Abstract
Heat shock proteins (Hsps) are a class of molecular chaperones that play an essential role in preserving cellular functions under stressful conditions. The over production of recombinant proteins often causes cellular stress that results in aggregation/misfolding of proteins, which sometimes leads to the formation of inclusion bodies. Here we report the cloning and characterization of heat-inducible PgHsp70 from Pennisetum glaucum, a heat and drought tolerant plant that showed stability and chaperone activity at elevated temperatures. The predicted amino acid sequence of PgHsp70 revealed a high homology with Hsp70 from other plants, and the overall 3D structure homology modeling is similar to that of the constitutively expressed bovine cytosolic Heat Shock Cognate (HSC)-70. The purified recombinant protein had an apparent molecular mass of 70 kDa and displayed optimal chaperone activity at 50°C, and pH 8.0. Under these conditions, the T1/2 of PgHsp70 increased from 10 to 15 h in the presence of glycerol. The PgHsp70 exhibited a higher chaperone activity towards glutamate dehydrogenase than alcohol dehydrogenase. The expression of recombinant carbonic anhydrase (CA ) in E. coli in a catalytically active soluble form rather than in inclusion bodies was made feasible by co-expression of PgHsp70. Circular dichroism (CD) studies of the recombinant PgHsp70 did not reveal any discernible changes in the α-helix content, with increase in temperature from 35 to 85°C, thus suggesting a critical role of α-helix content in maintaining the chaperone activity.
Key words: abiotic stress, carbonic anhydrase, chaperone, circular dichroism, heat shock proteins (Hsp70), Pennisetum glaucum, pH stable
Introduction
All living organisms respond to sudden change in temperature, which causes the induction of a set of highly conserved proteins called heat shock proteins (Hsps).1,2 These Hsps function as molecular chaperones and are essential for the cellular metabolic homeostasis.3,4 As molecular chaperones, they ensure the stability of other cellular proteins and protect them from various physiological and environmental insults.5,7 They have a broad range of functions ranging from the prevention of protein aggregation, refolding of misfolded proteins, degradation of unstable proteins and dissolution of protein complexes, besides some act as transcription factors.4,8
Hsps are found in different cellular compartments and have been broadly classified according to their molecular weights such as, Hsp-100, 90, 70, 60 and small Hsps. Hsp70 (DnaKDnaJ-GrpE) and Hsp100 (ClpA and ClpB) chaperones have been shown to play a role in refolding and increased thermal stability of bacterial luciferases in Escherichia coli.9 The Hsp70 belongs to a highly conserved family of heat shock proteins, is present in various sub-cellular compartments and helps in combating diverse forms of cellular stresses. It is known to protect various substrate proteins such as, citrate synthase, malate dehydrogenase, α-lactalbumin and insulin from thermal or chemical aggregation.8,10 Eukaryotic Hsp70s conventionally have three functional domains: an N-terminal ATPase domain (∼45 kDa), a substrate binding domain (∼15 kDa), and a C-terminal domain (∼25 kDa).8 The C-terminal domain has a terminal sequence motif EEVD, which has been implicated in binding proteins such as Hop (Hsp70 Hsp90 organizing protein) through their tetratricopeptide motifs.11
Chaperones of some of the Hsp70, 60, 40 and small heat shock proteins are unable to refold their substrates without ATP.12,13 In their ATP bound forms, Hsp70s bind and release substrates quickly, while, in the ADP bound state, substrate peptides are bound in a hydrophobic pocket covered by an α-helical lid, which must open to allow substrate binding or release.14,15 The importance of α-helical lid in Hsp70 was explained clearly by Slepenkov et al.16 The genes for various HSPs have been cloned and characterized and overexpression of some of these has resulted in developing transgenic plants showing tolerance to various stresses.17,18 However, detailed biochemical analysis and the role of plant HSPs as chaperones have been investigated only in a few cases.17,19–22 The overexpression of tobacco NtHSP70-1 confers drought-stress tolerance in plants.23 The expression of AtHSP70 and AtHSP101 genes was shown to be upregulated in calmodulin-binding protein phosphatase AtPP7 overexpression lines after heat shock, suggesting a possible role for AtPP7 in regulating the expression of (HSP) genes.24 Recently a transcriptional profiling of Arabidopsis heat shock proteins and transcription factors revealed an extensive overlap between heat and non-heat stress response pathways.25 In another study, Su and Li26 have recently reported that Arabidopsis stromal Hsp70s [cpHsc70-1 (At4g24280) and cpHsc70-2 (At5g49910)] are essential for plant development and important for thermotolerance of germinating seeds.
Protein folding has been recognized as one of the central problems in biology.8,27 Many proteins in a living cell will not fold properly without the assistance of molecular chaperones. The improper folding of linear amino acid sequences into non-native protein conformation leads to illegitimate interaction with other cellular components and subsequent protein aggregations.8,27 The overexpression of recombinant proteins under artificially induced conditions puts an additional demand for protein folding and protein quality control system assisted by molecular chaperones.8
The present investigation deals with the in-vitro chaperone activity of a thermostable inducible Hsp70 (PgHsp70) from a heat tolerant Pennisetum glaucum (Pearl millet). The purified recombinant PgHsp70 was characterized for elucidating its structural and biochemical features. We have shown that the maintenance of alpha helix conformation is essential for the ATP-independent chaperone activity of PgHsp70. When the carbonic anhydrase (CA) gene from Pennisetum glaucum was overexpressed in E. coli using an isopropyl-β-D-thiogalactopyranoside (IPTG) inducible protein expression system, the recombinant protein aggregated into inclusion bodies. The simultaneous overexpression of PgHsp70 in the same cells, however, resulted in the production of catalytically active soluble CA.
Results and Discussion
Cloning and in silico analysis of PgHsp70.
Pennisetum glaucum, a Poaceae family member, is a heat and drought-tolerant crop that grows at high temperatures (>45°C). During differential screening of stress induced subtractive cDNA libraries among many other genes, a set of HSPs was observed.28 The C-terminus of Hsp70s from various species show highly conserved motifs like the cytosolic group is EEVD, for ER it is HDEL, for mitochondria it is PEAEYEEAKK and for plastids it is PEGDVIDADFTDSK.35 HDEL motif in BiP is an ER retention signal. The EEVD motif in cytosolic Hsp70 is essential for interaction with Hsp70 interacting proteins such as Hip/p48 and p60/Hop.36 Deletion of EEVD from a rat Hsc70 resulted in a dramatic increase in ATPase activity suggesting a role for EEVD in modulating ATPase activity of Hsc70.37 The PgHsp70 has a signal peptide and lacks the EEVD motif, and this might be the property of the chloroplast homologues. The full length cDNA of PgHsp70 is 2,377 bp in length and contains an open reading frame of 2,039 bp and 128 bp 5′ and 210 bp 3′ untranslated regions (UTR) [Acc.No. EF495353]. The ORF encodes for a 649 amino acids long protein with an apparent molecular weight of 70 kDa and an estimated isoelectric point of 5.0. The deduced amino acid sequence of PgHsp70 [Pg] possessed an overall 88–90% amino acid sequence identity with chloroplast Hsps picked up from data base such as Oryza sativa [Os] (Acc.No. ABA97211, showing 88% similarity), Pisum sativum [Ps] (Acc.No. CAA49147, showing 90% similarity), Cucumix sativus [Cs] (Acc.No. ABM92419, showing 89% similarity) and Arabidopsis thaliana [At] (Acc.No. NP194159 are showing 88% similarity) (Fig. 1). The phylogenetic tree shows protein sequence variation among various plant Hsps.
Figure 1.
