Abstract
This communication describes the consensus multi-locus typing scheme established by the Cryptococcal Working Group I (Genotyping of Cryptococcus neoformans and C. gattii) of the International Society for Human and Animal Mycology (ISHAM) using seven unlinked genetic loci for global strain genotyping. These genetic loci include the housekeeping genes CAP59, GPD1, LAC1, PLB1, SOD1, URA5 and the IGS1 region. Allele and sequence type information are accessible at http://www.mlst.net/.
Keywords: Cryptococcus neoformans, Cryptococcus gattii, Genotyping, Multi Locus Sequence Typing
Introduction
Cryptococcus neoformans, the agent of cryptococcosis, had been considered a homogeneous species until 1949 when the existence of four serotypes was revealed based on the antigenic properties of its polysaccharide capsule [1]. Such heterogeneity of the species, however, remained obscure until the two morphologically distinct teleomorphs of C. neoformans were discovered during the mid 1970s [2,3]. The teleomorph Filobasidiella neoformans was found to be produced by strains of serotype A and D [2] while F. bacillispora was found to be produced by strains of serotype B and C [3]. Ensuing studies revealed numerous differences between the anamorphs of the two Filobasidiella species with regards to their ecology, epidemiology, pathobiology, biochemistry and genetics.
Presently, the etiologic agent of cryptococcosis is classified into two species [4], C. neoformans, with two varieties: C. neoformans var. grubii (serotype A) [5] and C. neoformans var. neoformans (serotype D) [6], as well as an AD hybrid, and C. gattii (serotypes B and C) [7]. Intra-species genetic diversity has also been revealed as more genotyping methods have been applied for each serotype. In addition inter-species hybrid strains of AB and BD serotypes have been described [8,9]. As a result, the number of scientifically valid species within C. neoformans has become a controversial issue because of the differing opinions among taxonomists as to the appropriate definition of a species. There are several research groups focusing on the molecular determination of the number of genetically diverse sub-groups within each serotype. The molecular methods employed by each group to define these sub-groups vary from DNA fingerprinting [10,11] and PCR fingerprinting based on microsatellite- (M13) or minisatellite-specific primers (e.g., (GACA)4 or (GTG)5) [12–16], over random amplification of polymorphic DNA (RAPD) analysis [17–20], amplified fragment length polymorphism (AFLP) analysis [21–23], restriction fragment length polymorphism (RFLP) analysis of the URA5 [16,24] and PLB1 genes [25], the use of IGS sequences [26], multigene sequence analysis [27, Meyer et al. unpublished data], to multi-locus sequence typing (MLST) [23,28] and multi-locus microsatellite typing (MLMT) [29,30]. This research has revealed associations between geographic origin and particular genotypes, implying an epidemiologic significance of certain genotypes. Different methods have resulted in various numbers of sub-groups or different nomenclature of those sub-groups. However, due to the lack of a cross-reference consensus between the results obtained by different genotyping method, there is currently no concordance on a universally acceptable genotyping method for this important human pathogen.
Recognizing the urgent need for a standardized globally acceptable typing method, a Cryptococcus working group I, ‘Genotyping of Cryptococcus neoformans and C. gattii’, was formed under the umbrella of the International Society of Human and Animal Mycoses (ISHAM) in the beginning of 2007 which united all the major research groups that were involved in molecular strain typing of C. neoformans complex. The members of this ISHAM working group met at the 3rd Trends in Medical Mycology (TIMM3) Meeting in Torino, Italy in October 2007, and reviewed all the typing techniques in use. The group selected multi-locus sequence typing (MLST) as the method of choice for future strain typing in light of its high discriminatory power as well as reproducibility between different laboratories. The working group also chose standard reference strains representing the eight known major molecular types of the agent of cryptococcosis as well as the nomenclature of each genotype.
Consensus genotype nomenclature
As a result of the Torino meeting, the working group recognized that the different genotyping methods used by the different research groups lead to corresponding major genotypes for the agents of cryptococcosis (Table 1). Principally, the two main typing systems being used are: PCR fingerprinting using primers specific for microsatellite (M13) [14,16] or minisatellite (GACA)4 DNA [13,15] and AFLP analysis [21]. In both typing schemes, over 2000 isolates were grouped into eight major molecular types. With some exceptions [26,31], the molecular types of C. neoformans are correlated with the serotypes: C. neoformans var. grubii, serotype A, consists of molecular types VNI=AFLP1 and VNII=AFLP1A; the hybrid serotype AD comprises VNIII=AFLP3; and C. neoformans var. neoformans, serotype D, corresponds to VNIV=AFLP2. C. gattii consists of VGI=AFLP4, VGII = AFLP6, VGIII=AFLP5, and VGIV=AFLP7, which all correspond to both serotypes B or C [16,21, unpublished data]. Based on these findings, it was agreed by all cryptococcal working group members present in Torino to use the VNI–VNIV and VGI–VGIV nomenclature [16] since it correlated with the current concept of two species and represents the global population structure based on more than 2000 C. neoformans and C. gattii isolates among which C. neoformans var. grubii (serotype A=VNI) being the most prevalent molecular type world-wide.
Table 1.
