Abstract
Non-LTR retrotransposons – including LINE-1 (or L1), Alu and SVA elements – have proliferated during the past 80 million years of primate evolution and now account for approximately one third of the human genome. These transposable elements are now known to affect the human genome in many different ways: generating insertion mutations, genomic instability, alterations in gene expression and also contributing to genetic innovation. As the sequences of human and other primate genomes are analyzed in increasing detail, we are begining to understand the scale and complexity of the past and current contribution of non-LTR retrotransposons to genomic change in the human lineage.
Also known as “jumping genes”, transposable elements (TEs) are discrete pieces of DNA that can move from site to site within (and sometimes between) genomes. Although their discovery dates back to the 1940s1, it took about half a century before we began to understand how TEs interact with their genomic environment. A crucial stage was reached with the completion of the first human genome sequence, which revealed that nearly half of our genome is derived from TEs2,3 (FIG. 1a). Actually, this is likely to be an underestimate, as many ancient TEs inserted in the human genome have probably diverged beyond recognition3. The scale of the contribution of TEs to the human genome is all the more remarkable when considering that protein-coding regions account for just 1.5% of the human genome3.
Figure 1. The transposable element content of the human genome.
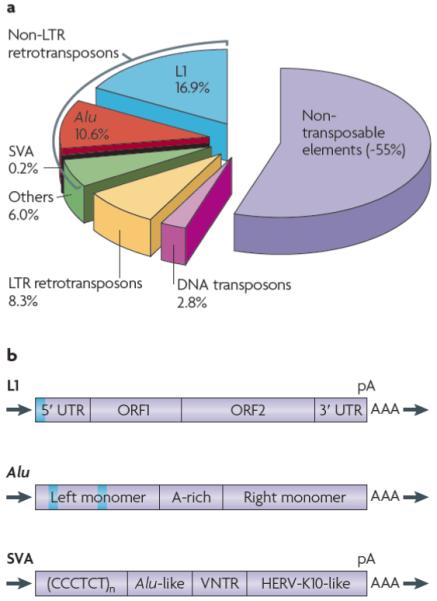
a | The transposable element content of the human genome. About 45% of the human genome can currently be recognized as being derived from transposable elements, the vast majority of which are non-LTR retrotransposons such as L1, Alu and SVA elements. L1, LINE-1; LTR, long terminal repeat. b | The canonical LINE-1 (L1) element consists of two open reading frames (ORF1 and ORF2) flanked by 5′ and 3′ untranslated regions (UTR). The 5′ UTR possesses an internal RNA polymerase II promoter (grey box). The element ends with an oligo dA-rich tail (AAA) preceded by a polyadenylation signal (pA). c | The canonical Alu element consists of two related monomers separated by an A-rich linker region (with consensus sequence A5TACA6). The left monomer contains A and B boxes (grey boxes) which are promoters of transcription by RNA polymerase III. The element ends with an oligo dA-rich tail (AAA) that can be up to one hundred bp in length. d | The canonical SVA element has a composite structure consisting of (from the 5′ end to 3′ end): (i) a (CCCTCT)n hexamer repeat region, (ii) an Alu-like region consisting of two antisense Alu fragments and an additional sequence of unknown origin, (iii) a variable number of tandem repeats region made of 35-50 bp-long units, and (iv) a region derived from the env gene and the 3′ LTR of the endogenous retrovirus HERV-K10. The element ends with an oligo dA-rich tail (AAA) preceded by a polyadenylation signal (pA). L1, Alu and SVA elements are typically flanked by target site duplications (black arrows) generated upon integration. Elements are not drawn to scale.
TEs can be separated into two major classes: DNA transposons and retrotransposons. DNA transposons, which make up ~3% of the human genome (FIG. 1a), are able to excise themselves from the genome, move as DNA and paste themselves into new genomic sites4. Although they are currently not mobilizing in the human genome, they were active during early primate evolution, until ~37 million years (My) ago5. Retrotransposons duplicate via RNA intermediates that are reverse-transcribed and inserted at new genomic locations4. Retrotransposons can be subdivided into two groups, distinguished by the presence or absence of long terminal repeats (LTRs). Human LTR elements are endogenous retroviruses which account for ~8% of the genome (FIG. 1a). Most endogenous retroviruses inserted in the human genome >25 My ago, and their activity is presently very limited in humans, if occurring at all3,6. By contrast, the vast majority of human TEs result from the present and past activity of non-LTR retrotransposons, typified by LINE-1 (or L1), Alu and SVA elements, that collectively account for about one third of the human genome3 (FIG. 1a). L1, Alu and SVA non-LTR retrotransposons are the only TEs unequivocally shown to be currently active in humans, as demonstrated by more than 60 reported cases of de novo insertions responsible for genetic disorders7-11.
The extremely high density of TEs in our genome poses the question: what is their evolutionary significance and impact during human evolution? The development of innovative molecular methodologies such as retrotransposition assays in cultured cells12,13 and computational techniques for comparative genomics, in conjunction with the availability of multiple primate genome sequences (such as the human3, chimpanzee14 and macaque15 genomes), have resulted in a progressive shift of the focus of TE research to how diverse and profound the impact of TE activity on genomic evolution is. The past years have witnessed a number of important discoveries regarding ways in which TEs affect human genome evolution, so it is now possible to literally quantify the overall impact TE activity has had on shaping our genome. For example, it has long been recognized that recombination between TEs can trigger genomic deletions in humans, as these deletions have caused several genetic disorders8. However, only recently have genome-wide comparisons of human and other primate genomes permitted us to determine the magnitude and significance of TE recombination-mediated deletions at an evolutionary scale16-18.
In this review, we focus on the evolutionary impact of non-LTR retrotransposons, which are by far the most abundant TEs in the human genome and the most active TEs during recent human evolution. First, we briefly describe the structure of non-LTR retrotransposons and mechanisms by which they move. Then, we explore the evolutionary dynamics of non-LTR retrotransposons, that is, what has made them so evolutionary successful in the human genome. Addressing this question helps us to understand the ways in which and to what extent TEs in general - and non-LTR retrotransposons in particular - have impacted human genome evolution. This impact turns out to be tremendously diverse and considerable, ranging from local instability (for example, through insertion mutagenesis and seeding of microsatellites) to large-scale structural variation (for example, through ectopic recombination and transduction of flanking sequences) to contributions to genetic innovation (for example, through new gene formation and exonization) and alterations in gene expression (for example, through alternative splicing and epigenetic regulation). Finally, we conclude with potential future research directions.
Human non-LTR retrotransposons
L1 elements
There are >500,000 L1 copies in the human genome, resulting from their continued mobilization activity for the past 150 My3. L1 elements constitute ~17% of the human genome, which makes them the most successful TEs in the human genome by mass (FIG. 1a). The canonical, full-length L1 element is ~6 kilobases (kb) in length and it consists of a 5′ untranslated region (UTR) containing an internal RNA polymerase II promoter19, two open reading frames (ORF1 and ORF2) and a 3′ UTR containing a polyadenylation signal ending with an oligo dA-rich tail of variable length20 (FIG. 1b). ORF1 encodes an RNA-binding protein and ORF2 encodes a protein with endonuclease and reverse-transcriptase activities20. This molecular machinery allows the retrotransposition process known as target-primed reverse transcription (TPRT) to occur (BOX 1), thus making L1 elements the only autonomous TEs in the human genome. However, not all L1 copies are competent for retrotransposition. Indeed, as a result of the TPRT process and decay over time, most L1 copies are inactivated by truncations, internal rearrangements and mutations3,21. Out of the >500,000 L1 elements inserted in the human genome, less than 100 copies are intact22.
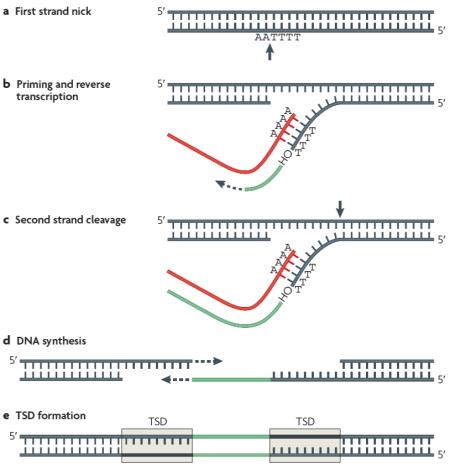
Box 1 | The retrotransposition cycle.
The increase in copy numbers of non-LTR retrotransposons occurs via an RNA-based duplication process termed retrotransposition. The first step in L1 retrotransposition involves RNA polymerase II-mediated transcription of a genomic L1 locus from an internal promoter that directs transcription initiation at the 5′ boundary of the L1 element19,129. Carrying an internal promoter makes sense for a retrotransposon if it is to generate autonomous duplicate copies at multiple locations in the genome. The L1 RNA is exported to the cytoplasm where ORF1 (encoding an RNA-binding protein) and ORF2 (encoding a protein with endonuclease and reverse-transcriptase activities) are translated. Both proteins exhibit strong cis-preference27; consequently, they preferentially associate with the L1 RNA transcript that encoded them, to produce a ribonucleoprotein (RNP) particle. The RNP is then transported back into the nucleus by a mechanism that is poorly understood.