Alignment of HSP70 proteins from selected crop species: Protein sequences were analyzed using DNA analysis software DNASTAR. For protein alignment, Clustal V of MegAlign was used in which clusters were aligned as pairs, then collectively as sequence groups to produce the overall alignment. Comparison of PgHsp70 with deduced amino acid sequences of chloroplast of Oryza sativa (accession no. ABA97211), Pisum sativum (accession no. CAA 49147), Cucumix sativus (accession no. ABM92419) and Arabidopsis thaliana (accession no. Np194159).
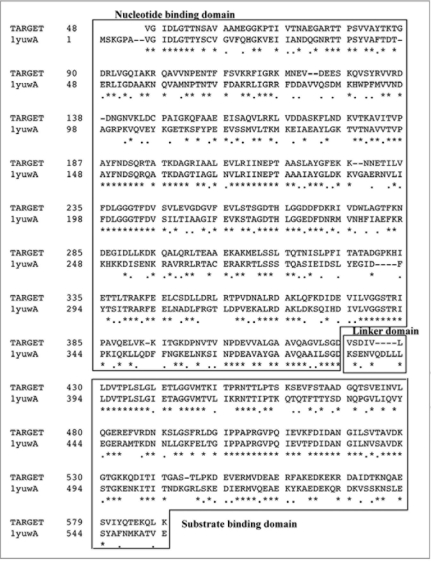
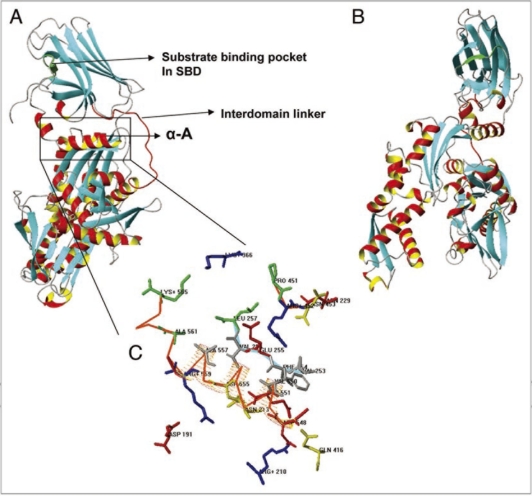
The PgHsp70 and bovine template sequence (1YUW) share a similarity of about 52%. The target PgHsp70 was modeled from sequence 48 to 589, based on sequence alignment shown in Figure 2. A close inspection of the comparative model (Fig. 3) and the pair wise alignment helps in identifying important residues involved in interdomain interactions and residues important for the protein function. As expected, the overall predicted 3D structure is similar to the bovine HSP 70,32 and an exposed linker connects the two domains, Substrate Binding Domain (SBD) and Nucleotide Binding Domain (NBD) (Figs. 2 and 3A-C). The PgHsp70 SBD is formed by sequence 431–589 and NBD is formed by sequence 48–424 and the linker is formed by a relatively shorter sequence 424–430 (red colored Cα trace). Substrate binding site is formed by 574–579 (Green Cα trace). The interaction between the domains is stabilized by a number of interacting residues. There is a salt bridge between R559 and D191. A salt bridge corresponding to 1YUW (bovine), K325:E530 is absent in PgHSP70, as the residue corresponding to K530 is substituted in PgHSP70 by K565. The interaction is also stabilized by long range ionic interactions like amoung E255 and R452 as well as hydrophobic interactions between the residues: V253, F254, V256, V550, V554 and A557. The overall comparison of the template with PgHSP70 displays conservation of important residues in space.
Figure 2.
Sequence alignment of PgHsp70 and protein sequence corresponding to the template (1YUW). The domain regions such as nucleotide binding, linker and substrate binding are boxed and marked. Stars and dots in the sequences indicate the positions of identical amino acids and semi conserved substitutions, respectively.
Figure 3.
(A and B) Ribbon representation of PgHsp70 comparative model, Helix A highlighted in a box. The region above Helix A corresponds to the substrate binding domain (SBD) and the region below, to the nucleotide binding domain (NBD). Interdomain linker represented as red Cα trace. (C) Detailed view of interactions between SBD (Cα trace in red orange) and NBD (Cα trace in Cyan) and few discontinuous residues without Cα trace, side chains of residues identical or conserved to the template in blue, red, yellow or dark grey for +, −, polar, or nonpolar. PgHsp70 residues not conserved with respect to the template residues are shown in green.
Expression profile of PgHsp70 gene under different stress conditions.
To establish detailed expression data of PgHsp70, a comprehensive RT-PCR (reverse transcriptase polymerase chain reaction) analysis was carried out from RNA samples isolated from different stress (temperature, cold, drought, ABA [abscisic acid], salicylic acid [SA] and salt) treated Pennisetum glaucum seedlings at various time points. The control seedlings were not subject to any stress. The relative transcript changes and those which showed at least more than two-fold changes as compared to the control condition were taken as significant. Table 1 represents the summarized data of biochemical and physical stress, which revealed that in all other stresses, except heat stress and SA treatment, the level of transcript was decreased. However, in the case of heat stress, the transcript level of PgHsp70 was upregulated upto 10- to 15-fold (Table 1).
Table 1.
The relative fold change of PgHsp70 transcript under different abiotic stress conditions
| Gene | Control (water, rt) | Heat (48°C) | Drought (withholding water, 34 h) | Salt (250 mM NaCl) | Cold (4°C) | ABA (100 μM) | SA (1 mM) |
| PgHsp70 | NS | +++ | − | NS | − | t | + |
The fold change between 2 to 5 are given a single (+) or (−) sign for upregulation and downregulation respectively; Similarly, (−−) denotes 5 to 10 fold decrease, (+++) denotes 10- to 15-fold increase and NS denotes for not significant change and in the expression values; rt, room temperature.
Production of recombinant PgHsp70 protein.
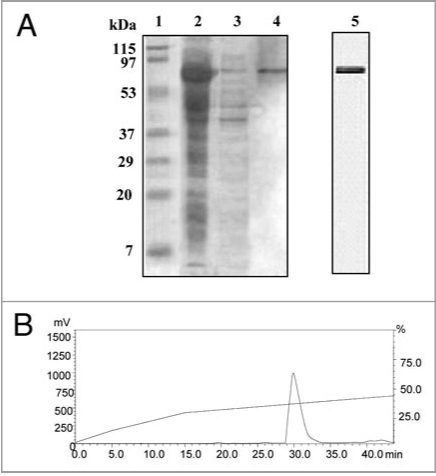
The PgHsp70 was overexpressed in E. coli BL21 (DE3) after addition of IPTG in the late exponential and early stationary phases of growth. The protein profile of the E. coli BL21 (DE3) carrying the pETPgHsp70 was analyzed by 10% SDS-PAGE gels. Analysis of crude bacterial extracts revealed high levels of expression of an approximately 70 kDa recombinant protein in E. coli BL21 (DE3) carrying the pET-PgHsp70 construct. Further analysis revealed that the majority of the recombinant protein was in the soluble fraction (Fig. 4A). The recombinant hexa-histidinetagged PgHsp70 protein was purified by affinity chromatography on a Ni-NTA column in a single step, which yielded ⩾95% purity. The HPLC analysis showed one dominant peak (retention time: 30 min) (Fig. 4B).
Figure 4.
(A) SDS-PAGE analysis of purified recombinant PgHsp70 protein. PgHsp70 carrying a N-terminal hexahistidine tag was overproduced in pET-28a(+) and purified by Ni2+ NTA affinity chromatography. Lane 1 is molecular mass markers; the sizes (in kDa) are indicated adjacent (left side) to the gel. Lane 2 IPTG-induced supernatant fraction containing enriched PgHsp70 protein (70 kDa). Lane 3 is un-induced protein PgHsp70, Lane 4 is Ni2+ NTA purified PgHsp70. The gel containing lanes 1–4 is stained with Coomassie Brilliant Blue. Lane 5: the purified PgHsp70 protein was run on another gel and silver stained. (B) Reconfirmation of PgHsp70 purity by the reverse-phase-HPLC analysis (detector: 280 nm) on C18 (Phenomenex, C18, 5 μM 1.D. 250 × 4.6 nm) using aceto-nitrile (0.1% TFA)/water (0.1% TFA) gradient 0−15 min at the rate of 2% min−1 solvent; 15 to 45 min 0.5% min−1 solvent at a flow rate of 1.0 ml min−1.