Concordance of different molecular typing methods used for Cryptococcus neoformans and Cryptococcus gattii
| Species/Variety/ Hybrid |
Serotype | PCR- fingerprinting molecular type Meyer et al. [14,16,39] |
PCR- fingerprinting molecular type Viviani et al. [13] |
AFLP genotype Boekhout et al. [21] |
AFLP genotype Litvintseva et al. [23] |
URA5 RFLP type Meyer et al. [16] |
PLB1 RFLP type Latouche et al. [25] |
IGS genotype Diaz et al. [26,31] |
ITS genotype Katsu et al. [40] |
|---|---|---|---|---|---|---|---|---|---|
|
C. neoformans var. grubii |
A | VNI | VN6 (VN5) | AFLP1 | VNI | VNI | A1 | 1A/1B | ITS1 |
| A | VNII | AFLP1A/AFLP1B | VNB | VNII | 1A | ITS1 | |||
| A | VNII | VN7 | AFLP1A/AFLP1B | VNII | VNII | A2 | 1C | ITS1 | |
| AD Hybrid | AD | VNIII | VN3/VN4 | AFLP3 | VNIII | A3 | 2C | ITS1/ITS2 | |
|
C. neoformans var. neoformans |
D | VNIV | VN1 (VN2) | AFLP2 | VNIV | A4 | 2A/2B /2C |
ITS2 | |
| C. gattii | B/C | VGI | AFLP4A/AFLP4B | VGI | A5 | 4 | ITS3/ITS7 | ||
| B/C | VGII | AFLP6 | VGII | A6 | 3 | ITS4 | |||
| B/C | VGIII | AFLP5A/AFLP5B/ AFLP5C |
VGIII | A7 | 5 | ITS5 | |||
| B/C | VGIV | AFLP7 | VGIV | A8 | 6 | ITS6 |
Consensus standard strains
To enable global standardization, the working group also agreed to use a set of standard strains representing each of the eight major molecular types. This included the molecular type strains used in PCR fingerprinting or URA5-RFLP analysis [16] plus additional strains representing type cultures or strains, which are used in major cryptococcal genome projects (Table 2). All standard strains are publicly available from the CBS-Fungal Biodiversity Centre (CBS) (http://www.cbs.knaw.nl), the American Type Culture Collection (ATCC) (http://www.atcc.org) or the Fungal Genetic Stock Center (FGS) (http://www.fgsc.net). The corresponding collection numbers are listed in Table 2.
Table 2.
Standard/reference strains for Cryptococcus neoformans and Cryptococcus gattii strain typing
| CBS # | ATCC# | FGS# | Other numbers | MAT & Serotype |
Comments | References |
|---|---|---|---|---|---|---|
| Cryptococcus neoformans | ||||||
| Cryptococcus neoformans var. grubii | ||||||
| VNI (Meyer et al. [14,16])=AFLP1 (Boekhout et al. [21])=VN6 (VN5) (Viviani et al. [13]) | ||||||
| CBS 10085 | ATCC MYA-4564 |
10415 | WM 148; W10; Brown | αA | 1989, Australia, NSW, Sydney, clinical, CSF, HIV–, isolated by Sharon Chen |
[14,18] |
| CBS 8710 | ATCC 48922 | 9487 | DUMC 135.97; H99; NYSD 1649; CBS 10515; WM 04.15 |
αA | 1978, USA, NC, Durham, clinical, CSF, patient with Hodgkin’s lymphoma, isolated by John Perfect/Wiley Schell, type culture of C. neoformans var. grubii , genome sequence strain |
[5] |
| VNII (Meyer et al. [14/16])=AFLP1A (Boekhout et al. [21])=VN7 (Viviani et al. [13]) | ||||||
| CBS 10084 | ATCC MYA-4565 |
10416 | WM 626, W20; Cetin | αA | 1993, Australia, NSW, Sydney, clinical, CSF, HIV–, isolated by Sharon Chen |
[14,18] |
| AD hybrid | ||||||
| VNIII (Meyer et al. [14,16])=AFLP3 (Boekhout et al. [21])=VN33VN4 (Viviani et al. [13]) | ||||||
| CBS 10080 | ATCC MYA-4566 |
10417 | WM 628; 88B5400; Zapf | αA/aD | 1988, Australia, VIC, Melbourne, clinical, CSF, HIV+, isolated by Bryan Speed |
[14,18] |
| CBS 132 | ATCC 32045 |
- | CCRC 20528; DBVPG 6010; IFO 0608; IGC 3957; NRRL Y-2534 |
αA/aD | 1894, Italy, environmental, fermenting fruit juice, isolated by F. Sanfelice, type culture for C. neoformans |
[41] |
| Cryptococcus neoformans var. neoformans | ||||||
| VNIV (Meyer et al. [14,16])=AFLP2 (Boekhout et al. [21])=VN1 (VN2) (Viviani et al. [13]) | ||||||
| CBS 10079 | ATCC MYA-4567 |
10418 | WM 629; B 87455, Borg, F 14 |
αD | 1987, Australia, VIC, Melbourne, clinical, blood, HIV+, isolated by Bryan Speed |
[14] |
| CBS 6900 | ATCC 34873 |
10423 | B-3501; DBVPG 6228; CBS 7697 |
αD | 1975, USA, MD, Bethesda, NIH, crossing of NIH 12 x NIH 433, isolated by June Kwon-Chung |
[42] |
| Cryptococcus gattii | ||||||
| VGI (Meyer et al. [16])=AFLP4 (Boekhout et al. [21]) | ||||||
| CBS 10078 | ATCC MYA-4560 |
10419 | WM 179; Bryon; H33.1; MH56 |
αB | 1993, Australia, NSW, Sydney, clinical, CSF, HIV −, isolated by Sharon Chen |
[16,18] |
| CBS 6289 | ATCC 32269 |
