of the transcription factor SOX9, the proteoglycan aggrecan and MMP-13, amongst other genes, but changes in gene expression of the major cartilage collagen (type II) differed amongst the group. Some were in the normal range, but others showed strong increases more that 2 standard deviations above the normal range. There was also increased abnormal expression of collagen type I in a different subset of patients. This clearly showed that gene expression patterns differed amongst OA patients, suggesting there was not just one route to cartilage damage in OA, but many. The pattern of gene expression, particularly in those with low SOX9 and aggrecan expression, but high MMP-13 and also high type II collagen expression, is not explained by the known effects on chondrocytes of anabolic factors, such as TGFβ/IGF-1, or of inflammatory cytokines IL-1/TNFα, or of altered compressive loading. Clearly there are gaps in our understanding of the heterogeneity of OA, its varied growth factor/cytokine/chemokine environment, the altered biomechanical context (Abramson & Attur 2009; Blain 2009; Ramage et al. 2009) and newly discovered cellular processes, such as unfolded protein responses (Boot-Handford & Briggs 2010) and autophagy (Carames et al. 2010), that in combination drive different patterns of chondrocyte response.
An equally interesting result from our study came from comparing within each joint, cartilage gene expression from an intact unloaded site (posterior lateral condyle) and a damaged heavily loaded site (inferior medial condyle) (Brew et al. 2010). This showed that in spite of the difference in the physical damage, intact cartilage and heavily fibrillated cartilage in the same joint shared major changes in expression of prominent cartilage genes. All the cartilage in an OA joint was thus abnormal and the changes in gene expression were not a consequence of physical damage, but possibly predisposed the cartilage to damage. These analyses also revealed no evidence of chondrocyte hypertrophy, which has been reported as a major pathogenic mechanism in human OA and in some animal models of OA. In this cross section of late OA knees, MMP-13 expression was increased, but there was no up-regulation of type X collagen, or of matrilin-1, PTHrP, BCL-2 or VEGF all genes associated with chondrocyte hypertrophy in growth plate. So this analysis suggested that there were OA patient subsets present with high type II collagen expression, or high type I collagen expression, but that further differentiation of chondrocytes to hypertrophy was not a common event detected in most late human OA cartilage.
Cartilage has a very central and fundamental biomechanical function in the joint. However, OA joint failure affects all tissues and the pathology needs to be understood with a holistic approach (Figure 2) (Hardingham 2008; Abramson & Attur 2009; Bay-Jensen et al. 2010). One often ignored feature is that OA-like pathology can be initiated experimentally in many different ways. The fact that you can predispose a knee joint to OA by severing the cruciate ligament, or removing the meniscus, to upset the joint biomechanics, but also by inherited gene mutations in cartilage matrix proteins (e.g. collagen and aggrecan); or experimentally by poisoning the joint with iodoacetate; these together show that many initial events may compromise joint function and leads to OA-like pathology, but clearly each starts by different mechanisms. Thus although the OA pathology is accompanied by cartilage damage, in most instances cartilage changes are not the cause, but the consequence of OA. The pathology also involves changes in most other joint tissues, including subchondral bone, joint capsule, ligaments, muscles and tendons and marginal osteophytes, but cartilage damage is the most common feature and it is the weak ability of cartilage to repair itself that appears to make it most vulnerable to progressive damage and loss. Cartilage damage and loss in OA therefore appears as a common end-point to a heterogeneous range of inherited risk factors and joint insults that compromise the function of our joints and result in degeneration (Figure 2).
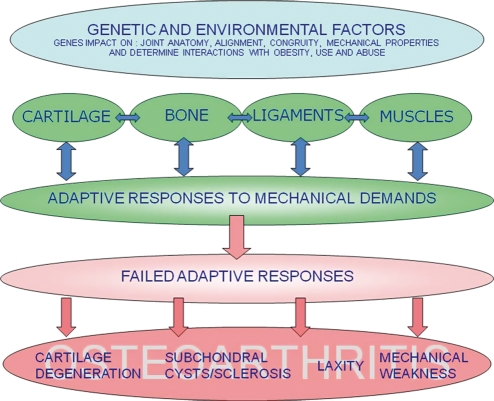
Figure 2.