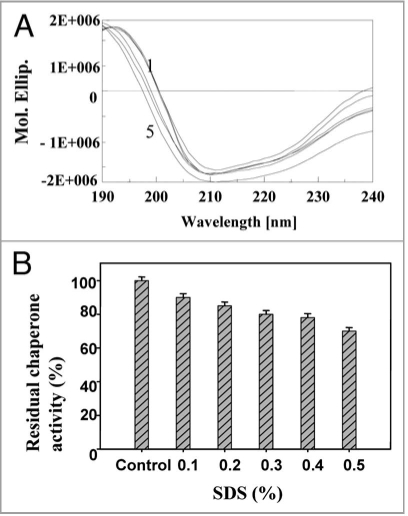
Chaperone activity of PgHsp70.
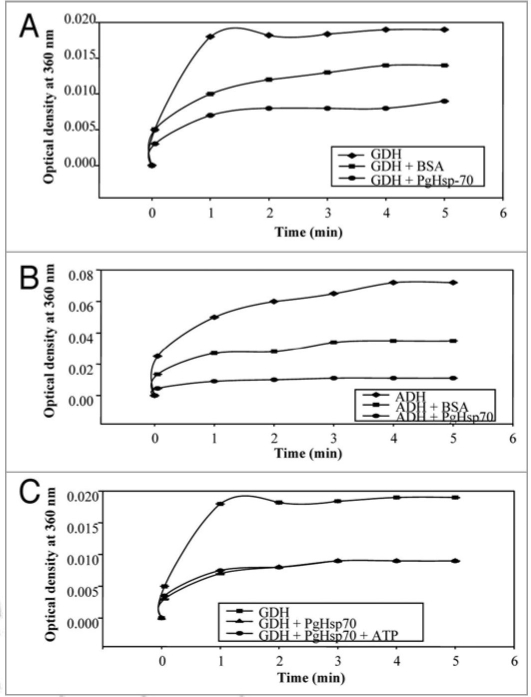
The recombinant PgHsp70 was assayed for chaperone activity by studying its ability to prevent thermal unfolding of glutamate dehydrogenase (GDH) and alcohol dehydrogenase (ADH). In thermal denaturation experiments, the protein substrates, GDH and ADH were protected efficiently by recombinant protein PgHsp70 but not efficiently by BSA (Fig. 5A and B) as assayed by light scatter measurements. On a comparative basis for the largest binding capacity for heat-denatured proteins and its early refolding property, ADH proved to be a better affinity substrate for chaperone activity of PgHsp70 as compared to GDH. These data also suggest that if the PgHsp70 protein is expressed with transit peptide and without an Hsp70 escort protein, it can also be active in contrast to the previous reports.38–40
Figure 5.
Thermally induced aggregation of model proteins such as glutamate dehydrogenase/or alcohol dehydrogenase with/without PgHsp70 at 50°C was Light scatter assayed by incubating the reaction mixture (1 ml) containing 20 mM Tris buffer (pH 8.0), 100 mM sodium chloride, 0.2 mg ml−1 glutamate dehydrogenase/or alcohol dehydrogenase and 0.2 mg ml−1 of the recombinant heat shock protein PgHsp70 (with or without ATP 5 mM) or BSA (0.2 mg ml−1) aggregation was monitored by the measuring the O.D. at 360 nm. (A) Light scatter assay was done using substrate protein GDH to assess the chaperone activity of PgHsp70 in the presence and absence of BSA (B) Light scatter assay was done using substrate protein ADH to assess the chaperone activity of PgHsp70 in the presence and absence of BSA. (C) Light scatter assay was done using substrate protein GDH to assess the chaperone activity of PgHsp70 in the presence and absence of ATP. The error bars have not been added in this figure because the SD in all the cases was less than 10%.
PgHsp70 chaperone activity (in-vitro) was not affected by the addition of ATP in the reaction mixture thus suggesting that recombinant PgHsp70 is independent of ATP for its chaperone activity (Fig. 5C). Here it is to note that there were no nucleotides present in the buffer in which the PgHsp70 was dissolved. Similar findings have been reported in Borrelia burgdorferi for Hsp60 and 70.41 Earlier, Cho and Bae21 have also reported ATPindependent thermo-protective activity of tobacco Hsp70. The tobacco Hsc18, however, showed inhibition in the chaperone activity in the presence of ATP.10 PgHSP70 was much better than BSA in terms of chaperone activity (Fig. 5).
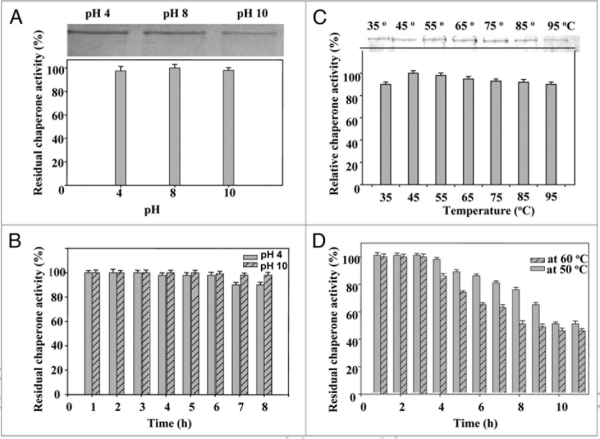
Although thermostability is an intrinsic property dictated by the primary structure of a protein, many external factors including ions influence the thermostability and chaperone activity. Majority of heat sock proteins (e.g., Hsp70) are active and stable at slightly acidic or neutral pH.37 However, PgHsp70 was found to be active over a broad pH range (Fig. 6A), which is nearly similar to that reported from Hsp70 of Plasmodium falciparum.38 The recombinant PgHsp70 was stable at pH 8.0 for 10 h at 50°C. Approximately 98% of the residual activity was recorded when the recombinant PgHsp70 was exposed to pH 4.0 and pH 10.0 for 6 and 7 h, respectively (Fig. 6B).
Figure 6.
Effect of pH (A) on chaperone activity of PgHsp70; (B) on stability of PgHsp70, (C) effect of temperature on chaperone activity (experimental conditions as mentioned in Fig. 5), and (D) temperature stability of PgHsp70.
PgHsp70 chaperone activity enhanced consistently with an increase in temperature from 35 to 45°C, and thereafter remained consistent (90%) up to 85°C, before showing a slight decline at 95°C (Fig. 6C). Thus PgHsp70 exhibits very high thermostability. Half-life values of the PgHsp70 at 50° and 60°C were 10 and 8 h, respectively (Fig. 6D), which are similar to those reported for thermophilic catalysts of some bacteria and fungi.42
Secondary structure and thermostability.
The contribution of aromatic residues in maintaining the secondary structure of proteins is well documented.43 In the context of PgHsp70, it is of particular interest to study the role of the aromatic residues at N-terminal e.g., Tyrosines [(Y) at 31, 35, 132, 188 and 222] and Tryptophans [(W) at 37 and 277] for their possible role in stability and chaperone activity.