The integration of the L1 element into the genome likely occurs via a process termed target-primed reverse transcription13,130,131 (TPRT), which was originally described for the R2 non-LTR retrotransposon of the silkworm Bombyx mori132. During TPRT, it is thought that the L1 endonuclease cleaves the first strand of target DNA, generally at 5′-TTTT/AA-3′ consensus sites133 (a). The free 3′ hydroxyl generated by the nick is then used to prime reverse transcription of L1 RNA (red) by the L1 reverse transcriptase (b). The second strand of the target DNA is cleaved (c) and used to prime second-strand synthesis (d), through poorly understood mechanisms. Hallmarks of the integration process include frequent 5′ truncations, presence of an oligo dA-rich tail at the 3′end, and 2-20 bp-long duplications of the target site3,21 (TSD) (e).
Alu and SVA retrotransposition also likely occurs via TPRT, through the hijacking of the L1 retrotransposition machinery12,29,30. The mechanism of Alu and SVA trans-mobilization by L1 proteins remains elusive. RNA polymerase III-mediated Alu transcripts are exported to the cytoplasm and bound to SRP9/14 proteins to form stable RNPs134,135. It has been hypothesized that Alu RNPs interact with ribosomes, thereby positioning Alu transcripts in close vicinity of nascent L1 ORF2 proteins12,42 (ORF1 protein enhances, but is not strictly required for, Alu retrotransposition12,136). However, it remains unclear whether Alu RNPs gain access to the L1 retrotransposition machinery in the cytoplasm or in the nucleus, as Alu RNPs might recruit L1 ORF2 proteins in the nucleus and immediately proceed with TPRT137.
Alu elements
There are >1 million Alu copies in the human genome3, resulting from their continued mobilization activity throughout the past ~65 My23. This makes Alu elements the most successful TEs in the human genome in terms of copy number. The typical full-length Alu element is ~300 base pairs (bp) in length and it exhibits a dimeric structure formed by fusion of two monomers derived from the 7SL RNA gene24 (a component of the signal recognition particle), which are separated by an A-rich linker region (FIG. 1c). The 5′ region contains an internal RNA polymerase III promoter (A and B boxes) and the element ends with an oligo dA-rich tail of variable length23. As Alu elements do not possess RNA polymerase III termination signals, Alu transcripts extend into the downstream flanking sequence until a terminator (typically a run of four or more consecutive Ts) is found25,26. Alu elements have no coding capacity and are, therefore, non-autonomous TEs. Instead, they borrow the retrotransposition molecular machinery encoded by L1 elements12 despite the fact that L1 ORF1 and ORF2 proteins exhibit strong cis-preference for L1 RNA27 (BOX 1), which is the reason why Alu elements are sometimes referred to as “a parasite’s parasite”28.
SVA elements
There are ~3,000 SVA copies in the human genome, resulting from continued activity throughout the ~25 My of hominoid evolution29,30. The typical full-length SVA element is ~2 kb in length and it is composed of an hexamer repeat region, an Alu-like region, a variable number of tandem repeats region, a HERV-K10-like region and a polyadenylation signal ending with an oligo dA-rich tail of variable length29,30 (FIG. 1d). Several lines of evidence suggest that SVA elements are transcribed by RNA polymerase II29,30. However, SVA elements apparently contain no internal promoter and they might rely, at least partly, on promoter activity in flanking regions29,30. Similar to Alu elements, SVA elements are non-autonomous TEs presumably trans-mobilized by the L1 retrotransposition machinery29,30 (BOX 1).
Other non-LTR retrotransposons
In addition to the L1, Alu and SVA elements, which are currently active, there are additional families of old, inactive non-LTR retrotransposons that comprise a total of ~6% of the human genome (FIG. 1a). Although far less numerous than L1 and Alu elements, these old elements represent a rich molecular fossil record testifying to the long-term relationship between TEs and the human genome3. This record indicates for example that before the autonomous L1 element and its Alu parasite expansions, the genome experienced retrotransposition of the autonomous L2 element and its MIR parasite3. These old elements may have substantially impacted human genome evolution31-34.
Evolutionary dynamics
The impact of non-LTR retrotransposons on human genome evolution largely results from their extremely high copy numbers (for example, there is one Alu insertion every ~3 kb on average3) and continued activity over tens of My. These two features are particularly striking when considering the various cellular processes that control retrotransposon activity (BOX 2). At an evolutionary scale, the vertical persistence of non-LTR retrotransposons, not only in primates but in mammals in general, sets them apart from most other TEs in mammals and other eukaryotes3,5,35. In this section, we discuss the evolutionary dynamics that have made non-LTR retrotransposons so prolific during primate genome evolution.
Box 2 | Cellular factors influencing retrotransposition.
TEs can be seen as selfish genetic entities whose spread can be deleterious to the host cell due to the genomic instability that is induced by a massive increase in copy number. As a result of the conflicting interests of TEs and the host genome, the cell has developed various processes to control retrotransposon activity, as predicted by the Red Queen hypothesis138. Below we provide examples of how L1 and Alu retrotransposition activity is regulated in host cells (for more detailed discussion, see refs. 11,139).
Regulation of L1 retrotransposition can occur at the transcription level. For example, novel regulatory regions have been frequently recruited during L1 evolution38; the current L1 5′ UTR contains several transcription factor-binding sites important for transcription activation or initiation140-142. In addition, DNA methylation at the promoter is known to repress L1 expression124,143. L1 elements are also subject to post-transcriptional regulation. For example, RNA-induced silencing through RNA interference has been suggested to reduce L1 retrotransposition in cultured cells144,145. The A-rich coding strand of the full-length human L1 contains 19 potential canonical and noncanonical polyadenylation signals that lead to truncation of full-length L1 transcripts by premature polyadenylation, thus ultimately contributing to the attenuation of L1 activity110. Furthermore, cells produce proteins such as those of the APOBEC3 family that can inhibit L1 and Alu retrotransposition146.
Alu activity is influenced by its primary sequence in that the accumulation of mutations across time may alter important motifs such as the internal RNA polymerase III promoter or SRP9/14 binding motifs26,42. The accumulation of mutations is facilitated by the high density of CpG dinucleotides that are prone to mutation as a result of the deamination of 5-methylcytosine residues125. Overall, it has been estimated that when an Alu copy reaches ~10% divergence from its subfamily consensus sequence, the likelihood that it continues to be active is remote42. The length and homogeneity of the oligo dA-rich tail also appear to be important for activity 147,148. The genomic environment in which Alu copies insert is crucial for retrotranspositional activity149-151 and the distance between the oligo dA-rich tail at the 3′ end of the Alu sequence and the RNA polymerase III terminator located in the downstream sequence, which determines the overall length of Alu transcripts26, is also important.
Subfamily structure and source elements
A key concept relevant to the evolutionary dynamics of L1, Alu and SVA sequences is that they can all be divided into subfamilies or “clades” of related elements based on diagnostic nucleotide substitutions and insertions/deletions exclusively shared by all subfamily members. For example, more than 200 Alu subfamilies are currently recognized in the human genome36, but only six subfamilies of the younger SVA family exist30. Not only are subfamilies different in age, but the diagnostic sequence mutations or changes that define subfamilies tend to accumulate hierarchically23,37. In other words, instead of two subfamilies being independently derived from an ancestral subfamily, most subfamilies represent an ongoing linear sequential evolution pattern where a series of subfamilies have each been successively derived one from the other. For example, it has been shown that during the past ~40 My, all L1 subfamilies in the human genome are derived from a single lineage from which they arose sequentially38. Similar patterns of subfamily evolution have been reported for Alu23 and SVA30 elements. These observations can be explained if one assumes that only a few elements (so-called ‘source’ or ‘master’ elements) are involved in the retrotransposition process and are responsible for the formation of all other subfamily members37.
The “master gene” model of retrotransposon amplification37 has been refined, in particular by quantifying the number of retrotransposition-competent elements within the human genome. Analysis of the >200 Alu subfamilies in the human genome suggested the existence of at least 143 Alu source elements36 and it has been estimated that an average human genome carries 80-100 retrotransposition-competent L1 copies, six of which (hot L1s) are probably responsible for the bulk of L1 retrotransposition22,39,40. These results further indicate that several source elements may exist within a subfamily because all six hot L1 elements belong to the L1-Ta subfamily22. A network-based analysis also revealed that human-specific Alu subfamilies typically contain ~15% of secondary source elements that contributed ~30% of subfamily members, in addition to a main master element41. Thus, there may be hundreds of active Alu ‘core’ sequences in the human genome42. Although they only represent a tiny fraction of all human non-LTR retrotransposons, source elements can be considered as the ultimate drivers of evolutionary change in the human genome because they are responsible for most L1, Alu and SVA elements inserted in our genome.