- | MUCL 30449, RV 20186; CBS 8273 |
αB | 1966, Congo, Kinshasa, clinical, CSF, isolated by E. Gatti/R. Eeckels, type strain of C. neoformans var. gattii, |
[43] |
| CBS 10510 | - | - | WM 276; TCS -SC1 | αB | 1993, Australia, NSW, Mt Annan National Park, environmental, Eycalyptus tereticornis woody debris, isolated by Tania Sorrell/Sharon Chen, genome sequence strain |
[16] |
| VGII (Meyer et al. [16])=AFLP6 (Boekhout et al. [21]) | ||||||
| CBS 10082 | ATCC MYA-4561 | 10420 | WM 178; 49435; Colter; IFM 50894 | αB | 1991, Australia, NSW, Sydney, clinical, CSF, HIV −, isolated by Sharon Chen |
[16] |
| CBS 10514 | - | - | CDC R265; WM 02.32 | αB | 2001, Canada, BC, Duncan, Vancouver Island, clinical, bronchial wash, isolated by British Columbia CDC, high virulent Vancouver Island outbreak strain, VGIIa, genome sequence strain |
[44] |
| VGIII (Meyer et al. [16])=AFLP5 (Boekhout et al. [21]) | ||||||
| CBS 10081 | ATCC MYA- 4562 |
10421 | WM 175; WM 161; E698; 689; TP 0689; D1.13H |
αB | 1992, USA, California, San Diego, Blind Recreation Center/Park Boulevard UPAS street, environmental, Eucalyptus spp. woody debris, isolated by Tania Pfeifer/David Ellis |
[16,19] |
| CBS 6955 | ATCC 32608 | 10424 | DBVPG 6225; MUCL 30454; NIH 191; CBS 6916 |
αC | Before 1970, USA, San Fernando, California, clinical, CSF. |
[45] |
| VGIV (Meyer et al. [16])=AFLP7 (Boekhout et al. [21]) | ||||||
| CBS 10101 | ATCC MYA- 4563 |
10422 | WM 779; King Cheetah; IFM 50896 |
αC | 1994, South Africa, Johannesburg, veterinary, Cheetah, isolated by Valarie Davis |
[16,46] |
Consensus multi-locus sequence typing loci
To overcome problems arising from inter-laboratory reproducibility associated with the two commonly used typing techniques, such as PCR fingerprinting or AFLP analysis, the working group decided to use multi-locus sequence typing (MLST) as the method of choice for future cryptococcal strain typing. MLST has become the number one typing approach for epidemiological investigations of microorganisms [32]. MLST, originally developed for bacteria [32], indexes the sequence variation in approximately 400–500 bp of five to ten genes composed primarily of housekeeping genes. This technique has proven to be highly discriminatory for a number of human pathogenic fungi: C. albicans [33], C. glabrata [34], C. tropicalis [35], Coccidioides spp. [36] and Histoplasma capsulatum [37]. Most of the published MLST schemes are developed as tools for the wider scientific community, by being made publicly available as online databases at http://www.mlst.net/ and http://pubmlst.org/. In the case of the Cryptococcus species complex, two different MLST typing schemes have been introduced to type isolates of C. neoformans [23], and C. gattii [28], using twelve and eight unlinked loci respectively.
In the first study, 12 unlinked polymorphic loci: MPD1, TOP1, MP88, CAP59, URE1, PLB1, CAP10, GPD1, TEF1, SOD1, LAC1 and the IGS1 ribosomal RNA intergenic spacer region, which are dispersed on nine different chromosomes, were used to type 102 globally obtained serotype A strains [23]. MLST differentiated three major groups among the studied isolates, corresponding to VNI, VNII and VNB, a Botswana specific genotype closely related to VNI. In connection with this study a central web based database was created at www.mlst.net (http://cneoformans.mlst.net/) allowing for an online determination of the alleles and sequence types of C. neoformans serotype A strains.
The second study used eight unlinked polymorphic loci: SXIa or SXIα, IGS1, TEF1, GPD1, LAC1, CAP10, PLB1, and MPD1, of which two are mating type locus specific and can not be amplified for all strains, to type 202 C. gattii strains. These loci were supplemented for a more detailed analysis of 9 closely related strains by 22 additional gene loci: HOG1, BWC1, CNB1, TOR1, CAC1, CRG1, URE1, FHB1, BWC2, CNA1, CBP1, TSA1, STE7, FTR1, PAK1, CAP59, ICL1, GPA1, GPB1, RAS1, CCP1, and TRR1 to investigate the origin of the Vancouver Island outbreak isolates [28]. MLST differentiated all four major molecular types of C. gattii (VGI, VGII, VGIII and VGIV) and highlighted two possible origins (Australia or South America) for the outbreak strains.