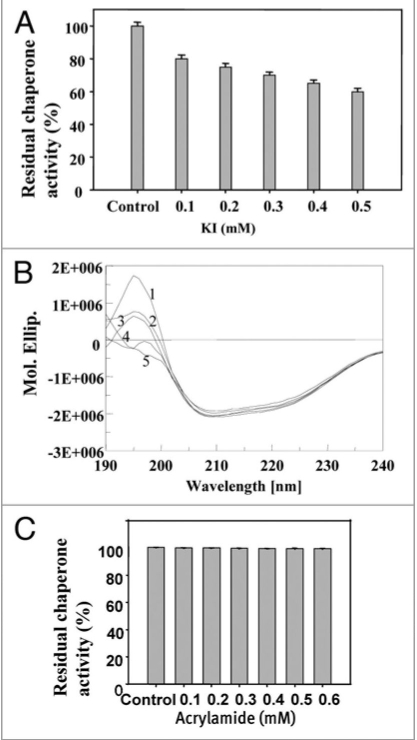
Modifications of crucial amino acids such as Y and W in the substrate binding site are believed to bring about important changes in their chaperone activity as reported earlier by Bernd Bukau (www.zmbh.uni-heidelberg.de/Bukau/images/Bukau.pdf). With a view to probe into the nature of molecular forces that stabilize PgHsp70, the structural investigation of PgHsp70 was carried out in the presence of perturbants of different kinds: acrylamide (neutral), I− (anionic), Cs+ (cationic) and denaturants Gdn.HCl, SDS and urea. The spectrum of native PgHsp70 indicated the presence of 80% {α}-helix, 5.1% {β}-turn, and 14.4% remainder (analyzed by Dichroweb software see in supplementary data). The secondary structure of PgHsp70 appeared entirely different from that reported for Bovine Hsp70, which had 15% {alpha}-helix, 24% {β}-sheet, 24% {beta}-turn, and 38% remainder. High percent of α-helices of PgHsp70 makes it very different from other reported Hsps. It, however, showed some similarity to the Salmonella Hsp70, which consists of 85% α-helices.44 The coiled α-helices structure in proteins confers a variety of functional capabilities;45 this property thus might be responsible for the thermostability of PgHsp70 (>45°C).
In order to understand the charge, stability effect of PgHsp70 (in the presence of 0.2 mM KI), a 15% decrease in α-helix content was recorded. The KI treatment resulted in the slight loss of both chaperone activity (Fig. 7A) and secondary structure (Fig. 7B). Charged quenchers are known to probe only the surface-exposed amino acid residues, while the polar and uncharged acrylamide can diffuse into the protein core and change the conformation.46 A decrease (10%) in the α-helix of PgHsp70 was observed at 0.1 mM acrylamide, and further addition resulted in only minimal changes. Surprisingly, there was no quantitative change in the chaperone activity of the PgHsp70 even at high concentration of acrylamide (0.5 mM) (Fig. 7C). As suggested by Eftink and Ghiron,46 we have concluded that, the acrylamide interacts by means of simple collision process and does not apparently interfere with the protein substrate-binding site of PgHsp70. The addition of different concentrations of CsCl (0.1–0.5 mM) to the reaction mixture also had only slight effect on both secondary structure (10% decreases in α-helix) and chaperone activity of PgHsp70 (results not shown).
Figure 7.
Effect of anion charger KI: (A) on chaperone activity of native PgHsp70 in 0.1 M phosphate buffer (pH 8.0) at 50°C for 5 min. (B) on secondary structure of PgHsp70 [spectra 1 & 2–5 represents native PgHsp70, different concentration of KI (0.2–0.5 mM) with native PgHsp70] (C) Effect of acrylamide on chaperone activity experimental conditions as mention in Figure 5. The error bars have not been added in this figure because the SD in all the cases was less than 10%.
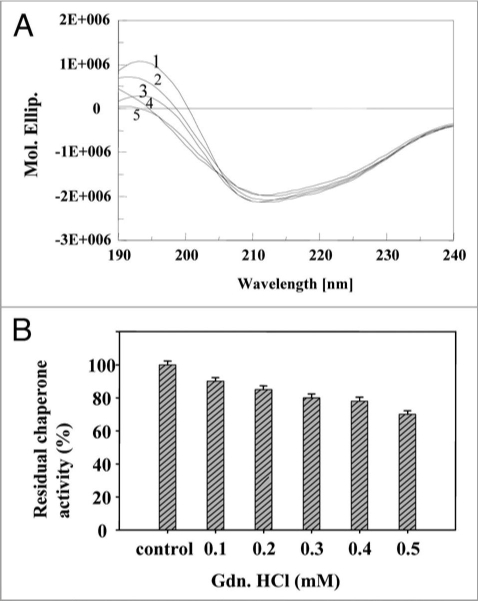
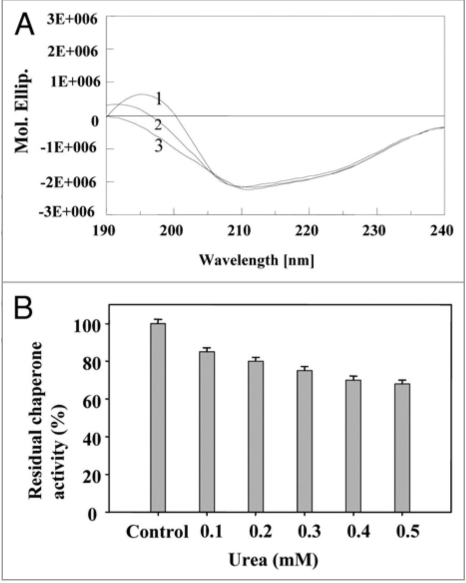
Gdn.HCl is known to interact reversibly with the carboxyl groups of amino acids.47 At low concentrations, Gdn.HCl (0.1 mM) is believed to shield the carboxyl groups in the vicinity of amino acid residues in the chaperone active site. As the Gdn.HCl concentration was increased, significant loss of conformational occurred (40% decrease in α-helix content and increase in random coil) (Fig. 8A), and also the chaperone activity decreased at 0.1 to 0.5 mM Gdn.HCl (Fig. 8B). Stabilization of secondary structure of PgHsp70 was carried out by incorporating glycerol (0.001% v/v) to a fixed concentration (0.1 mg ml−1) of PgHsp70 and recording the conformational changes and chaperone activity of PgHsp70 (Table 2); this confirmed the protective nature of glycerol against denaturation [Gdn.HCl (0.5 mM)]. Glycerol is thought to interact non-covalently with proteins and bring about protection against denaturation with Gdn.HCl. In a similar manner, Hsp70 chaperon activity was found to be stabilized by addition of ADP.38 The CD spectra in the presence of urea clearly revealed the unfolding of PgHsp70, which led to a 40% decrease in α-helix content (Fig. 9A). These conformational changes occurring in PgHsp70 could be responsible for a decline in the chaperone activity (Fig. 9B). A 38% inactivation of PgHsp70 chaperone activity was observed at 0.5 mM urea. These experimental results suggest that hydrophobic aromatic residues, located in the protein-binding site may be involved in maintaining the secondary structure as observed in other proteins of Bacillus sp.48 The CD spectra in the presence of SDS clearly revealed that the PgHsp70 is slightly changed in conformation (80% α-helix, 20% unordered) (Fig. 10A) and its chaperone activity (Fig. 10B).
Figure 8.
Effect of inhibitor Gdn.HC l: (A) on secondary structure of PgHsp70 [spectra 1–5 represent PgHsp70, PgHsp70 with different concentrations of Gdn.HC l (0.2 to 0.5 mM)], (B) on chaperone activity of PgHsp70 (experimental conditions as mentioned in Fig. 5).
Table 2.