Stealth drivers and long-term evolution
Another distinguishing feature of human retrotransposons is their persistent activity over tens of My of evolution. How have active retrotransposons been maintained over this time? Reconstruction of the evolutionary history of the Alu Yb lineage showed that it originated during early hominoid evolution, 18-25 My ago43. Strikingly, the Alu Yb lineage has dramatically expanded to ~2,000 copies within the past few My specifically in the human genome, as non-human hominoid primates carry only a handful of Alu Yb elements43-45. Therefore, the Alu Yb lineage remained in the genome with no or little retrotransposition for 15-20 My, while preserving the ability to generate a high number of new copies in a species-specific manner. These results suggest that long-lived, low-activity source elements may act as “stealth drivers” that occasionally produce elements, some of which may become highly active. While highly active “master” elements may be deleterious and negatively selected, low-activity stealth drivers may allow the Alu lineage to persist on the long term43. Attenuation of mobilization activity may be a common evolutionary strategy of various retrotransposons46,47. Therefore, the ability to maintain low to moderate levels of retrotransposition activity may be an important feature that allowed human retrotransposons to maintain long-term activity.
Impact on genome evolution
Amplification rates
Because of their continued activity and accumulation in the genome over tens of My, L1, Alu and SVA elements have had a tremendous impact on the evolution of primate genomes, both in terms of structure and function. To assess the impact of these elements on genome evolution we can first consider how frequently retrotransposition occurs in the germline. The current rate of Alu retrotransposition has been estimated as one insertion every ~20 births in humans, based on both the frequency of disease-causing de novo insertions compared to nucleotide substitutions48 and evolutionary comparisons of the human and chimpanzee genomes48 and of multiple human genome sequences49. The current rate of L1 retrotransposition has also been estimated as one insertion every ~20 births in humans based on disease-causing de novo insertions50 but as one insertion every ~200 births based on genome comparisons49. The difference between the two estimates might lie in the underlying assumptions of the methods, but no such bias is observed for Alu elements using the same approaches. Alternatively, the difference may reflect recent variation in the L1 retrotransposition rate or intense negative selection against L1 insertions. The current SVA retrotransposition rate has tentatively been estimated as one insertion every ~900 births based on genome comparisons49. However, there is more uncertainty around this rate due to the smaller datasets available for analysis. Although new heritable retrotransposition events take place in the germline, retrotransposition also occurs in somatic tissues with an impact ranging from cancer to a possible role in brain development8,51,52. Retrotransposon-induced somatic variation is a fascinating area of investigation that is likely to provide new insight into TE biology and their impact on human beings.
Amplification rates have not been uniform over time. For example, the vast majority of Alu elements were inserted by ~40 My ago, following a peak of amplification during which there was approximately one new Alu insertion in every birth53. Similarly, during the past ~70 My of evolution, variation in the L1 amplification rate has been observed, with the most prolific L1 subfamilies having amplified 12-40 My ago38. Genome-wide comparisons of the human and chimpanzee genomes provide additional evidence for recent variation in L1, Alu and SVA retrotransposition rates, as judged by the different numbers of species-specific elements that have inserted since the divergence of the two species ~6 My ago14,54,55. Such fluctuation in amplification rates on a short time-scale suggests influences at the host population level40,54.
Changes in copy number
Perhaps one of the most intuitive consequences of TE accumulation is their contribution to genome size increase56: L1 and Alu elements alone have contributed ~750 million bases (Mb) to the human genome sequence3 (FIG. 1). This increase in genome size is an ongoing process, as the human genome has accumulated ~2,000 L1, ~7,000 Alu and ~1,000 SVA copies within the past ~6 My of human evolution, a combined addition of >8 Mb14. Equally importantly, the ongoing expansion of retrotransposons has also created significant inter-individual variation in retrotransposon content; several hundred new mobile element insertions have been detected between multiple human genomic sequences49,57-59. These human-specific retrotransposons insertions are often polymorphic (present or absent) at orthologous loci among human individuals and they constitute highly informative genetic markers that are being used to investigate human evolutionary history, population structure and demography (BOX 3).
Box 3 | Retrotransposon insertion polymorphisms as genetic markers.
As revealed by pioneering studies on humans152-154, primates155 and non-primate groups156,157, retrotransposons afford several advantages that make them very poweful tools as genetic markers for studying human and non-human primate evolutionary history23,157,158. They are essentially homoplasy-free characters in that individuals which share retrotransposon copies at orthologous sites have inherited them from a common ancestor, that is precise excision of retrotransposons is extremely rare158,159. Their ancestral state is known as the absence of the element at any locus of interest, which makes it possible to include hypothetical ancestors for rooting of phylogenetic trees153. As there are only two possible character states for each locus - presence or absence of the element - genotyping of individuals for retrotransposon insertions is technically straight-forward. Furthermore, the vast majority of retrotransposon insertions are neutral residents of the genome160, and the gradual accumulation of elements over time makes it possible to determine loci most suitable to a range of time points in primate history. As a result, retrotransposon insertion polymorphisms (most notably Alu elements) have been used to decipher the phylogenetic relationships of various primate groups161,162, including the resolution of the human-chimpanzee-gorilla trichotomy demonstrating the close relationship between humans and chimpanzees163. Some retrotransposons have inserted so recently during human evolution that they are polymorphic for presence or absence among human populations and individuals23,49,164. In particular, Alu elements have proved highly informative for the study of human origins, by providing strong evidence for an African origin of anatomically modern humans153,154. More recently Alu element insertion polymorphisms have been used to investigate human population structure and demography154,165,166. Retrotransposon insertion polymorphisms are also being used as forensic tools, for example for species-specific DNA detection and quantitation, analysis of complex biomaterials, human gender determination and the inference of geographic origin of human samples167.
Local genomic instability
There are many ways through which retrotransposons can generate genomic instability. In this section, we consider the effects of retrotransposons at a local genomic scale, linked to insertions and their protein products, as well as consequences affecting retrotransposon sequences at a deeper time scale.
Insertion mutagenesis
The most straightforward way in which a retrotransposon can impact upon genome function, and thereby potentially influence genome evolution, is by inserting into protein-coding or regulatory regions (FIG. 2a). Due to the immediate phenotypic impact of such insertions, they were the first to be detected7. Examples of human genetic disorders caused by de novo L1, Alu and SVA insertions continue to accumulate, with 65 cases reported to date shown to cause various heritable diseases such as haemophilia, cystic fibrosis, Apert syndrome, neurofibromatosis, Duchenne muscular dystrophy, β-thalassemia, hypercholesterolemia and breast and colon cancers8,9,11. Overall, it has been estimated that ~0.3% of all human mutations are attributable to de novo L1, Alu and SVA insertions10. Interestingly, L1 (and to a lesser extent Alu and SVA) disease-causing insertions appear to be enriched on the X chromosome8,9,11. This observation may partly be attributable to ascertainment bias, as X-linked genetic disorders are often dominant in males and thus more easily detected. Alternatively, L1 elements may preferentially insert in the X chromosome, perhaps in relation to their possible involvement in X inactivation60,61.
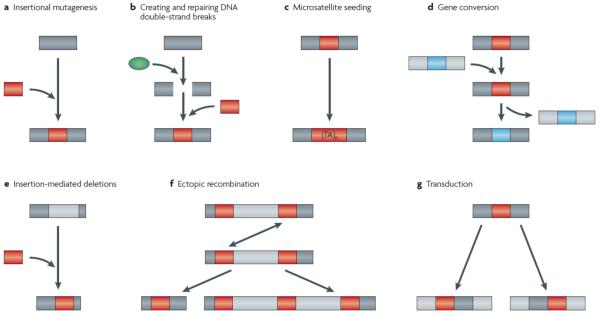
Figure 2. Impact of retrotransposons on human genome structure.
a | Typical insertion of an L1, Alu or SVA retrotransposon (red box) at a new genomic site (blue area). If the new genomic site is a genic region, the retrotransposon may cause insertional mutagenesis. b | The protein products of an L1 element (red circle) may create DNA double-strand breaks (broken blue area). Alternatively, an existing double-strand break may be repaired via non-classical endonuclease-independent insertion of a retrotransposon (red box). c | Microsatellites (e.g. (TA)n) may arise from the homopolymeric tracts endogenous to retrotransposons. d | Gene conversion may alter the sequence compositions of homologous retrotransposon copies (red and green boxes). e | Insertion of retrotransposons (red box) is sometimes associated with concomittant deletion of target genomic sequences (light blue box). f | Ectopic recombination (double arrowheaded line) between non-allelic homologous retrotransposons (red boxes) may result in genomic reaarangements such as deletions (left) or duplications (right) of intervening genomic sequences. g | 3′ and 5′ transduction can result in the co-retrotransposition of downstream or upstream flanking genomic sequence, respectively, along with a retrotransposon (left and right, respectively).
Creating and repairing DNA double-strand breaks
It has recently been shown that the ORF2 protein encoded by L1 elements, which has endonuclease activity, generates many more DNA double-strand breaks (DSBs) than the number associated with actual L1 insertions in mammalian cell lines62 (FIG. 2b). The extent to which this mechanism contributes to human genomic instability remains unknown, since the levels of L1 expression under these experimental conditions were much higher than those expected under normal cellular conditions. However, the repair of L1-mediated DSB lesions would leave no particular signature of L1 involvement. Thus, it is possible that a substantial fraction of the genomic instability associated with DSBs, which are highly mutagenic and recombinogenic, is ultimately attributable to L1 activity.