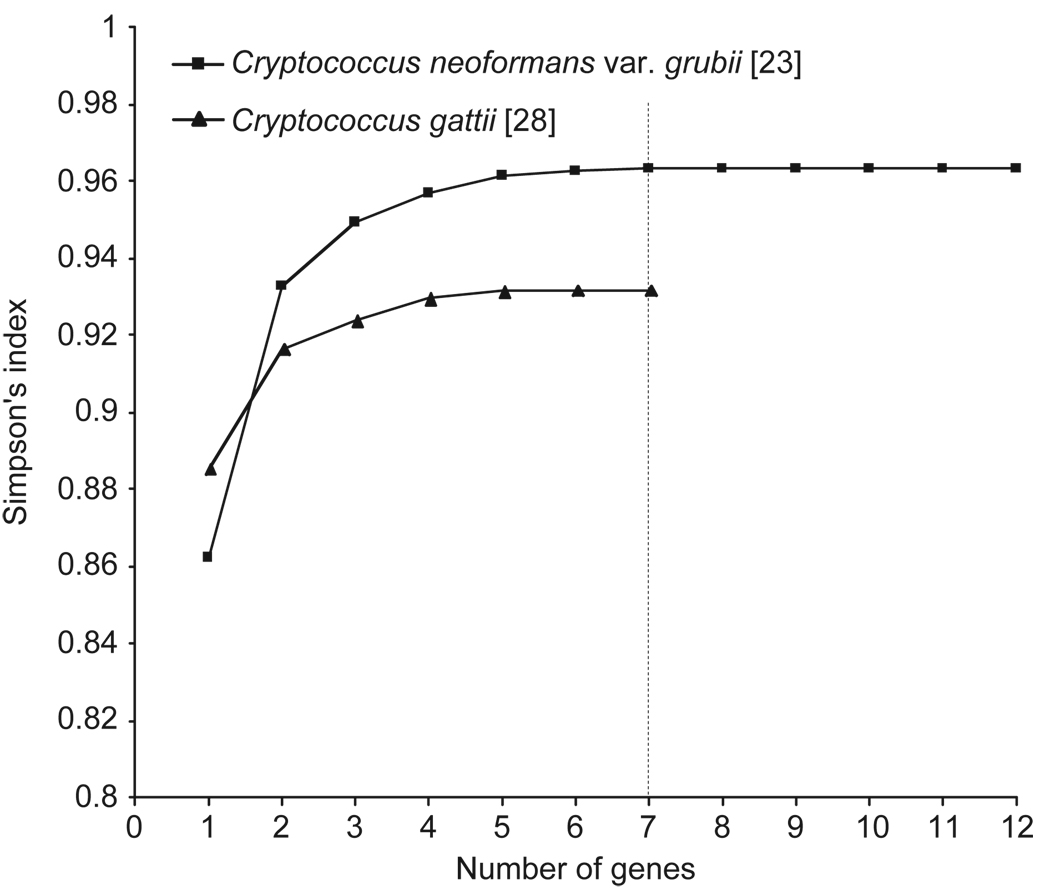
Statistical analysis using the Simpsons’s index of diversity [38] revealed that for both previously studied MLST data sets, a minimum of seven loci are required to differentiate between the sequence types of all strains (Fig. 1). For the Litvintseva et al. [23] MLST data set, the following loci resulted in the highest discrimination of the investigated strains: CAP59, IGS1, GPD1, LAC1, PLB1, MP88 and SOD1, with a Simpson’s index of diversity of 0.9632. For the Fraser et al. [28] MLST data set, the most discriminatory loci were: GPD1, IGS1, TEF1, LAC1, MPD1, CAP10 and PLB1, which resulted in a Simpson’s index of diversity of 0.9319.
Fig. 1.
Number of genes necessary to differentiate all sequence types obtained in the two previously published cryptococcal MLST schemes based on Simpson’s index of diversity [38]. For the Fraser et al. [28] data set the mating type locus specific genes, SXIa or SXIα, have been excluded for this analysis since they can’t be amplified from all isolates.
Both MLST schemes utilized highly polymorphic loci, which resulted in stable and reproducible typing systems that were able to distinguish between closely related strains. While using as many genetic loci as possible would enhance the discriminatory power of the MLST scheme, it would be pragmatic to achieve the maximal level of differentiation with a minimal set of genetic loci. The ideal MLST scheme for the Cryptococcus species complex should fulfill two criteria: (i) it should amplify and type the same genes from all five serotypes/eight molecular types using the same set of primers, and (ii) the selected genes should contain sufficient sequence diversity to produce a discriminatory typing scheme. Taking these facts into account, the working group has selected a set of seven gene loci for a cryptococcal consensus MLST scheme based on the results obtained in the previously published studies by Litvintseva et al. [23], Fraser et al. [28], and additional unpublished data obtained by Meyer et al. and Fisher et al. Special emphasis was placed on using loci that exhibited the largest number of different allele types, as well as the potential to use the same primers with all eight major molecular types identified previously for C. neoformans and C. gattii. These gene loci included six housekeeping genes CAP59, GPD1, LAC1, PLB1, SOD1, URA5, from which three genes code for cryptococcal virulence factors: the polysaccharide capsule (CAP59), melanin synthesis (LAC1) and cell invasion (PLB1), and the intergenic spacer, IGS1, which was selected based on its high allelic diversity.