Effect of different physical (temperature and pH) and chemical agents (denaturants and inhibitors) on secondary structure of PgHsp70
| Effect | α-helix | 3/10 helix | Parallel β-sheet | Antiparallel β-sheet | Beta turn | Unordered | Total |
| Control (PgHsp 70) | 0.582 | 0.222 | −0.001 | 0.000 | 0.051 | 0.144 | 0.996 |
| Temp. 35°C–85°C | 0.582 | 0.222 | −0.001 | 0.000 | 0.051 | 0.144 | 0.996 |
| pH (4, 8 and 10) | 0.582 | 0.222 | −0.001 | 0.000 | 0.051 | 0.144 | 0.996 |
| PgHsp 70 + Gdn.HC l (0.1 mM) | 0.069 | 0.138 | 0.172 | 0.069 | 0.190 | 0.363 | 1.000 |
| PgHsp 70 + Gdn.HC l (0.1 mM) + glycerol (0.001%) | 0.582 | 0.222 | −0.001 | 0.000 | 0.051 | 0.144 | 0.996 |
| PgHsp 70 + KI (0.1 mM) | 0.006 | 0.086 | 0.300 | 0.161 | 0.110 | 0.340 | 1.000 |
| PgHsp 70 + acrylamide (0.1 mM) | 0.542 | 0.262 | −0.001 | 0.000 | 0.051 | 0.144 | 0.996 |
| PgHsp 70 + Urea (0.1 mM) | 0.059 | 0.238 | 0.223 | 0.223 | 0.044 | 0.214 | 1.000 |
Figure 9.
Effect of urea: (A) on secondary structure of PgHsp70 spectra 1–3 represents different concentrations of urea (0.2 to 0.4 mM) with PgHsp70 (B) on Chaperone activity of native PgHsp70 (experimental conditions as mentioned in Fig. 5).
Figure 10.
Effect of SDS: (A) on secondary structure of PgHsp70 [spectra 1–5 represents PgHsp70 and different concentration of SDS (0.2 to 0.5%) with PgHsp70 (B) on chaperone activity (experimental conditions as mentioned in Fig. 5).
The excitation spectrum of native PgHsp70 showed λmax excitation at 325 nm, when emission was fixed at 340 nm (data not shown). When the fluorescence spectra of PgHsp70 were recorded as a function of pH, the fluorescence intensity did not change with shifting of pH from neutral to acidic and basic range (Fig. 11). As the pH was changed from 8 to 10 and 8 to 4, there was no significant change in the α-helix content (Table 2) and further the chaperone activity of the PgHsp70 was not affected at extreme pH. These observations also suggested that by preserving the secondary structure the chaperone activity can be retained.
Figure 11.
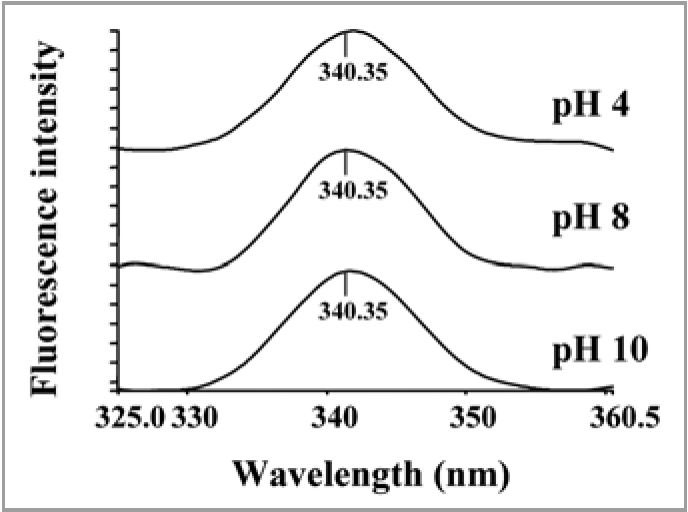
Effect of pH (A) on fluorescence emission spectra of PgHsp70 at 325 nm excitation.
The PgHsp70 chaperone activity enhanced with an increase in temperature from 35 to 45°C, and thereafter it changed slightly (Fig. 6C). This suggested that the chaperone activity of PgHsp70 is retained even at elevated temperatures.
PgHsp70 helps solubilization of recombinant carbonic anhydrase.
Carbonic anhydrases are zinc-containing enzymes catalyzing the reversible hydration of CO2. They are abundant in plants and algae, where they are essential for photosynthetic CO2 fixation.49 When we expressed carbonic anhydrase in E. coli, the partially folded protein intermediates were found to aggregate into inclusion bodies. Inclusion bodies produced in cytosol of all bacterial expression systems are composed of densely packed denatured protein molecules in the form of particles.27 Refolding of inclusion body proteins into biologically active forms requires treatment with some conventional chemical agents such as Tween-20, 40, 60 and 80; high concentration of urea and Triton X-100.27 However, these treatments are highly costly, time consuming, and the recovery is low. We were able to obtain the recombinant carbonic anhydrase in soluble fraction when it was co-expressed along with a molecular chaperone PgHsp70 (Fig. 12). These results clearly suggest that simultaneous co-expression of molecular chaperones like PgHsp70 can meet the additional demand for protein folding support and protein quality control system, under conditions when E. coli molecular chaperone system alone is unable to cope with the rate of recombinant carbonic anhydrase protein synthesis under IPTG induced condition. The recombinant carbonic anhydrase protein was available as a soluble fraction from 100 μl of E. coli lysate. It is shown to have conserved enzymatic activity determined at 220 U ml−1.
Figure 12.
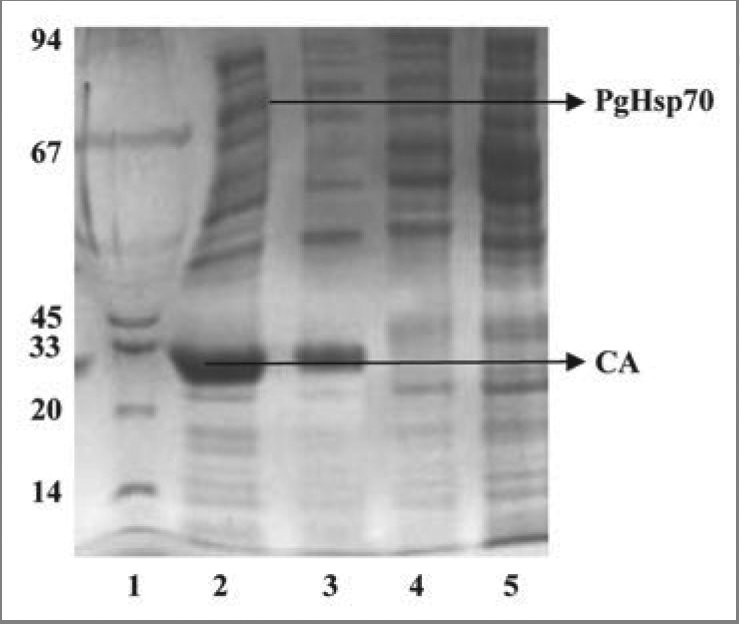
Coomassie brilliant blue-stained SDS-PAGE gel. Electrophoretic fractionation of simultaneous overexpression of PgHsp70 along with CA prevents the aggregation and accordingly prevents the formation of inclusion bodies in E. coli: Lane 1 is molecular weight of marker lane; the sizes (in kDa) are indicated adjacent (left side) to the gel. Lane 2 is co-expression of CA with PgHsp70 showing the high level of overproduction of CA in supernatant after after 3 h induction with IPTG (IPTG induced supernatant fraction). Lane 3 is induced CA in the pellet fraction (without PgHsp70); i.e., without co-expression the CA protein was going to pellet fraction. Lane 4 is a pellet fraction of only PgHsp70. Lane 5 is pellet fraction after co-expression of CA with PgHsp70; the CA and PgHsp70 proteins are not appearing. Lanes 4 and 5 were run in another gel and Coomassie Brilliant Blue stained.
Materials and Methods
Substrates and components for the chaperone assay.