L1 and Alu elements have also been linked to DSB repair. Evidence from L1 retrotransposition assays in cultured cells demonstrated that L1 insertions can occur independently of endonuclease in cell lines that lack the ability to perform non-homologous end joining, a major mechanism of DSB repair63 (FIG. 2b). Endonuclease-independent (ENi) L1 insertions lack the hallmarks of TPRT (BOX 1), thereby suggesting that L1 elements can integrate into and repair DSB DNA lesions63. In addition, dysfunctional telomeres can serve as substrates for ENi L1 retrotransposition and endonuclease-deficient LINE-like (Penelope) elements are present at the telomeres of several eukaryotes, suggesting that ENi retrotransposition may be an ancestral mechanism of RNA-mediated DNA repair associated with non-LTR retrotransposons used before the acquisition of an endonuclease domain64,65. Recent analyses of the human genome have shown that 0.5-0.7% of all L1 and Alu insertions have non-canonical structures and may have resulted from ENi retrotransposition66,67, suggesting that non-LTR retrotransposons in general, not just L1 elements, may serve as a ‘fail-safe’ mechanism in maintaining human genome integrity.
Sources of microsatellites
Because of their abundance in the genome and because they contain homopolymeric tracts, non-LTR retrotransposons have the ability to generate microsatellites at many loci in the genome (FIG. 2c). This has been studied in particular for Alu elements68,69, each new copy of which provides two potential sources of microsatellites: the linker region in the middle of the element and the 3′ oligo dA-rich tail (FIG. 1c). These homopolymeric repeats can experience various mutational forces such as nucleotide substitutions and replication slippage, which may ultimately result in microsatellites of varying length and complexity. Consequently, it is not surprising that ~20% of all microsatellites (including ~50% of mononucleotide microsatellites) shared by the human and chimpanzee genomes lie within Alu elements70. In addition, there are at least two examples of expansions of microsatellites that arose from A-rich regions of Alu elements causing genetic disorders71,72.
Gene conversion
Several studies indicate that Alu elements undergo gene conversion73,74 (FIG. 2d), a type of recombination defined as the non-reciprocal transfer of information between homologous sequences. Gene conversion may play a role in the evolution of Alu elements by inactivating active copies or resurrecting inactivated copies23. For example, it has recently been shown that the master element of the Alu Yh3a3 subfamily has been inactivated by gene conversion in humans, thus preventing further amplification of this subfamily75. In addition, because Alu elements make up >10% of the human genome, Alu-mediated gene conversion might have a substantial impact on the overall nucleotide diversity of our genome. Also it might impair the use of SNPs located within Alu sequences as genetic markers, since gene conversion would make these SNPs identical by state rather than identical by descent23. However, the significance of this phenomenon has not formally been tested and the development of second-generation sequencing approaches and personal genomics opens new avenues to resolving this question.
Genomic rearrangements
In addition to local genomic instability, retrotransposons can also generate genomic rearrangements such as deletions, duplications and inversions. In this section, we discuss three ways in which retrotransposons can create structural variation in the genome.
Insertion-mediated deletions
The insertion of L1 and Alu elements at new genomic sites sometimes results in the concomitant deletion of adjacent genomic sequence (FIG. 2e). This phenomenon was first observed during the analysis of L1 integrations within cultured human cells, where ~20% of L1 insertions were associated with structural rearrangements, including concomitant deletions at the insertion site, ranging in size from 1 bp to possibly >130 kb76-78. These deletions apparently can arise by endonuclease-dependent and ENi mechanisms78. L1 and Alu insertion-mediated deletions have subsequently been shown to occur naturally in the human and chimpanzee genomes, although they are usually shorter (<800 bp on average) and they occur at a much lower frequency than in cultured cells (~2% and ~0.3% of L1 and Alu insertion events, respectively)79,80. This may reflect, at least partly, negative selection against large, disruptive insertion-mediated deletions. Consistent with these observations, a 46 kb-long L1 insertion-mediated deletion event in the PDHX gene has recently been implicated in pyruvate dehydrogenase complex deficiency81 and human-chimpanzee genome comparisons identified a single insertion-mediated deletion event that caused functional gene loss within the past ~6 My79.
It has also been noted that ~90% of non-classical, ENi L1 and Alu insertions are associated with deletions of flanking sequence ranging in size from 1 bp to 14 kb, including one deletion that removed an olfactory receptor gene from the human and chimpanzee genomes66,67. Altogether, it has been estimated that during primate evolution, as many as ~45,000 insertion-mediated deletions may have removed >30 Mb of genomic sequences18.
Ectopic recombination
Due to their extremely high copy numbers, L1 and Alu elements can also create structural genomic variation at the post-insertion stage, through recombination between non-allelic homologous elements (FIG. 2f), including between elements that have been inserted in the genome for a long time. Ectopic recombination can result in various types of genomic rearrangements such as deletions, duplications and inversions.
It has long been recognized that Alu recombination-mediated deletions (RMD) occur in the human genome, as shown by the >70 reported cases of Alu RMDs being responsible for various forms of cancer and genetic disorders8,10. By contrast, only three disease-causing L1 RMD events have been reported17. Genome-wide comparisons identified 492 Alu compared with 73 L1 RMD events in the human genome since the human-chimpanzee divergence16,17. L1 RMDs are larger on average than Alu RMDs and they occur more frequently in gene-poor regions of the genome than Alu RMDs. These results are suggestive of negative selection against long, deleterious L1 RMDs in gene-rich regions of the genome18,82,83. Thus, Alu and L1 RMD events detectable by comparative genomics approaches largely represent the fraction of all RMDs that have escaped negative selection. Yet, based on human and chimpanzee genome comparisons, these events have collectively removed nearly 1 Mb of genomic sequence from the human genome within the past few My16-18, thereby underscoring their important evolutionary impact on the human genome.
The human genome contains many large (>10 kb in length) and highly similar (>90% sequence identity) duplicated genomic regions, termed segmental duplications. Interestingly, the boundaries of human segmental duplications are significantly enriched in Alu elements, that is, they comprise ~24% of boundary sequences but only ~11% elsewhere in the human genome84. Considering that ~5% of the human genome has been duplicated within the past ~40 My, recombination between Alu elements may represent an important mechanism for the origin and expansion of segmental duplications in our genome84.
The contribution of L1 and Alu elements to chromosomal inversions has also been recently investigated by comparative genomics. Nearly half of the inversions that took place in the human and chimpanzee genomes since their divergence involve L1 and Alu elements, and ~20% of all inversions can clearly be identified as products of L1-L1 or Alu-Alu recombination events85. Although this type of rearrangement does not result in gain or loss of genomic sequence, it contributes to genomic variation sometimes with functional significance, since several events are involved in the inversion of exons85.
Transduction of flanking sequences
In addition to duplicating themselves, L1 and SVA elements can sometimes carry upstream or downstream flanking genomic sequences with them (termed 5′ and 3′ transduction, respectively) (FIG. 2g). In 3′ transduction, the RNA transcription machinery skips the weak retrotransposon polyadenylation signal and terminates transcription by using an alternative polyadenylation signal located downstream in the 3′ flanking sequence. Similarly, 5′ transduction occurs when a promoter located upstream of the retrotransposon is used to transcribe the sequence down to the retrotransposon86,87. The transcript containing the retrotransposon along with the extra genomic sequence is subsequently integrated back into the genome via retrotransposition. Initially characterized using cell culture-based methods88, 3′ transduction has subsequently been shown to occur frequently in the human genome: ~10% of both L1 and SVA insertions are associated with 3′ transduction events30,89-91.
Genetic innovation
Variation in the number of genes among species indicates that new genes are continuously generated over evolutionary time. Comparative genomic studies have confirmed the notion of “evolutionary tinkering”92 according to which new genes most commonly arise by rearrangements between pre-existing genetic structures. In this section, we explore mechanisms by which retrotransposons have fostered genetic innovations in the human lineage.
Transduction-mediated gene formation
Retrotransposon-mediated transduction (discussed above) can lead to the duplication of coding sequences fortuitously located in the transduced flanking genomic sequence. The potential of L1 retrotransposons to mediate exon shuffling via 3′ transduction has been experimentally confirmed using cell culture assays88. This mechanism has subsequently been shown to have mediated the formation of a new gene family during recent human evolution, via multiple SVA-mediated transduction events of the AMAC1 gene89 (BOX 4).
Box 4 | Birth of a gene family by retrotransposon-mediated transduction.
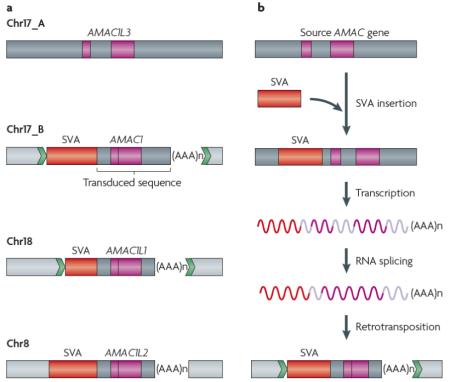
It has been experimentally demonstrated using cell culture assays that L1 retrotransposons can mediate exon shuffling via 3′ transduction88. Subsequent analyses of the human genome have confirmed that L1-mediated transduction indeed took place during human genome evolution and that it may account for 0.6-1% of human DNA3,90,91. However, whether it contributes to evolving new gene function remained an open question. A recent analysis of SVA retrotransposons has demonstrated the evolutionary significance of retrotransposon-mediated 3′ transduction, by showing that SVA-mediated transduction is responsible for the creation of the AMAC1 gene family that comprises four copies in the human genome89.