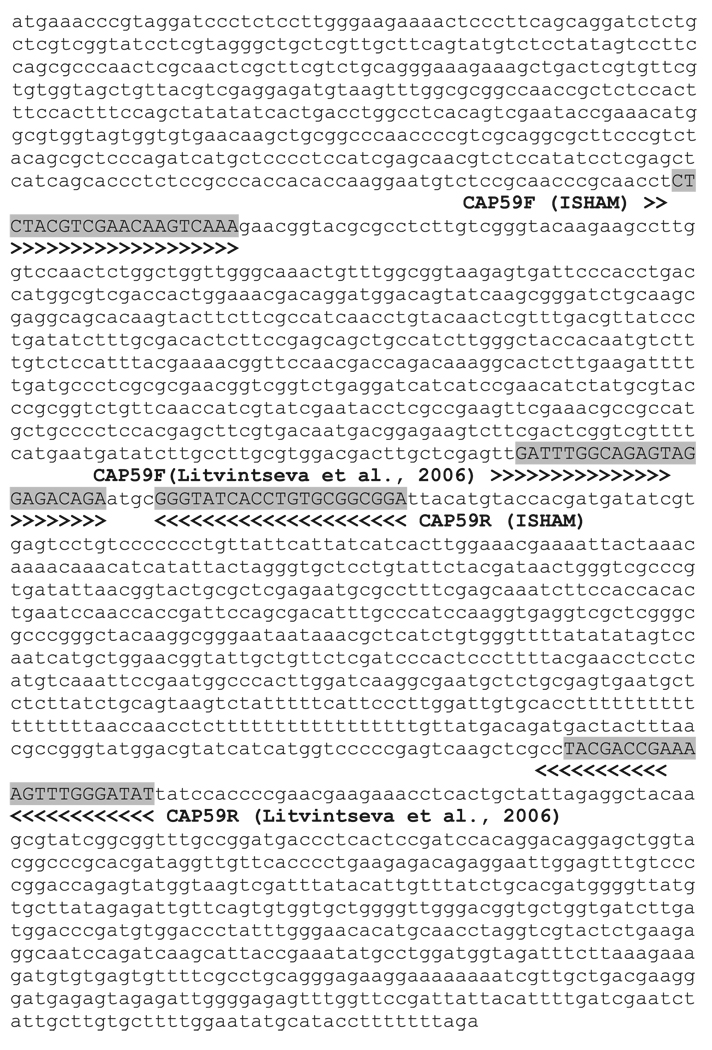
All the herein proposed MLST loci, except for the CAP59 locus, are similar to the ones used previously enabling the incorporation of, and comparisons with all previously obtained data. The region of the CAP59 locus proposed for the consensus MLST scheme represents a different fragment of the CAP59 gene used by Litvintseva et al. [23] (Fig. 2). This new locus was chosen based on the fact that it can be amplified from all eight molecular types using the same primers.
Fig. 2.
CAP59 genomic sequence of strain H99 (Cryptococcus neoformans var. grubii, VNI, http://www.broad.mit.edu) used as master sequence, indicating the location of the Cryptococcal Working Group (ISHAM) proposed consensus primers used in Fraser et al. [28] and the location of the primers used in Litvintseva et al. [23].
An additional locus, TEF1, which also showed high discriminatory power when used for C. neoformans var. grubii and for C. gattii molecular type VGII, was excluded from the consensus typing scheme. This was based on the fact that sequence data are only available for C. neoformans var. grubii and technical problems had been encountered when amplifying this locus. However, this locus may offer additional discrimination in some of the eight major molecular types.
To enable amplification of all seven loci from the eight major molecular types of C. neoformans and C. gattii, the previously published primers were tested on all eight major molecular types in three of the six laboratories (Teun Boekhout’s laboratory at the CBS, June Kwon Chung’s laboratory at the NIH, Matthew Fisher’s laboratory at the Imperial College, Wieland Meyer’s laboratory at the University of Sydney, Tom Mitchell’s laboratory at Duke University, and Maria Anna Viviani’s laboratory at the Università degli Studi di Milano) that collaborated in the development of the herein presented consensus MLST scheme. Satisfactory amplifications were obtained for all loci except for the SOD1 locus, where two different sets of primers were finally used to amplify either VNI–VNIV for C. neoformans or VGI–VGIV for C. gattii (Table 3). The specific primers and the suggested amplification conditions to amplify the seven gene loci are given in Table 3. Primer directions are listed according to the orientation in the genome sequence of the strain H99 at the Broad Institute (http://www.broad.mit.edu). Variations in the quality of the amplification products, resulting from either the Taq DNA polymerase enzyme or the PCR machine and PCR conditions used, were observed between participating laboratories. For that reason, the amplification conditions given in Table 3 should only serve as a guideline that may be optimized by individual laboratories.
Table 3.