Glutamate dehydrogenase, alcohol dehydrogenase, KI, CsCl, acrylamide, urea, SDS, p-nitrophenylacetate and Gdn.-HCl were purchased from Sigma (USA), Ni2+-NTA resin (Qiagen). Buffer solutions were prepared with ultra-high quality water (ELGA UGQ, UK). All other chemicals were of analytical grade. Pennisetum glaucum seeds (line 863B; a drought tolerant parental line) were procured from International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, India. Seeds were surface sterilized and then germinated as described earlier.28
Construction of P. glaucum cDNA Library.
A cDNA library was constructed from 5 μg of Poly A RNA in Uni-ZAP XR vector using Zap-cDNA synthesis kit (Stratagene, La Jolla, CA) following the manufacturer’s protocol. The resulting phage library contained 1 × 109 plaque forming units per milliliter. Sequence analysis of one of the clones identified a full-length cDNA that showed a highest (99%) with BLASTx chloroplast-Hsp70 of Cucumix sativus [Cs] (Acc. No. ABM92419) homology to the PgHsp70 (Acc.No. CD726127).
Cloning, expression and purification of recombinant PgHsp70.
To clone and express PgHsp70 in E. coli as a N-terminal His-tagged recombinant protein of a prokaryotic expression vector, pET-28a(+) was used for PCR amplification of coding region. Two oligo nucleotides were designed; one for the N-terminus region, 5′-CGA GTC CAT ATG GCC GCG AAG GGA GAC GG-3′ and the other for the C-terminus region, 5′-CTA GGA TCC TTT AGT CGA CTT CCT CGA TCT TG-3′ of PgHsp70. The 5′ and 3′ untranslated regions were removed and an Nde I site at the translation initiation site and a Not I site just downstream of the translation termination codon were introduced. Using these primers the complete coding sequence for the PgHsp70 was PCR amplified (150 ng of each primer, 200 mM dNTPs, 2.5 units Taq DNA polymerase in a 50 μl reaction; 94°C 1 min; 55°C 1 min and 72°C 1 min; 30 cycles) using total cDNA as template. The amplified product was gel purified, digested with Nde I and Not I and cloned into Nde I and Not I sites of pET-28a(+) expression vector.
Overnight bacterial culture of E. coli BL21 transformed with the respective expression plasmids were inoculated to fresh LB broth containing 50 μg ml−1 kanamycin and incubated at 200 rpm and 37°C to obtain an optical density of 0.7. at 600 nm. The production of recombinant protein PgHsp70 was induced in the bacterial cells by the addition of IPTG to a final concentration of 1 mM in the culture broth. After 3 h of incubation the bacterial cells were harvested by centrifugation at 8,000 xg for 10 min at 4°C. The bacterial cell pellet was resuspended in lysis buffer containing 20 mM Tris (pH 8.0), 100 mM sodium chloride and lysed by treatment with 0.5 mg ml−1 lysozyme for 30 min at 37°C followed by sonication. The lysate was cleared by centrifugation at 8,000 xg for 20 min and then supernatant loaded onto Ni-NTA column pre-equilibrated with lysis buffer containing 5 mM imidazole. The column was washed with lysis buffer, followed by elution of protein fractions with increasing concentrations of imidazole (50, 100, 150 mM) in the lysis buffer (50, 100 and 150 mM). The collected fractions were analyzed by measuring the optical density at 280 nm and this was followed by SDS-PAGE analysis (we have loaded the same amount (0.1 mg ml−1) of protein loaded on each lane). The purified PgHsp70 was dialyzed extensively, with at least four or five changes of dialysis buffer (0.1 mM phosphate buffer, pH 8.0) to remove Tries. The concentration of PgHsp70 protein was estimated by using bovine serum albumin (BSA) as the standard according to the standard method of Bradford.
Real-time PCR analysis of Pghsp70 transcript.
The real time quantitative PCR amplification of Pghsp70 along with Pg-β tubulin as an internal housekeeping standard was performed with specific oligonucleotide primers (FORWORD-5′-CGC ATT GAA CCA GGA AGT CAT G-3′, REVERSE-5′-CCC CTC ATT TGC TGT CGG TAA-3′), using the first strand cDNA synthesized from RNA samples collected from 12-d old Pennisetum seedlings exposed to different stress [heat (48°C), salt (250 mM), drought (withholding water up to 34 h), cold (4°C), ABA (100 μM) and SA (1 mM)] along with control seedlings, in the presence of SYBR-GreenR using Icycler (BioRad, USA). At the end of the PCR cycles, the products were put through a melt curve analysis to check the specificity of PCR amplification. The experiments were repeated at least five times independently and the data were averaged. The relative change in expression levels of Pghsp70 in response to different stress conditions and application of ABA and heat stress was estimated using REST software.29 with β-tubulin as a reference gene.
HPLC analysis. The protein PgHsp70 (0.1 mg ml−1) sample was filtered through 0.22 μm filter (Pall Gellman Lab, Ann Arbor, MI) before applying onto the column with an auto sampler. The reverse-phase-HPLC analysis (detector: 280 nm) on C18 (Phenomenex, C18, 5 μμ 1.D. 250 × 4.6 μm) using solvent of aceto-nitrile (0.1% TFA)/water (0.1% TFA) G gradient column for 0–15 min at the rate of 2% min−1 solvent; 15 to 45 min at the rate of 0.5% min−1 flow rate of 1.0 ml min−1.
Chaperone activity assay of PgHsp70 by spectrophotometry and densitometery.
The ability of PgHsp70 to prevent the thermal aggregation of protein substrates such as glutamate dehydrogenase and alcohol dehydrogenase at 50°C temperature was assayed by incubating the reaction mixture (1 ml) containing 20 mM Tris buffer (pH 8.0), 100 mM sodium chloride, 0.2 mg ml−1 glutamate dehydrogenase or alcohol dehydrogenase and 0.2 mg ml−1 of the recombinant heat shock protein PgHsp70 (with or without ATP 5 mM) or BSA (0.2 mg ml−1). The assay was carried out according to the method of Jaenicke and Rudolph30 using a Peltier-controlled cell and the aggregation of the protein substrates monitored by measuring the light scatter over different time intervals at 360 nm in a JASCO-810 Spectropolarimeter. Protein aggregation was also measured by determining the amount of protein that remained soluble or pelleted at 10,000 xg for 30 min at 50°C, followed by SDS-PAGE to determine the relative amounts of the GDH or ADH that remained in solution or aggregated in the presence or absence of the recombinant PgHsp70.
100% chaperone activity means: The activity which was performed at 50°C by incubating reaction mixture 1 ml containing of 20 mM Tris buffer (pH 8.0), 100 mM sodium chloride, 0.2 mg ml−1 glutamate dehydrogenase and 0.2 mg ml−1 of the recombinant heat shock protein PgHsp70 (without ATP) or BSA and any denaturants considered as control of 100% chaperone activity.
Effect of temperature and pH on chaperone activity of PgHsp70.
Effect of temperature on the recombinant PgHsp70 chaperone activity was assayed at pH 8.0 (phosphate buffer) by pre-incubating PgHsp70 at various temperatures (35–95°C) for 2 h, followed by relative PgHsp70 chaperone activity, was determined by using the reaction mixture incubated at 50°C. The effect of pH on chaperone activity of PgHsp70 was evaluated by pre-incubating the PgHsp70 in different 0.1 M buffers [Glycine-HCl, pH 4.0; phosphate (K2HPO4-KH2PO4), pH 8.0; Glycine-NaOH, pH 10.0] for 2 h and relative PgHsp70 chaperone activity was determined by using the reaction mixtures at pH 8.0 for 5 min.
Thermostability, half-life and pH stability of PgHsp70.