As part of a genome-wide analysis of SVA-mediated transduction, Xing et al. 89 identified 143 events that transduced sequences ranging in size from a few dozen bp to almost 2 kb. Interestingly, three transduced sequences located on chromosomes 8, 17 and 18 were found to originate from the same source locus located elsewhere on chromosome 17 (a). In the figure, flanking sequences of the original locus are shown as blue boxes and the flanking sequences of the transduced loci are shown as light blue boxes. Target site duplications are shown as green arrows. SVA elements are depicted as red bars and the transduced sequences are shown as blue bars with coding regions shown as purple bars. SVA element oligo dA-rich tails are shown as “(AAA)n”. Analysis of the four paralogous sequences identified four copies of the AMAC1 gene. The ancestral AMAC1L3 gene copy at the source locus consisted of two exons separated by an intron. By contrast, the three transduced copies were intronless versions of AMAC1L3, as a result of the splicing of the intron during the retrotransposition process (b). Evolutionary analyses demonstrated that the three transduction events all took place ~7-14 My ago, as human and African great apes share all four AMAC1 copies, whereas orangutan and other primate and non-primate species that have been analyzed only possess the ancestral AMAC1L3 gene. Experimental studies indicated that, in addition to AMAC1L3, at least two of the three transduced AMAC1 genes are expressed in human tissues. RNA transcript sequence analyses of the expressed AMAC1 duplicates further revealed that the promoter sequence had been duplicated along with the AMAC1 coding sequence as part of the 3′ transduction process. This indicates that retrotransposon-mediated gene transduction can duplicate not only coding regions of genes but also their regulatory regions, thus retaining functional potential after duplication. Hence, this retrotransposon-mediated duplication mechanism can lead to rapid generation of functional gene families.
Gene retrotransposition
In contrast to transduction, gene retrotransposition only duplicates gene sequences and no retrotransposon sequence is co-duplicated in the process. This is because gene retrotransposition is based on the hijacking of the L1 retrotransposition machinery by host mRNA transcripts93, similar to Alu and SVA retrotransposition. As a result, gene retrotransposition generally does not duplicate upstream regulatory regions, thus requiring duplicated genes to fortuitously acquire new regulatory regions to be functional. Therefore, gene retrotransposition was long thought to generate non-functional duplicate gene copies termed retropseudogenes. However, genome-wide searches have confirmed the importance of gene retrotransposition in the emergence of new primate genes94-96 and it has been estimated that at least one new retrogene per My emerged in the human lineage during the past ~65 My97 (for more detailed discussion, see ref. 96).
Exonization
Alternative splicing is a widespread mechanism that occurs in 40-60% of human genes3,98. By producing more than one type of mRNA from a single gene, alternative splicing significantly contributes to human proteome variation98. Interestingly, retrotransposon sequences are sometimes recruited as exons that become integrated to genes, a process termed exonization (FIG. 3a). It was initially estimated based on transcript sequence data that ~4% of human protein-coding sequences contained TEs (mostly Alu and L1)99. However, a recent analysis at the protein level more conservatively suggested that this proportion is closer to ~0.1%100. Exonization is thought to be facilitated by the fact that many TEs carry cryptic donor and acceptor splice sites. For example, a typical Alu sequence contains nine GT dinucleotides and 14 AG dinucleotides that represent as many cryptic donor and acceptor splice sites, respectively101,102. Consistently, Alu exonization has occurred repeatedly during primate evolution103. It has been estimated that ~5% of alternatively spliced exons are derived from Alu elements in humans and that most – if not all – Alu exons are alternatively spliced presumably because constitutively expressed Alu exons are deleterious and negatively selected101. Consistently, the three reported cases of exonized Alu elements becoming constitutively expressed are all associated with genetic disorders98.
Figure 3. Impact of retrotransposons on human gene expression.
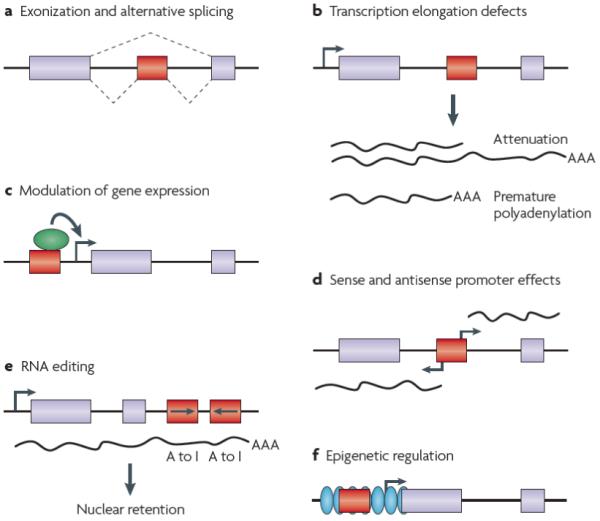
a | Retrotransposon sequence (red box) can be recruited as coding sequence and integrated into a gene (made up here of two exons, blue boxes). This is often associated with alternative splicing (dashed lines). b | Presence of a retrotransposon (red box) in the intron of a gene (sequence between the two blue boxes representing exons) can result in transcription elongation defects such as attenuation or premature termination. c | Retrotransposons (red box) carry transcription factor (green ovals) binding sites that can up- or down-regulate (green arrow) the expression of neighboring genes (horizontal arrow and blue boxes). d | Retrotransposons (red box) carry sense and anti-sense promoters (horizontal arrows) that can initiate downstream and upstream transcription. e | Presence of two Alu elements in opposite orientation (red boxes) in gene transcripts can lead to A-to-I editing, which can result in suppression of expression through nuclear retention of edited RNA transcripts. f | Retrotransposon sequences (red boxes) can be methylated, which may initiate and spread heterochromatin formation (green ovals), thereby altering expression of neighboring genes (horizontal arrow and blue boxes).
Non-LTR retrotransposons have also been involved in facilitating the molecular domestication of other TEs. This is exemplified by SETMAR, a chimeric primate gene resulting from fusion of a SET histone methyltransferase gene to the transposase gene of an Hsmar1 DNA transposon104. The birth of SETMAR might have never occurred without the contribution of an Alu element that inserted in and partially deleted the 5′-terminal inverted repeat of the Hsmar1 element104. Because both terminal inverted repeats of DNA transposons are necessary for transposition, the Alu insertion may have contributed to the recruitment of the Hsmar1 transposon as part of SETMAR by immobilizing it at a period when the Hsmar1 family was experiencing high levels of transposition in primate genomes5. Overall, it is striking that non-LTR retrotransposons seem to directly contribute a disproportionately small number of domesticated genes compared to other TEs (such as DNA transposons), despite the fact that they are the most numerous TEs in the human genome105,106.
Impact on gene expression
As described above, retrotransposons have dramatically impacted human evolution at the DNA level. Evidence is also accumulating that retrotransposons significantly shape human evolution at the RNA level through various mechanisms, which we discuss in this section.
Modulation of gene expression
Retrotransposons impact the expression of nearby genes through a variety of mechanisms. Similar to Alu elements, L1 sequences can provide new splice sites that may promote exonization and alternative splicing107,108 (FIG. 3a). In addition, intronic L1 elements can interfere with transcriptional elongation of the host gene due to reduced ability of RNA polymerase II to read through L1 sequences109 (FIG. 3b). Furthermore, retrotransposon sequences can provide polyadenylation signals inducing termination of gene transcripts110-112 (FIG. 3b). It has also been shown that Alu elements carry transcription factor-binding sites that may serve to modulate gene expression113,114 (FIG. 3c). The functional promoter sequences of L1 and Alu elements can also initiate sense or anti-sense transcription through other genes115-117 (FIG. 3d).
The potential of L1 endogenous promoter and polyadenylation signals to create transcriptome diversity in humans is illustrated by 15 human genes that were apparently split by L1 elements inserted in antisense orientation in intronic sequences118. In each of these genes, a transcript containing exons upstream of the insertion site terminates at the L1 3′ antisense polyadenylation signal; a second transcript derived from the L1 5′ antisense promoter drives expression of a transcript that includes the downstream exons of the gene. These observations provide a mechanistic basis for the emergence of new gene structures by gene fission.
RNA editing
RNA editing is a process by which RNA nucleotide sequences are co- or post-transcriptionally modified, as exemplified by the conversion of adenosine to inosine (A-to-I) in double-stranded RNA (FIG. 3e). A-to-I editing is widespread in humans and >90% of all A-to-I substitutions occur within Alu sequences embedded in mRNA transcripts119-122. Editing within Alu elements might be favored by the dimeric structure of these elements and the occasional occurrence of pairs of Alu elements in head-to-tail orientation. A-to-I editing can eliminate splice sites and therefore might affect alternative splicing of exonized Alu sequences. Furthermore, it has recently been shown that A-to-I editing of pairs of inverted Alu elements in 3′ UTRs can suppress expression through nuclear retention of mRNA transcripts123.