MLST loci information
| Gene locus |
Gene product |
Chromo- some locationa |
Primer name and sequence (If not specified differently primers listed will work for C.n. and C.g.) |
Amplification conditions | No. of bases analysed (bp)a |
Analysed sequence fragment, start (5′) and end (3′) pointsa |
Ref. |
|---|---|---|---|---|---|---|---|
| CAP59 | Capsular associated protein |
1 |
CAP59F 5′ CTCTACGTCGA GCAAGTCAAG 3′ CAP59R 5′ TCCGCTGCA CAAGTGATACCC 3′ |
94°C 3min; 35 cycles: 94°C 30s, 568C 30s, 72°C 1min Alternative conditions: 30 cycles: 94°C 30s, 64°C 30s, 72°C 1min or: 30 cycles: 95°C 3min, 95°C 30s, 54°C 30s, 72°C 1min |
559 | 5′- ACGGTACGCGCCG GAGACAGAATG-3′ |
[28] |
|
Alternative primers: CAP59LF 5′ GTGAACAA GCTGCGGC 3′ CAP59LR 5′ GGATTCAG TGTGGTGGAAGA 3′ |
35 cycles: 94°C 30s, 60°C 30s, 72°C 1min |
[current study] |
|||||
| GPD1 | Glyceraldehyde- 3-phosphate dehydrogenase |
7 |
GPD1F 5′ CCACCGAACCC TTCTAGGATA 3′ GPD1R 5′ CTTCTTGGCA CCTCCCTTGAG 3′ |
94°C 3min; 35 cycles: 94°C 45s, 63°C 1min, 72°C 2min Alternative conditions: 12 cycles; 62 − 56°C step-down 2°C every 2 cycles 95°C 3 min; 95°C 30 sec, 62 −56°C 30 s, 72°C 1 min; followed by 25 cycles: 95°C 30 s, 56°C 30 s, 72°C 1 min |
543 | 5′- GGTTTCGGTACGG GACCCTGCCAA-3′ |
[28] |
| LAC1 | Laccase | 8 |
LAC1F 5′ AACATGTTCCCT GGGCCTGTG 3′ LAC1R 5′ ATGAGAATTG AATCGCCTTGT 3′ |
94°C 3min; 30 cycles: 94°C 30s, 58°C 30s, 72°C 1min Alternative conditions: 30 cycles: 95°C 30s, 50°C 30s, 72°C 1min |
469 | 5′- GTAAGTATCAGCT CAAGCTAAACA-3′ |
[28] |
| PLB1 | Phospholipase | 12 |
PLB1F 5′ CTTCAGGCGGA GAGAGGTTT 3′ PLB1R 5′ GATTTGGCGT TGGTTTCAGT 3′ |
94°C 3min; 30 cycles: 94°C 45s, 61°C 45s, 72°C 1min Alternative conditions: 12 cycles; 62 −56°C step-down 2°C every 2 cycles 95°C 3 min; 95°C 30 s, 62 −56°C 30 s, 72°C 1 min; followed by 25 cycles: 95°C 30 s, 56°C 30 s, 72°C 1 min |
532 | 5′- TGTTACTTGGATT CTGGAACATCG-3′ |
[23] |
| SOD1 | Cu, Zn superoxide dismutase |
5 |
Primers for C.n. SOD1CNF 5′AAGCCTCT CATCCATATCTT 3′ SOD1CNR 5′TTCAACCAC GAATATGTA 3′ Primers for C.g. SOD1CGF 5′ GATCCTCAC GCCATTACG 3′ SOD1CGR 5′ GAATGATG CGCTTAGTTGGA 3′ |
94°C 3min; 35 cycles: 94°C 30s, 52°C 30s, 72°C 1.5min |
700 | 5′ - CCACGTGCTCGCA CCTGTCAATGC-3′ |
[46] |
|
Alternative primers for C.n.: SOD1-f 5′ TCTAATCGAAA TGGTCAAGG 3′ SOD1-r 5′ CGCAGCTGTT CGTCTGGATA 3′ |
12 cycles; 62 −56°C step-down 2°C every 2 cycles 95°C 3 min; 95°C 30 sec, 62 −56°C 30 sec, 72°C 1 min; followed by 25 cycles: 95°C 30 sec, 56°C 30 sec, 72°C 1 min |
535 | 5′ -ATCGCTCACCGCT GCCCATTGTCA-3′ |
[23] | |||
| URA5 | Orotidine monophosphate pyrophosphorylase |
8 |
URA5F 5′ ATGTCCTCCCA AGCCCTCGAC 3′ URA5R 5′ TTAAGACCTCT GAACACCGTACTC 3′ |
94°C 3min; 35 cycles: 94°C 45s, 63°C 1min, 72°C 2min Alternative conditions: 30 cycles: 94°C 45s, 63°C 1min, 72°C 2min (C.n.) 26 cycles: 94°C 30 s, 68°C 30s, 72°C 30s (C.g.) or: 30 cycles: 95°C 3 min; 95°C 30 sec, 63°C 30 sec, 72°C 1min |
601 | 5′ - TTTTCGGCAACTCT TGGAAAGCTC-3′ |
[16] |
| IGS1 | Ribosomal RNA intergenic spacer |
2 |
IGSF 5′ ATCCTTTGCAGA CGACTTGA 3′ IGSR 5′ GTGATCAGTGC ATTGCATGA 3′ |
94°C 3min; 35 cycles: 94°C 30s, 60°C 30s, 72°C 1min Alternative conditions: 30 cycles: 94°C 30s, 56°C 30s, 72°C 1min |
723 | 5′ - TAAGCCCTTGTTAA AGATTTATTG-3′ |
[23] |
Note:
The sequences of the genome of strain H99 (C. neoformans var. grubii, VNI) at the Broad Institute (http://www.broad.mit.edu) were used as the master sequences. Nucleotide bases shown in bold typeface denote nucleotide bases that could vary between the different molecular types.
Automatic allele type and sequence type retrieval
Allele types for C. neoformans were assigned according to Litvintseva et al. [23] and for C. gattii according to by Fraser et al. [28], if applicable. The exact start- and endpoints for the sequence of each analyzed locus are given in Table 3 based on the H99 genome sequence at the Broad Institute (http://www.broad.mit.edu/), these may change over time if more strains are studied. The latest sequence cut points are listed at the webpage for each locus. To standardize the assignment of allele types (AT) and sequence types (ST), a centralized globally accessible MLST database will be established at www.mlst.net/. The online software NRDB (http://linux.mlst.net/nrdb/nrdb.htm) allows for an automatic retrieval of allele and sequence types and will assign a new allele and sequence type for any submitted unknown sequence. These are then uploaded to the database via a database curator. The designated curators are contactable via the website.
Conclusion
In conclusion the ISHAM working group on ‘Genotyping of Cryptococcus neoformans and C. gattii’ proposes the following set of genetic loci as an international standard for multi-locus sequence typing for C. neoformans and C. gattii: CAP59, GPD1, LAC1, PLB1, SOD1, URA5 and IGS1.