Thermostability and half-life of PgHsp70 were determined by incubating 5 ml of the suitably diluted (dilutions of 0.2 mg ml−1 were prepared in 0.1 M phosphate buffer at pH 8.0) protein sample at 50°C (with/without glycerol 0.001%), 90 and 80°C over a period of 10 h, and subsequently assayed for chaperone activity at 50°C. The pH stability of PgHsp70 (0.2 mg ml−1) was determined by subjecting it to different pH values (0.1 M acetate buffer, pH 4.0 and 5.0; 0.1 M phosphate buffer, pH 8.0; and 0.1 M Tris-glycine buffer, pH 10.0) over a period of 7 h, and subsequently assayed for chaperone activity at 50°C and pH 8.0.
Secondary structure determination of PgHsp70.
The secondary structure of the purified PgHsp70 (0.1 mg ml−1 prepared in 0.1 M phosphate of pH 8.0) was determined by CD spectroscopy carried out at 45°C on a JASCO-810 Spectropolarimeter equipped with Peltier controlled thermostat cell holder (PTC-423S). The path length of cuvette used was 0.2 cm. Temperature scans were performed by exposing the PgHsp70 to different temperatures (35 to 85°C) and the changes in the structural conformation were recorded at a scanning rate of 20 nm min−1 from 190 to 240 nm (the significant changes in the CD spectrum were analyzed in the region of 207 to 222 nm). The CD signal was converted into total molar ellipticity [θ], and the secondary structure of the purified PgHsp70 was determined by using the average of 10 scans with blank-subtraction and plotting as per Sreerama and Woody31 (Dichroweb software; London, UK).
Effect of KI, urea, SDS, Gdn-HCl and pH on the secondary structure and chaperone activity of PgHsp70.
The purified PgHsp70 (The concentration of protein 0.1 mg ml−1 prepared in 0.1 M phosphate of pH 8.0) was titrated at room temperature against different concentrations of KI, urea, Gdn.HCl and SDS. The changes in the structural conformation by CD and chaperone activity of PgHsp70 were recorded. The effect of pH on PgHsp70 was studied by using different pH buffers (Glycine-HCl for pH 4.0; phosphate for pH 8.0; Glycine-NaOH for pH 10.0) and changes in the secondary structural conformation of the protein and its in-vitro chaperone activity were recorded.
Effect of pH on PgHsp70 by fluorescence measurements.
The steady-state fluorescence measurements were performed in a Hitachi F-4500 fluorescence spectrofluorometer at room temperature using a slit width of 5/5 nm (exc/emi) with a slow scan of 15 nm min-1 and quartz cells of 1 cm path length. Fluorescence emission spectra were recorded at 340 nm by using excitation wavelengths of 325 nm for the tryptophan (W) and tyrosine (Y) fluorophore. The protein samples of PgHSP70 were prepared in different 0.1 M buffer systems (Glycine-HCl, pH 4; phosphate [K2HPO4-KH2PO4] pH 8.0; Glycine-NaOH pH 10.0) and changes in their intrinsic fluorescence intensity were recorded at 280 nm.
Co-expression of PgHsp70 with carbonic anhydrase.
Initially the carbonic anhydrous coding region was cloned in Nde I and BamH I sites of pET14b the resultant construct expressed the recombinant carbonic anhydrase in E. coli which completely partitioned into inclusion bodies. The same recombinant carbonic anhydrase expression cassette from pET14b was PCR amplified along with T7 promoter and T7 terminator using the following oligonucleotide forward 5′GCG GCC GCA TCA TGG CGA CCA CAC CCG TCC-3′ and reverse primer 5′GAG CTC CGA AGT GGC GAG CCC GAT CT3′ synthesized based on the sequence information of pET14b vector. The Not I and Sac I restriction enzyme sites were introduced in the primers. The PCR amplified carbonic anhydrous expression cassette was re cloned into Not I and Sac I sites of pET28a in which PgHsp70 gene was already cloned into Nde I and Not I sites as mentioned earlier. The recombinant carbonic anhydrase expression cassette from pET14b starting from T7 promoter to T7 terminator was PCR amplified using the following oligonucleotide primers with Not I and Sac I restriction enzyme sites (Not I: 5′GCG GCC GCA TCA TGG CGA CCA CAC CCG TCC-3′ and Sac I: 5′GAG CTC CGA AGT GGC GAG CCC GAT CT3′) and re cloned into Not I and Sac I sites of pET28a in which PgHsp70 gene was already cloned into Nde I and Not I sites as mentioned earlier. The simultaneous overexpression of both recombinant proteins was induced in E. coli for two hours in the presence of 1 mM IPTG. The presence of both recombinant carbonic anhydrase as well as recombinant HSP70 proteins was analyzed both in the soluble and insoluble fractions of the E. coli lysate.
Assay for carbonic anhydrase.
The carbonic anhydrase activity was determined by the method of Armstrong et al.32 by incubating the reaction mixture containing 1.0 ml of 5 mM p-Nitrophenyl acetate prepared in 0.1 M phosphate buffer (pH 8.0) with 0.2 ml of appropriately diluted carbonic anhydrase (0.2 mg ml−1) for 5 min at room temperature, and determining the hydrolyzed p-Nitrophenylacetate at 348 nm. One carbonic anhydrase unit is defined as one m mole of p-nitrophenylacetate hydrolyzed ml−1 min−1 under the assay conditions.
Homology modeling structure determination.
The 3D structure of PgHsp70 was generated using first approach mode of the comparative modeling tools available at the Swiss model server. The PDB crystal structure (1YUW, 3) of a bovine HSC70 (amino acids: 1–554, resolution 2.6 Å). Jiang et al.33 was used as template to generate the comparative protein model. The molecular graphics were generated using MOLMOL.34 All the experiments were performed in three times and mean of the three values with SD within 10% was taken.
Conclusions
The data presented in this study clearly suggests that Hsp70 from Pennisetum glaucum is a thermostable protein and retains its chaperone activity at higher temperatures. By comparing specific structural property in relation to the chaperone activity of PgHsp70, it was found that there was no change in the α-helix content or the chaperone activity with the increase in temperature (45–85°C); this hinted a critical role of α-helix in maintaining its chaperone activity. Though thermostable in nature, PgHsp70 showed an overall 3-D structure similar to that of bovine’s HSC-70 and exhibit positive ability to refold and help express recombinant protein (e.g., CA) in soluble form rather than in the inclusion bodies in E. coli. Finally the chaperone activity of PgHSP70 on different protein substrates, high thermostability, response to broader pH range and ATP-independent in-vitro chaperone activity reflect on its novel properties and suggest that it may be a good candidate for cell homeostasis at high temperature stress. Overall, these features will have important implications on the expression of recombinant proteins in soluble and active form.