Epigenetic regulation
The epigenetic regulation of retrotransposon activity through DNA methylation represents an important defence mechanism for the cell (BOX 2): the L1 promoter CpG island is typically highly methylated124, and Alu and SVA elements are enriched in CpG sites30,125 to the extent that one third of all human CpG sites are contained within Alu sequences126. Because L1, Alu and SVA elements are frequently found in or near genes, retrotransposon-mediated heterochromatin formation and spread could repress transcription of nearby genes (FIG. 3f). Consistently, Alu elements may be excluded from imprinted regions of the human genome due to their potential negative impact on methylation associated with imprinting127. Similarly, it has been proposed that the high density of L1 elements on the X chromosome may be explained by their involvement in X inactivation60,61. Under this hypothesis, L1 elements would serve as booster stations to propagate the signal that silences one of the two female X chromosomes. However, formal demonstration of retrotransposon-mediated epigenetic control of neighboring genes in humans and evaluation of the extent of this phenomenon at a genome-wide scale represent active topics of investigation in the field.
Conserved non-coding elements
Recent genome comparisons have revealed the occurrence of numerous conserved non-coding elements (CNEs) in the human genome. Strikingly, many CNEs appear to be derived from ancient TE sequences, in particular a class of non-LTR retrotransposons known as short interspersed elements (SINEs, to which Alu elements belong)31-33. These ancient SINE-derived sequences are currently evolving under strong negative selection and have apparently taken on regulatory functions31-33. It remains unclear whether the frequent recruitment of SINEs as CNEs reflects an endogenous functional property of these elements, is a by-product of their high copy numbers in mammalian genomes or results from their distinctive sequence architecture which makes them more readily identifiable as old retrotransposons106. In any event, the genome-wide contribution of this phenomenon to human evolution remains to be determined, but is likely to be important.
Conclusions and future directions
For tens or even hundreds of My, TEs (including non-LTR retrotransposons) have shaped the evolution of the genomes in which they reside128. Maintaining activity over extended periods of time is a distinguishing feature of non-LTR retrotransposons that was instrumental to their evolutionary success in the human lineage. Our understanding of the factors underlying this evolutionary success is still incomplete and the next few years will probably shed new light on this intriguing question. This intricate relationship does not mean that non-LTR retrotransposons have been maintained in the human genome on such a timescale because of various evolutionary advantages they have conferred to their host genome. On the contrary, we believe that the profound impact of retrotransposons on genome evolution is a by-product of, not the reason for, the evolutionary success of these selfish genetic elements.
This view is supported by the notion that retrotransposons often represent a threat to human health. While their involvement in causing genetic diseases through insertion mutagenesis as a result of their sustained mobilization activity has long been established, other mechanisms are less well understood. For example, investigating the contribution of L1 endonuclease to the generation of DSBs in germline and somatic tissues might have important implications for understanding the L1 integration process and interactions with DNA repair mechanisms, as well as for chromosomal damage and human health more generally. Although the contribution of retrotransposons to genomic deletions, such as insertion-mediated deletions and RMDs is well established, other types of genomic rearrangements such as retrotransposon recombination-mediated tandem duplications are less well understood, partly because they are more difficult to characterize through computational comparisons of genome sequences. Given that duplications are a key contributor to genetic innovation, the extent to which retrotransposons have contributed to the formation of new genes in the human genome might still be vastly underestimated. This is also true for many aspects of retrotransposon impact on gene expression. For example, there is growing evidence that TEs, not just non-LTR retrotransposons, have been a rich source of material for the assembly and evolution of regulatory networks106. By providing a wealth of genomic and transcriptomic sequence data, next generation sequencing and personal genomics will shed new light on the surprisingly dynamic nature of TEs and the roles that they play in shaping within- and inter-individual variation. This will allow researchers to dissect retrotransposon-induced variation at ever-increasing resolution. Such information is crucial if we are to better understand the overall TE impact on human health, genome evolution and the unique traits that make us human.
Online summary.
 LINE-1 (or L1), Alu and SVA elements belong to the non-LTR retrotransposon class of transposable elements and they account for about one third of the human genome.
LINE-1 (or L1), Alu and SVA elements belong to the non-LTR retrotransposon class of transposable elements and they account for about one third of the human genome. L1, Alu and SVA elements are the only transposable elements unequivocally shown to be currently active in humans, as demonstrated by de novo insertions responsible for genetic disorders.
L1, Alu and SVA elements are the only transposable elements unequivocally shown to be currently active in humans, as demonstrated by de novo insertions responsible for genetic disorders. The expansion of L1, Alu and SVA elements is characterized by the dispersal, in a series of subfamilies, of elements of different evolutionary age that share common nucleotide substitutions and following a “master gene” model of amplification.
The expansion of L1, Alu and SVA elements is characterized by the dispersal, in a series of subfamilies, of elements of different evolutionary age that share common nucleotide substitutions and following a “master gene” model of amplification. The evolutionary impact of L1, Alu and SVA elements on the human genome is tremendously diverse and considerable.
The evolutionary impact of L1, Alu and SVA elements on the human genome is tremendously diverse and considerable. L1, Alu and SVA elements generate instability at a local genomic scale, linked to insertions (insertion mutagenesis and DNA repair) and their protein products (generation of DNA double-strand breaks), as well as consequences affecting their sequences at a deeper time scale (seeding of microsatellites and gene conversion).
L1, Alu and SVA elements generate instability at a local genomic scale, linked to insertions (insertion mutagenesis and DNA repair) and their protein products (generation of DNA double-strand breaks), as well as consequences affecting their sequences at a deeper time scale (seeding of microsatellites and gene conversion). L1, Alu and SVA elements also generate genomic rearrangements such as deletions, duplications and inversions and thus create structural variation in the genome through insertion-mediated deletions, ectopic recombination and transduction of flanking sequence.
L1, Alu and SVA elements also generate genomic rearrangements such as deletions, duplications and inversions and thus create structural variation in the genome through insertion-mediated deletions, ectopic recombination and transduction of flanking sequence. L1, Alu and SVA elements have fostered genetic innovation during human and primate evolution through transduction-mediated gene formation, gene retrotransposition and exonization.
L1, Alu and SVA elements have fostered genetic innovation during human and primate evolution through transduction-mediated gene formation, gene retrotransposition and exonization. L1, Alu and SVA elements also significantly shape human evolution at the RNA level by modulating expression of nearby genes, RNA editing and epigenetic regulation.
L1, Alu and SVA elements also significantly shape human evolution at the RNA level by modulating expression of nearby genes, RNA editing and epigenetic regulation.
Highlighted references.
Ref. #3: A landmark paper analyzing the entire human genome sequence and revealing that transposable elements make up nearly half of our genome.
Ref. #12: The authors establish an experimental test of Alu retrotransposition in cultured cells and demonstrate that L1 ORF2 protein is required for Alu retrotransposition.
Ref. #13: A landmark paper presenting the development and characterization of an in vitro assay to measure L1 retrotransposition in cultured cells.
Refs. #16 & 17: References 16 and 17 report genome-wide analyses demonstrating that L1 and Alu recombination-mediated deletions have greatly impacted human genome evolution.
Ref. #22: This study indicates that most of L1 retrotransposition in humans may result from the activity of just six highly active L1 elements.
Ref. #41: The authors refine the master gene model of Alu amplification by showing that human-specific Alu subfamilies typically contain 10-20% retrotransposition-competent copies.
Ref. #49: The first genome-wide comparison of inter-individual structural variation attributable to transposable elements in humans.
Refs. #76 & 77: References 76 and 77 demonstrate that L1 retrotransposition can be associated with various forms of genomic instability in cultured cells.
Refs. # 88 & 89: Reference 88 shows that retrotransposon-mediated transduction can create new genes in cultured cells and reference 89 demonstrates the evolutionary significance of this phenomenon during human evolution.
Ref. #110: This study demonstrates that the L1 element contains many polyadenylation signals, resulting in truncated transcripts and attenuated L1 activity.
Ref. #118: This paper shows how anti-sense promoter and polyadenylation signals of L1 elements can lead to the formation of new genes by fission of pre-existing genes.
Refs. #153 & 154: References 153 and 154 elegantly show how Alu insertion polymorphisms can be used human evolutionary history and demography.
Acknowledgements
We apologize to colleagues whose work could not be discussed or cited due to space constraints. Our research on various aspects of mobile elements is supported by a Young Investigator ATIP Award from the Centre National de la Recherche Scientifique (CNRS) to R.C., as well as Louisiana Board of Regents Governor’s Biotechnology Initiative GBI (2002-005), National Science Foundation grant BCS-0218338, National Institutes of Health PO1 AG022064, and National Institutes of Health RO1 GM59290 all to M.A.B.
Glossary
- Long terminal repeats (LTR)
Sequences of 300-1,000 base pairs that are directly repeated at the 5′ and 3′ ends of LTR retrotransposons and retroviruses.
- Hominoids
The group of primates comprising humans and apes, which diverged from Old World monkeys about 25 million years ago.
- Trans-mobilization
The process by which non-autonomous retrotransposons such as Alu and SVA elements borrow the L1 retrotransposition machinery to perform their own retrotransposition.
- Red Queen hypothesis
Proposed by Van Valen in 1973, this hypothesis states that, for an evolutionary system, continuing development is needed just in order to maintain its fitness relative to the systems it is co-evolving with.