Schematic of pathogenic interactions in osteoarthritis (from Hardingham 2008). The genetic and environmental background (blue), influences how interactions amongst joint tissues adapt to joint biomechanics and maintain healthy function (green), but can lead to failed responses (pink), which may be progressive and result in pathology in different tissue compartments as present in the heterogeneous range of clinical OA. Cartilage damage is a common end-point as amongst all joint tissues it has the poorest ability to repair damage. The heterogeneity of OA is poorly characterized clinically or biochemically and the relative contribution of different pathogenic processes amongst OA patients is unknown.
There are important consequences if we accept that changes in many joint tissues can cause OA and that the active processes differ in different patients: (1) cartilage is only one of many valid targets for therapeutic intervention; (2) correcting, or repairing cartilage alone is unlikely to cure OA; and (3) there is a need to treat joint failure holistically and to tackle those facets of the pathology most active in each patient. So whilst the ability of chondrocytes from OA cartilage to re-initiate matrix production given appropriate signals is important for strategies to promote cartilage repair and regeneration, this may be futile in an OA joint, unless it is combined with treatments that make the joint environment more normal, including reducing inflammation, reducing joint laxity, improving muscle tone and correcting joint alignment and biomechanics. It remains to be seen, with improved means of early detection (Bauer et al. 2006), if there are patients for whom a treatment can be put in place early enough to extend the useful working life of failing joints.
Conclusions
The major advances in cell and matrix biology of cartilage in recent years have presented new research challenges. For future progress we need a closer marriage between biology, chemistry, physics and biomechanics to unravel how chondrocytes form and maintained cartilage in health and what goes wrong in disease, how cell phenotype and cell behaviour is generated and maintained by epigenetic changes affecting nuclear DNA and how molecular scale detail explains the composite physical and biomechanical properties of cartilage.
Acknowledgments
I extend thanks to the many who I have worked with and who have helped educate me in the ways of cartilage. I particularly acknowledge the help and support of members of UKCTE and colleagues in The Wellcome Trust Centre for Cell-Matrix Biology, University of Manchester supported by The Wellcome Trust. I would also like to thank Dick Heinegard for kind permission to use his illustration for Figure 1.
References
- Abramson SB, Attur M. Developments in the scientific understanding of osteoarthritis. Arthr. Res. Ther. 2009;11:227. doi: 10.1186/ar2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbero A, Ploegert S, Heberer M, Martin I. Plasticity of clonal populations of dedifferentiated adult human articular chondrocytes. Arthr. Rheum. 2003;48:1315–1325. doi: 10.1002/art.10950. [DOI] [PubMed] [Google Scholar]
- Bauer DC, Hunter DJ, Abramson SB, et al. Osteoarthritis biomarkers network: classification of osteoarthritis biomarkers: a proposed approach. Osteoarthr. Cartil. 2006;14:723–727. doi: 10.1016/j.joca.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Bay-Jensen AC, Hoegh-Madsen S, Dam E, et al. Which elements are involved in reversible and irreversible cartilage degradation in osteoarthritis? Rheumatol. Int. 2010;30:435–442. doi: 10.1007/s00296-009-1183-1. [DOI] [PubMed] [Google Scholar]
- Benya P, Shaffer JD. Dedifferentiated chondrocytes re-express the differentiated collagen phenotype when cultured in agarose gels. Cell. 1982;30:215–224. doi: 10.1016/0092-8674(82)90027-7. [DOI] [PubMed] [Google Scholar]