Acknowledgements
The authors would like to thank Matthew O’Sullivan for allowing us to use the software page developed as part of his PhD to determine the number of gene loci to be essential for an MLST scheme based on the Simpson’s index of diversity. This work was supported by an NH&MRC project grant #352303 to Wieland Meyer. June Kwon-Chung was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA. Matthew Fisher and David Aanensen were supported by the Wellcome Trust. Sitali Simwami was supported by the BBSRC, UK. Ferry Hagen was supported by funds from the Odo van Vloten Foundation. Anastasia P. Litvintseva and Thomas G. Mitchell were supported by a US Public Health Service NIH grant AI 25783. Luciana Trilles was supported by CAPES scholarship from the Ministério da Educação, Brazil. Sirada Kaocharoen was supported by the Chulalongkorn University Graduate Scholarship to commemorate the 72th anniversary of his majesty King Bhumibol Adulyadej, Thailand.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Evans EE. The antigenic composition of Cryptococcus neoformans. I. A serologic classification by means of the capsular and agglutination reactions. J Immunol. 1950;64:423–430. [PubMed] [Google Scholar]
- 2.Kwon-Chung KJ. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68:821–833. [PubMed] [Google Scholar]
- 3.Kwon-Chung KJ. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:943–946. [PubMed] [Google Scholar]
- 4.Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006;6:574–587. doi: 10.1111/j.1567-1364.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- 5.Franzot SP, Salkin IF, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999;37:838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon-Chung KJ. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- 7.Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M. (1557) Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. basillisporus (Basidiomycota, Hymenomycetes, Tremello-mycetidae) Taxon. 2002;51:804–806. [Google Scholar]
- 8.Bovers M, Hagen F, Kuramae EE, et al. AIDS patient death caused by novel Cryptococcus neoformans x C. gattii hybrid. Emerg Infect Dis. 2008;14:1105–1108. doi: 10.3201/eid1407.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bovers M, Hagen F, Kuranae EE, et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:599–607. doi: 10.1111/j.1567-1364.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- 10.Spitzer ED, Spitzer SG. Use of a dispersed repetitive DNA element to distinguish clinical isolates of Cryptococcus neoformans. J Clin Microbiol. 1992;30:1094–1097. doi: 10.1128/jcm.30.5.1094-1097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varma A, Kwon-Chung KJ. DNA probe for strain typing of Cryptococcus neoformans. J Clin Microbiol. 1992;30:2960–2967. doi: 10.1128/jcm.30.11.2960-2967.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer W, Mitchell TG. PCR fingerprinting to distinguish species and strains of yeast. In: Maresca B, Kobayashi GS, editors. Molecular biology of pathogenic fungi: A laboratory manual. New York: Telos Press; 1993. pp. 293–302. [Google Scholar]
- 13.Viviani MA, Wen H, Roverselli A, et al. Identification by polymerase chain reaction fingerprinting of Cryptococcus neoformans serotype AD. J Med Vet Mycol. 1997;35:355–360. [PubMed] [Google Scholar]
- 14.Meyer W, Marszewska K, Amirmostofian M, et al. Molecular typing of global isolates of Cryptococcus neoformans var neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA – a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790–1799. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1790::AID-ELPS1790>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 15.Cogliati M, Allaria M, Tortorano AM, Viviani MA. Genotyping Cryptococcus neoformans var. neoformans with specific primers designed from PCR-fingerprinting bands sequenced using a modified PCR-based strategy. Med Mycol. 2000;38:97–103. doi: 10.1080/mmy.38.2.97.103. [DOI] [PubMed] [Google Scholar]
- 16.Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruma P, Chen SC, Sorrell TC, Brownlee AG. Characterization of Cryptococcus neoformans by random DNA amplification. Lett Appl Microbiol. 1996;23:312–316. doi: 10.1111/j.1472-765x.1996.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen SCA, Brownlee A, Sorrell T, et al. Identification by random amplification of polymorphic DNA (RAPD) of a common molecular type of C. neoformans var neoformans in patients with AIDS. J Infect Dis. 1996;173:754–758. doi: 10.1093/infdis/173.3.754. [DOI] [PubMed] [Google Scholar]
- 19.Sorrell TC, Chen SC, Ruma P, et al. Concordance of clinical and environmental isolates of Cryptococcus neoformans var gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J Clin Microbiol. 1996;34:1253–1260. doi: 10.1128/jcm.34.5.1253-1260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boekhout T, van Belkum A, Leenders AC, et al. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol. 1997;47:432–442. doi: 10.1099/00207713-47-2-432. [DOI] [PubMed] [Google Scholar]
- 21.Boekhout T, Theelen B, Diaz M, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907. doi: 10.1099/00221287-147-4-891. [DOI] [PubMed] [Google Scholar]