- Homopolymeric tract
A DNA sequence made of the same nucleotide repeated in tandem.
- Microsatellite
A class of repetitive DNA that is made up of tandem repeats that are 1-8 base pairs in length.
- Identical by state
Alleles that have the same character state, but have not been inherited from a common ancestor, as a result of independent evolutionary changes.
- Identical by descent
Alleles that have the same character state because it was directly inherited from a common ancestor.
- Retrogene
Expressed and functional gene generated by retrotransposition, usually with an intact open reading frame consistent with that of the parental gene.
- Molecular domestication
The recruitment of a transposable element coding sequence into a new functional role by the genome.
- Imprinting
An epigenetic phenomenon by which certain genes are expressed in a parent-of-origin-specific manner.
- X inactivation
The process by which one of the two copies of the X chromosome present in female mammals is inactivated during early embryogenesis. The inactive X chromosome is silenced by packaging into transcriptionally inactive heterochromatin.
- Homoplasy
Similarity due to independent evolutionary change, that is, not inherited from a common ancestor.
Biographies
Richard Cordaux received his Ph.D. from the laboratory of Mark Stoneking at the Max Planck Institute for Evolutionary Anthropology in Leipzig (Germany). He carried out postdoctoral studies with Mark Batzer at Louisiana State University. He became a tenured investigator at the Centre National de la Recherche Scientifique (CNRS) and the University of Poitiers (France) in 2006. He was awarded the CNRS bronze medal in 2009. His group focuses on mobile elements, comparative genomics, population genetics in humans and bacterial endosymbionts.
Mark Batzer received his Ph.D. from the laboratory of William R. Lee at Louisiana State University. He carried out postdoctoral studies with Prescott Deininger at the LSU Health Sciences Center, and then with Pieter de Jong in the Human Genome Center at Lawrence Livermore National Laboratory. He became a faculty member at the LSU Health Sciences Center in 1995 and moved to a position as a professor of Biological Sciences at LSU in 2001. He is presently an LSU System Boyd Professor and Dr. Mary Lou Applewhite Distinguished Professor at LSU. His laboratory focuses on mobile elements, comparative genomics, population genetics, and human molecular genetics.
Footnotes
Online links
FURTHER INFORMATION
Cordaux Lab homepage: http://site.voila.fr/rcordaux
Batzer Lab homepage: http://batzerlab.lsu.edu/
dbRIP, a database of retrotransposon insertion polymorphisms: http://dbrip.brocku.ca/
CSHL Dolan DNA learning center human diversity module: http://www.geneticorigins.org/pv92/aluframeset.htm
Repbase, a database of eukaryotic transposable elements: http://www.girinst.org/repbase/index.html
Access to this interactive links box is free online.
Contributor Information
Richard Cordaux, Université de Poitiers, CNRS UMR 6556 Ecologie, Evolution, Symbiose, 40 Avenue du Recteur Pineau, 86022 Poitiers, France.
Mark A. Batzer, Department of Biological Sciences, Louisiana State University, 202 Life Sciences Building, Baton Rouge, LA 70803, USA
References
- 1.McClintock B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 2.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–63. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. American Society for Microbiology Press; Washington: 2002. [Google Scholar]
- 5.Pace JK, 2nd, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–32. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–91. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Kazazian HH, Jr., et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–6. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 8.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–93. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 9.Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet. 2005;117:411–27. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 10.Callinan PA, Batzer MA. Retrotransposable elements and human disease. Genome Dyn. 2006;1:104–115. doi: 10.1159/000092503. [DOI] [PubMed] [Google Scholar]
- 11.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–58. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 12.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–8. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 13.Moran JV, et al. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–27. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 14.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 15.Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–34. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 16.Sen SK, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han K, et al. L1 recombination-associated deletions generate human genomic variation. Proc Natl Acad Sci U S A. 2008;105:19366–71. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordaux R. The human genome in the LINE of fire. Proc Natl Acad Sci U S A. 2008;105:19033–4. doi: 10.1073/pnas.0810202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–29. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babushok DV, Kazazian HH., Jr. Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28:527–39. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 21.Szak ST, et al. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0052. research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brouha B, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–9. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 24.Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23:158–61. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Shaikh TH, Roy AM, Kim J, Batzer MA, Deininger PL. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J Mol Biol. 1997;271:222–34. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- 26.Comeaux MS, Roy-Engel AM, Hedges DJ, Deininger PL. Diverse cis factors controlling Alu retrotransposition: What causes Alu elements to die? Genome Res. 2009;19:545–55. doi: 10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei W, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–39. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiner AM. SINEs and LINEs: the art of biting the hand that feeds you. Curr Opin Cell Biol. 2002;14:343–50. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- 29.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–51. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, et al. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 31.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 32.Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–74. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamal M, Xie X, Lander ES. A large family of ancient repeat elements in the human genome is under strong selection. Proc Natl Acad Sci U S A. 2006;103:2740–5. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krull M, Petrusma M, Makalowski W, Brosius J, Schmitz J. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs) Genome Res. 2007;17:1139–45. doi: 10.1101/gr.6320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA. Loss of LINE-1 activity in the megabats. Genetics. 2008;178:393–404. doi: 10.1534/genetics.107.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price AL, Eskin E, Pevzner PA. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 2004;14:2245–52. doi: 10.1101/gr.2693004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deininger PL, Batzer MA, Hutchison CA, 3rd, Edgell MH. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–11. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- 38.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lutz SM, Vincent BJ, Kazazian HH, Jr., Batzer MA, Moran JV. Allelic heterogeneity in LINE-1 retrotransposition activity. Am J Hum Genet. 2003;73:1431–7. doi: 10.1086/379744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seleme Mdel C, et al. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc Natl Acad Sci U S A. 2006;103:6611–6. doi: 10.1073/pnas.0601324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordaux R, Hedges DJ, Batzer MA. Retrotransposition of Alu elements: how many sources? Trends Genet. 2004;20:464–7. doi: 10.1016/j.tig.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Bennett EA, et al. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–83. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han K, et al. Under the genomic radar: The Stealth model of Alu amplification. Genome Res. 2005;15:655–64. doi: 10.1101/gr.3492605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carter AB, et al. Genome-wide analysis of the human Alu Yb-lineage. Hum Genomics. 2004;1:167–78. doi: 10.1186/1479-7364-1-3-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibbons R, et al. Distinguishing humans from great apes with AluYb8 repeats. J Mol Biol. 2004;339:721–9. doi: 10.1016/j.jmb.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 46.Li TH, Schmid CW. Alu’s dimeric consensus sequence destabilizes its transcripts. Gene. 2004;324:191–200. doi: 10.1016/j.gene.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 47.Han JS, Boeke JD. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–8. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 48.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–7. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 49.Xing J, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009 doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kazazian HH., Jr. An estimated frequency of endogenous insertional mutations in humans. Nat Genet. 1999;22:130. doi: 10.1038/9638. [DOI] [PubMed] [Google Scholar]
- 51.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 52.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–12. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol Evol. 1991;33:311–20. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- 54.Hedges DJ, et al. Differential alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–75. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mills RE, et al. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 2006;78:671–9. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu G, et al. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 2003;13:358–68. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Badge RM, Alisch RS, Moran JV. ATLAS: a system to selectively identify human-specific L1 insertions. Am J Hum Genet. 2003;72:823–38. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–51. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boissinot S, Entezam A, Young L, Munson PJ, Furano AV. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 2004;14:1221–31. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–7. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 61.Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci U S A. 2000;97:6634–9. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–93. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morrish TA, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–65. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 64.Morrish TA, et al. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–12. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 65.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci U S A. 2007;104:9352–7. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sen SK, Huang CT, Han K, Batzer MA. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 2007;35:3741–51. doi: 10.1093/nar/gkm317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srikanta D, et al. An alternative pathway for Alu retrotransposition suggests a role in DNA double-strand break repair. Genomics. 2008 doi: 10.1016/j.ygeno.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arcot SS, Wang Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: a source for the genesis of primate microsatellites. Genomics. 1995;29:136–44. doi: 10.1006/geno.1995.1224. [DOI] [PubMed] [Google Scholar]
- 69.Jurka J, Pethiyagoda C. Simple repetitive DNA sequences from primates: compilation and analysis. J Mol Evol. 1995;40:120–6. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- 70.Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18:30–8. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Justice CM, et al. Phylogenetic analysis of the Friedreich ataxia GAA trinucleotide repeat. J Mol Evol. 2001;52:232–8. doi: 10.1007/s002390010151. [DOI] [PubMed] [Google Scholar]
- 72.Kurosaki T, Ninokata A, Wang L, Ueda S. Evolutionary scenario for acquisition of CAG repeats in human SCA1 gene. Gene. 2006;373:23–7. doi: 10.1016/j.gene.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 73.Kass DH, Batzer MA, Deininger PL. Gene conversion as a secondary mechanism of short interspersed element (SINE) evolution. Mol Cell Biol. 1995;15:19–25. doi: 10.1128/mcb.15.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roy AM, et al. Potential gene conversion and source genes for recently integrated Alu elements. Genome Res. 2000;10:1485–95. doi: 10.1101/gr.152300. [DOI] [PubMed] [Google Scholar]