- Blain EJ. Involvement of cytoskeletal elements in articular cartilage homeostasis and pathology. Int. J. Exp. Path. 2009;90:1–15. doi: 10.1111/j.1365-2613.2008.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot-Handford RB, Briggs MD. The unfolded protein response and its relevance to connective tissue diseases. Cell Tissue Res. 2010;339:197–211. doi: 10.1007/s00441-009-0877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brew CJ, Clegg PD, Boot-Handford RP, Andrew JP, Hardingham TE. Gene expression in human chondrocytes in late osteoarthritis is changed in both fibrillated and intact cartilage without evidence of generalised chondrocyte hypertrophy. Annal. Rheum. Dis. 2010;69:234–240. doi: 10.1136/ard.2008.097139. [DOI] [PubMed] [Google Scholar]
- Carames B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthr. Rheum. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JM, Olin AI, Murdoch AD, et al. Alternative splicing of aggrecan G3 domain influences interactions with tenascin-C and other extracellular matrix proteins. J. Biol. Chem. 2004;279:12511–12518. doi: 10.1074/jbc.M400242200. [DOI] [PubMed] [Google Scholar]
- Fosang AJ, Hardingham TE. Isolation of the N-terminal globular domains from cartilage proteoglycan: identification of G2 domain and its lack of interaction with hyaluronate and link protein. Biochem. J. 1989;261:801–809. doi: 10.1042/bj2610801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham TE. The role of link-protein in the structure of cartilage proteoglycans aggregates. Biochem. J. 1979;177:237–247. doi: 10.1042/bj1770237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham T. Extracellular matrix and pathogenic mechanisms in osteoarthritis. Curr. Rheumatol. Rep. 2008;10:30–36. doi: 10.1007/s11926-008-0006-9. [DOI] [PubMed] [Google Scholar]
- Hardingham TE, Muir H. The specific interaction of hyaluronic acid with cartilage proteoglycans. Biochim. Biophys. Acta. 1972;279:401–405. doi: 10.1016/0304-4165(72)90160-2. [DOI] [PubMed] [Google Scholar]
- Heinegard D. Proteoglycan and more – from molecules to biology. Int. J. Exp. Path. 2009;90:575–586. doi: 10.1111/j.1365-2613.2009.00695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadler KA, Baldock C, Bella J, Boot-Handford RP. Collagens at a glance. J. Cell Sci. 2007;120:1955–1958. doi: 10.1242/jcs.03453. [DOI] [PubMed] [Google Scholar]
- Katapodi T, Tew SR, Hardingham TE. Assembly of cartilage matrix by osteoarthritic human articular chondrocytes on Hyalograft matrices varies with donor, but is enhanced in hypoxic conditions. Biomaterials. 2009;30:535–540. doi: 10.1016/j.biomaterials.2008.09.064. [DOI] [PubMed] [Google Scholar]
- Koob TJ, Vogel K. Proteoglycan synthesis in organ-cultures of regions of bovine tendon subjected to different mechanical forces. Biochem. J. 1987;246:589–598. doi: 10.1042/bj2460589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Murdoch AD, Grady LM, Ablett MP, Katopodi T, Meadows RS, Hardingham TE. Chondrogenic differentiation of human bone marrow stem cells in transwell cultures: generation of scaffold-free cartilage. Stem Cells. 2007;25:2786–2796. doi: 10.1634/stemcells.2007-0374. [DOI] [PubMed] [Google Scholar]
- Pelttari K, Winter A, Steck E, et al. Premature induction of hypertrophy during in vitro chondrogenesis of human mesenchymal stem cells correlates with calcification and vascular invasion after ectopic transplantation in SCID mice. Arthr. Rheum. 2006;54:3254–3266. doi: 10.1002/art.22136. [DOI] [PubMed] [Google Scholar]
- Ramage L, Nuki G, Salter DM. Signalling cascades in mechanotransduction: cell-matrix interactions and mechanical loading. Scand. J. Med. Sci. Sports. 2009;19:457–469. doi: 10.1111/j.1600-0838.2009.00912.x. [DOI] [PubMed] [Google Scholar]
- Tew SR, Hardingham TE. Regulation of SOX9 mRNA in human articular chondrocytes involving p38 MAPK activation and mRNA stabilization. J. Biol. Chem. 2006;281:39471–39479. doi: 10.1074/jbc.M604322200. [DOI] [PubMed] [Google Scholar]
- Tew SR, Li Y, Pothacharoen P, Tweats LM, Hawkins RE, Hardingham TE. Retroviral transduction with SOX9 enhances re-expression of the chondrocyte phenotype in passaged osteoarthritic human articular chondrocytes. Osteoarthr. Cartil. 2005;13:80–89. doi: 10.1016/j.joca.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Tew SR, Murdoch AD, Rauchenberg RP, Hardingham TE. Cellular methods in cartilage research: primary human chondrocytes in culture and chondrogenesis in human bone marrow stem cells. Methods. 2008;45:2–9. doi: 10.1016/j.ymeth.2008.01.006. [DOI] [PubMed] [Google Scholar]