- 22.Halliday CL, Bui T, Krockenberger M, Malik R, Ellis DH, Carter DA. Presence of alpha and a mating types in environmental and clinical collections of Cryptococcus neoformans var gattii strains from Australia. J Clin Microbiol. 1999;37:2920–2926. doi: 10.1128/jcm.37.9.2920-2926.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var grubii (serotype A), including a unique population in Botswana. Genetics. 2006;172:2223–2238. doi: 10.1534/genetics.105.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velegraki A, Kiosses VG, Kansouzidou A, et al. Prospective use of RFLP analysis on amplified Cryptococcus neoformans URA5 gene sequences for rapid identification of varieties and serotypes in clinical samples. Med Mycol. 2001;39:409–417. doi: 10.1080/mmy.39.5.409.417. [DOI] [PubMed] [Google Scholar]
- 25.Latouche GN, Huynh M, Sorrell TC, Meyer W. PCR-restriction fragment length polymorphism analysis of the phospholipase B (PLB1) gene for subtyping of Cryptococcus neoformans isolates. Appl Environ Microbiol. 2003;69:2080–2086. doi: 10.1128/AEM.69.4.2080-2086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz MR, Boekhout T, Theelen B, Fell JW. Molecular sequence analyses of the intergenicspacer (IGS) associated with rDNA of the two varieties of the pathogenic yeast, Cryptococcus neoformans. Systemat Appl Microbiol. 2000;23:535–545. doi: 10.1016/S0723-2020(00)80028-4. [DOI] [PubMed] [Google Scholar]
- 27.Bovers M, Hagen F, Kuramae, Boekhout T. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Gen Biol. 2008;45:400–421. doi: 10.1016/j.fgb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Fraser JA, Giles SS, Wenink EC, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 29.Hanafy A, Kaocharoen S, Jover-Botella A, et al. Multi-locus microsatellite typing for Cryptococcus neoformans var grubii. Med Mycol. 2008;46:685–696. doi: 10.1080/13693780802027062. [DOI] [PubMed] [Google Scholar]
- 30.Karaoglu H, Man Ying Lee C, Carter D, Meyer W. Development of polymorphic microsatellite markers for Cryptococcus neoformans. Mol Ecol Res. 2008;8:1136–1138. doi: 10.1111/j.1755-0998.2008.02196.x. [DOI] [PubMed] [Google Scholar]
- 31.Diaz MR, Boekhout T, Kiesling T, et al. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic fungus: Cryptococcus neoformans. FEMS Yeast Res. 2005;5:1129–1140. doi: 10.1016/j.femsyr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Taylor JW, Fisher MC. Fungal multi-locus sequence typing - it’s not just for bacteria. Curr Opin Micro. 2003;6:1–6. doi: 10.1016/s1369-5274(03)00088-2. [DOI] [PubMed] [Google Scholar]
- 33.Bougnoux ME, Aanensen DM, Morand S, et al. Multi-locus sequence typing of Candida albicans: strategies, data exchange and applications. Infect Genet Evol. 2004;4:243–252. doi: 10.1016/j.meegid.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Dodgson Ar, Pujol C, Denning DW, Soil DR, Fox AJ. Multi-locus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Micro. 2003;41:5709–5717. doi: 10.1128/JCM.41.12.5709-5717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tavanti A, Davidson AD, Johnson EM, et al. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J Clin Micro. 2005;43:5593–5600. doi: 10.1128/JCM.43.11.5593-5600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koufopanou V, Burt A, Taylor J. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci USA. 1994;94:5478–5482. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasuga T, White TJ, Koenig G, et al. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol Ecol. 2003;12:73–84. doi: 10.1046/j.1365-294x.2003.01995.x. [DOI] [PubMed] [Google Scholar]
- 38.Simpson EH. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- 39.Feng X, Yao Z, Ren D, Liao W. Simultaneous identification of molecular and mating types within the Cryptococcus species complex by PCR-RFLP analysis. J Med Microbiol. 2008;57:1481–1490. doi: 10.1099/jmm.0.2008/003665-0. [DOI] [PubMed] [Google Scholar]
- 40.Katsu M, Kidd S, Ando A, et al. The internal transcribed spacers and 5.8S rRNA gene show extensive diversity among isolates of the Cryptococcus neoformans species complex. FEMS Yeast Res. 2004;4:377–388. doi: 10.1016/S1567-1356(03)00176-4. [DOI] [PubMed] [Google Scholar]
- 41.Sanfelice F. Contributo alla morfologia e biologia dei blastomiceti che si sviluppano nei succhi di alcuni frutti. Ann Igien. 1894:463–495. [Google Scholar]
- 42.Varma A, Kwon-Chung KJ. Restriction fragment polymorphism in mitochondrial DNA of Cryptococcus neoformans. J Gen Microbiol. 1989;135:3353–3362. doi: 10.1099/00221287-135-12-3353. [DOI] [PubMed] [Google Scholar]
- 43.Gatti F, Eeckels H. An atypical strain of Cryptococcus neoformans (Sanfelice) Vuillemin. Part I: Description of the diseases and of the strain. Ann Soc Belge Méd Trop. 1970;50:689–694. [PubMed] [Google Scholar]
- 44.Kidd S, Hagen F, Tscharke R, et al. A rare genotype of Cryptococcus gattii caused the Cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) PNAS. 2004;107:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon-Chung KJ, Bennett JE, Theodore TS. Cryptococcus bacillisporus sp. nov. serotype B-C of Cryptococcus neoformans. Int J Syst Bacteriol. 1978;28:616–620. [Google Scholar]
- 46.Bolton LA, Lobetti RG, Evezard DN, et al. Cryptococcosis in captive cheetah (Acinonyx jubatus): two cases. J S Afr Vet Assoc. 1999;70:35–39. doi: 10.4102/jsava.v70i1.748. [DOI] [PubMed] [Google Scholar]
- 47.D’Souza CA, Hagen F, Boekhout T, Cox GM, Heitman J. Investigation of the basis of virulence in serotype A strains of Cryptococcus neoformans from apparently immunocompetent individuals. Curr Genet. 2004;46:92–102. doi: 10.1007/s00294-004-0511-y. [DOI] [PubMed] [Google Scholar]