- 75.Styles P, Brookfield JF. Source gene composition and gene conversion of the AluYh and AluYi lineages of retrotransposons. BMC Evol Biol. 2009;9:102. doi: 10.1186/1471-2148-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–25. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 77.Symer DE, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–38. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 78.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–95. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Callinan PA, et al. Alu Retrotransposition-mediated Deletion. J Mol Biol. 2005;348:791–800. doi: 10.1016/j.jmb.2005.02.043. [DOI] [PubMed] [Google Scholar]
- 80.Han K, et al. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 2005;33:4040–52. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mine M, et al. A large genomic deletion in the PDHX gene caused by the retrotranspositional insertion of a full-length LINE-1 element. Hum Mutat. 2007;28:137–42. doi: 10.1002/humu.20449. [DOI] [PubMed] [Google Scholar]
- 82.Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci U S A. 2006;103:9590–4. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song M, Boissinot S. Selection against LINE-1 retrotransposons results principally from their ability to mediate ectopic recombination. Gene. 2007;390:206–13. doi: 10.1016/j.gene.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 84.Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003;73:823–34. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee J, Han K, Meyer TJ, Kim HS, Batzer MA. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS ONE. 2008;3:e4047. doi: 10.1371/journal.pone.0004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hancks D, Ewing A, Chen JE, Tokunaga K, Kazazian H. Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009 doi: 10.1101/gr.093153.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raiz J, et al. 5′-transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009 doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moran JV, DeBerardinis RJ, Kazazian HH., Jr. Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–4. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 89.Xing J, et al. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci U S A. 2006;103:17608–13. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goodier JL, Ostertag EM, Kazazian HH., Jr. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–7. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 91.Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–5. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jacob F. Evolution and tinkering. Science. 1977;196:1161–6. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 93.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–7. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 94.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–73. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 95.Babushok DV, et al. A novel testis ubiquitin-binding protein gene arose by exon shuffling in hominoids. Genome Res. 2007;17:1129–38. doi: 10.1101/gr.6252107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of Young Human Genes after a Burst of Retroposition in Primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kreahling J, Graveley BR. The origins and implications of Aluternative splicing. Trends Genet. 2004;20:1–4. doi: 10.1016/j.tig.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 99.Nekrutenko A, Li WH. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–21. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- 100.Gotea V, Makalowski W. Do transposable elements really contribute to proteomes? Trends Genet. 2006;22:260–7. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 101.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–7. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lev-Maor G, Sorek R, Shomron N, Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–91. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 103.Krull M, Brosius J, Schmitz J. Alu-SINE exonization: en route to protein-coding function. Mol Biol Evol. 2005;22:1702–11. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 104.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci U S A. 2006;103:8101–6. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–22. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 106.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–21. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–74. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 110.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–6. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 111.Lee JY, Ji Z, Tian B. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res. 2008;36:5581–90. doi: 10.1093/nar/gkn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen C, Ara T, Gautheret D. Using Alu elements as polyadenylation sites: A case of retroposon exaptation. Mol Biol Evol. 2009;26:327–34. doi: 10.1093/molbev/msn249. [DOI] [PubMed] [Google Scholar]
- 113.Shankar R, Grover D, Brahmachari SK, Mukerji M. Evolution and distribution of RNA polymerase II regulatory sites from RNA polymerase III dependant mobile Alu elements. BMC Evol Biol. 2004;4:37. doi: 10.1186/1471-2148-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–85. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 117.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 118.Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–8. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–25. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–5. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 122.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–3. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 123.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. Embo J. 2008;27:1694–705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–34. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 125.Xing J, et al. Alu element mutation spectra: molecular clocks and the effect of DNA methylation. J Mol Biol. 2004;344:675–82. doi: 10.1016/j.jmb.2004.09.058. [DOI] [PubMed] [Google Scholar]
- 126.Rubin CM, VandeVoort CA, Teplitz RL, Schmid CW. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic Acids Res. 1994;22:5121–7. doi: 10.1093/nar/22.23.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Greally JM. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc Natl Acad Sci U S A. 2002;99:327–32. doi: 10.1073/pnas.012539199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet. 2007;8:241–59. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- 129.Lavie L, Maldener E, Brouha B, Meese EU, Mayer J. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. 2004;14:2253–60. doi: 10.1101/gr.2745804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Feng Q, Moran JV, Kazazian HH, Jr., Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–16. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 131.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. Embo J. 2002;21:5899–910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 133.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A. 1997;94:1872–7. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sinnett D, Richer C, Deragon JM, Labuda D. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J Biol Chem. 1991;266:8675–8. [PubMed] [Google Scholar]
- 135.Chang DY, Hsu K, Maraia RJ. Monomeric scAlu and nascent dimeric Alu RNAs induced by adenovirus are assembled into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res. 1996;24:4165–70. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wallace N, Wagstaff BJ, Deininger PL, Roy-Engel AM. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kroutter EN, Belancio VP, Wagstaff BJ, Roy-Engel AM. The RNA polymerase dictates ORF1 requirement and timing of LINE and SINE retrotransposition. PLoS Genet. 2009;5:e1000458. doi: 10.1371/journal.pgen.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Van Valen L. A new evolutionary law. Evolutionary Theory. 1973;1:1–30. [Google Scholar]
- 139.Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 140.Tchenio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–5. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–40. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–55. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–9. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 144.Soifer HS, Zaragoza A, Peyvan M, Behlke MA, Rossi JJ. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 2005;33:846–56. doi: 10.1093/nar/gki223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang N, Kazazian HH., Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 146.Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35:637–42. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- 147.Roy-Engel AM, et al. Active Alu element “A-tails”: size does matter. Genome Res. 2002;12:1333–44. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dewannieux M, Heidmann T. Role of poly(A) tail length in Alu retrotransposition. Genomics. 2005;86:378–81. doi: 10.1016/j.ygeno.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 149.Chesnokov I, Schmid CW. Flanking sequences of an Alu source stimulate transcription in vitro by interacting with sequence-specific transcription factors. J Mol Evol. 1996;42:30–6. doi: 10.1007/BF00163208. [DOI] [PubMed] [Google Scholar]
- 150.Aleman C, Roy-Engel AM, Shaikh TH, Deininger PL. Cis-acting influences on Alu RNA levels. Nucleic Acids Res. 2000;28:4755–61. doi: 10.1093/nar/28.23.4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roy AM, et al. Upstream flanking sequences and transcription of SINEs. J Mol Biol. 2000;302:17–25. doi: 10.1006/jmbi.2000.4027. [DOI] [PubMed] [Google Scholar]
- 152.Perna NT, Batzer MA, Deininger PL, Stoneking M. Alu insertion polymorphism: a new type of marker for human population studies. Hum Biol. 1992;64:641–8. [PubMed] [Google Scholar]
- 153.Batzer MA, et al. African origin of human-specific polymorphic Alu insertions. Proc Natl Acad Sci U S A. 1994;91:12288–92. doi: 10.1073/pnas.91.25.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Stoneking M, et al. Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa. Genome Res. 1997;7:1061–71. doi: 10.1101/gr.7.11.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ryan SC, Dugaiczyk A. Newly arisen DNA repeats in primate phylogeny. Proc Natl Acad Sci U S A. 1989;86:9360–4. doi: 10.1073/pnas.86.23.9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shimamura M, et al. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature. 1997;388:666–70. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- 157.Shedlock AM, Okada N. SINE insertions: powerful tools for molecular systematics. Bioessays. 2000;22:148–60. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 158.Ray DA, Xing J, Salem AH, Batzer MA. SINEs of a nearly perfect character. Syst Biol. 2006;55:928–35. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- 159.van de Lagemaat LN, Gagnier L, Medstrand P, Mager DL. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res. 2005;15:1243–9. doi: 10.1101/gr.3910705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cordaux R, Lee J, Dinoso L, Batzer MA. Recently integrated Alu retrotransposons are essentially neutral residents of the human genome. Gene. 2006;373:138–44. doi: 10.1016/j.gene.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 161.Roos C, Schmitz J, Zischler H. Primate jumping genes elucidate strepsirrhine phylogeny. Proc Natl Acad Sci U S A. 2004;101:10650–4. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ray DA, et al. Alu insertion loci and platyrrhine primate phylogeny. Mol Phylogenet Evol. 2005;35:117–26. doi: 10.1016/j.ympev.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 163.Salem AH, et al. Alu elements and hominid phylogenetics. Proc Natl Acad Sci U S A. 2003;100:12787–91. doi: 10.1073/pnas.2133766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cordaux R, Srikanta D, Lee J, Stoneking M, Batzer MA. In search of polymorphic Alu insertions with restricted geographic distributions. Genomics. 2007;90:154–8. doi: 10.1016/j.ygeno.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Watkins WS, et al. Genetic variation among world populations: inferences from 100 Alu insertion polymorphisms. Genome Res. 2003;13:1607–18. doi: 10.1101/gr.894603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bamshad MJ, et al. Human population genetic structure and inference of group membership. Am J Hum Genet. 2003;72:578–89. doi: 10.1086/368061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ray DA, Walker JA, Batzer MA. Mobile element-based forensic genomics. Mutat Res. 2007;616:24–33. doi: 10.1016/j.mrfmmm.2006.11.019. [DOI] [PubMed] [Google Scholar]