Acknowledgements
We thank to Dr. Pavan Umate, Mr. A. Mishra and Mr. Dang Quang Hung (International Centre for Genetic Engineering and Biotechnology, New Delhi) for their help in preparation of the manuscript. We also thank to Prof. T. Satyanarayana for critical reading of the manuscript (University of Delhi, South campus New Delhi). This work was financially supported by the Department of Biotechnology (Ministry of Science and Technology, Government of India) research grants.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10547
References
- 1.Ritossa F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia. 1962;18:571–573. [Google Scholar]
- 2.Studer S, Narberhaus F. Chaperone activity and homo- and hetero-oligomer formation of bacterial small heat shock proteins. J Biol Chem. 2000;275:37212–37218. doi: 10.1074/jbc.M004701200. [DOI] [PubMed] [Google Scholar]
- 3.Becker J, Craig EA. Heat-shock proteins as molecular chaperones. Eur J Biochem. 1994;219:11–23. doi: 10.1007/978-3-642-79502-2_2. [DOI] [PubMed] [Google Scholar]
- 4.Hartl FU, Hlodan R, Langer T. Molecular chaperones in protein folding the art of avoiding sticky situations. Trends Biochem Sci. 1994;19:20–25. doi: 10.1016/0968-0004(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 5.Hendrick JP, Hartl FU. The role of molecular chaperones in protein folding. The Fed Am Soci Exper Biol J. 1995;9:1559–1569. doi: 10.1096/fasebj.9.15.8529835. [DOI] [PubMed] [Google Scholar]
- 6.Mosser DD, Caron AW, Bourget L, Denis-Larose C, Massie B. Role of the human heat shock protein Hsp70 in protection against stress-induced apoptosis. Mol Cell Biol. 1997;17:5317–5327. doi: 10.1128/mcb.17.9.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fink AL. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 8.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1988;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 9.Zavilgelsky GB, Kotova VY, Mazhul’ MM, Manukhov IV. Role of Hsp70 DnaK-DnaJ-GrpE and Hsp100 ClpA and ClpB chaperones in refolding and increased thermal stability of bacterial luciferases in Escherichia coli cells. Biochem Mosc. 2002;67:986–992. doi: 10.1023/a:1020565701210. [DOI] [PubMed] [Google Scholar]
- 10.Smykal P, Masin J, Hrdy I, Konopasek I, Zarsky V. Chaperone activity of tobacco HSP18 a small heatshock protein is inhibited by ATP. Plant J. 2000;23:703–713. doi: 10.1046/j.1365-313x.2000.00837.x. [DOI] [PubMed] [Google Scholar]
- 11.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, et al. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;10:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 12.Ziemienowicz A, Zylicz M, Floth C, Hubscher U. Calf thymus Hsc 70 protein protects and reactivates prokaryotic and eukaryotic enzymes. J Biol Chem. 1995;270:15479–15484. doi: 10.1074/jbc.270.26.15479. [DOI] [PubMed] [Google Scholar]
- 13.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding refolding and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 14.Flynn GC, Chappell TG, Rothma JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 15.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gotesman ME, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slepenkov SV, Witt SN. Kinetic analysis of interdomain coupling in a lidless variant of the molecular chaperone DnaK: DnaK’s lid inhibits transition to the low affinity state. Biochem. 2002;41:12224–12235. doi: 10.1021/bi0263208. [DOI] [PubMed] [Google Scholar]
- 17.Grover A. Molecular biology of stress responses. Cell Stress Chaperones. 2002;7:1–5. doi: 10.1379/1466-1268(2002)007<0001:mbosr>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katiyar-Agarwal S, Agarwal M, Grover A. Heattolerant basmati rice engineered by overexpression of hsp101. Plant Mol Biol. 2003;51:677–686. doi: 10.1023/a:1022561926676. [DOI] [PubMed] [Google Scholar]
- 19.Sun W, Montagu MV, Verbruggen N. Small heat shock proteins and stress tolerance in plants. Biochim Biophy Acta. 2002;1577:1–9. doi: 10.1016/s0167-4781(02)00417-7. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Willmund F, Golecki JR, Cacace S, Hess B, Market C, et al. The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas. Plant J. 2007;50:265–277. doi: 10.1111/j.1365-313X.2007.03047.x. [DOI] [PubMed] [Google Scholar]
- 21.Cho EK, Bae SJ. ATP-independent thermoprotective activity of Nicotiana tabacum heat shock protein 70 in Escherichia coli. J Biochem Mol Biol. 2007;40:107–112. doi: 10.5483/bmbrep.2007.40.1.107. [DOI] [PubMed] [Google Scholar]
- 22.Tiroli AO, Ramos CH. Biochemical and Biopysical characterization of small heat shock proteins from sugarcane. Involvement of a specific region located at the N-terminus with substrate specificity. The Inter J Biochem Cell Biol. 2007;398:18–831. doi: 10.1016/j.biocel.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Cho EK, Hong CB. Overexpression of tobacco NtHSP70-1 contributes to drought-stress tolerance in plants. Plant Cell Rep. 2006;25:349–358. doi: 10.1007/s00299-005-0093-2. [DOI] [PubMed] [Google Scholar]
- 24.Liu HT, Li GL, Chang H, Sun DY, Zhou RG, Li B. Calmodulin-binding protein phosphatase PP7 is involved in thermotolerance in Arabidopsis. Plant Cell Environment. 2007;30:156–164. doi: 10.1111/j.1365-3040.2006.01613.x. [DOI] [PubMed] [Google Scholar]
- 25.Swindell WR, Huebner M, Weber AP. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BioMed Cen Gen. 2007;8:125. doi: 10.1186/1471-2164-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su PH, Li HM. Arabidopsis stromal Hsp70s are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 2008;146:1231–1241. doi: 10.1104/pp.107.114496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh SM, Panda AK. Solubilization and refolding of bacterial inclusion body proteins. J Biosci Bioengin. 2005;99:303–310. doi: 10.1263/jbb.99.303. [DOI] [PubMed] [Google Scholar]
- 28.Mishra RN, Reddy PS, Nair S, Markandeya G, Reddy AR, Sopory SK, et al. Isolation and characterization of expressed sequence tags ESTs from subtracted cDNA libraries of Pennisetum glaucum seedlings. Plant Mol Biol. 2007;64:713–732. doi: 10.1007/s11103-007-9193-4. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaenicke R, Rudolph R. A Practical Approach in Protein Structure Creighton TE. In: Creighton TE, editor. A Practical Approach in Protein Structure. Oxford: IRL Press; 1989. pp. 191–223. [Google Scholar]
- 31.Sreerama N, Woody RW. PolyProII helices in globular proteins: Identification and circular dichroic. Anal Biochem. 1994;33:10022–10025. doi: 10.1021/bi00199a028. [DOI] [PubMed] [Google Scholar]
- 32.Armstrong JMcD, Myers DV, Verpoorte JA, Edsall JT. Purification and properties of human erythrocyte carbonic anhydrases. J Biol Chem. 1966;241:5137–5149. [PubMed] [Google Scholar]
- 33.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell. 2005;20:513–524. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph Model. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 35.Guy CL, Li QB. The organization and evolution of the spinach stress 70 molecularchaperone gene family. Plant Cell. 1998;10:539–556. doi: 10.1105/tpc.10.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu FH, Wu SJ, Hu SM, Hsiao CD, Wang C. Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J Biol Chem. 1999;274:34425–4432. doi: 10.1074/jbc.274.48.34425. [DOI] [PubMed] [Google Scholar]
- 37.Boice JA, Hightower LE. A Mutational Study of the peptide-binding domain of Hsc70 guided by secondary structure prediction. J Biol Chem. 1997;272:24825–24831. doi: 10.1074/jbc.272.40.24825. [DOI] [PubMed] [Google Scholar]
- 38.Ramya TNC, Surolia N, Surolia A. 15-Deoxyspergualin modulates Plasmodium falciparum heat shock protein function. Biochem Biophy Res Comm. 2006;348:585–592. doi: 10.1016/j.bbrc.2006.07.082. [DOI] [PubMed] [Google Scholar]
- 39.Archer EK, Keegstra K. Analysis of chloroplast transit peptide function using mutations in the carboxyl-terminal region. Plant Mol Biol. 1993;23:1105–1115. doi: 10.1007/BF00042345. [DOI] [PubMed] [Google Scholar]
- 40.Sichting M, Mokranjac D, Azem A, Neupert W, Hell K. Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J. 2005;24:1046–1056. doi: 10.1038/sj.emboj.7600580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scopio A, Johnson P, Laquerre A, Nelson DR. Subcellular localization of chaperone activities of Borrelia burgdorferi Hsp60 and Hsp70. J Biol Chem. 1994;176:6449–6456. doi: 10.1128/jb.176.21.6449-6456.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satyanarayana T, Noorwez SM, Kumar S, Rao JLUM, Ezhilvannan M, Kaur P. Development of an ideal starch saccharification process using amylolytic enzymes from thermophiles. Biochem Soci Tran. 2004;32:276–278. doi: 10.1042/bst0320276. [DOI] [PubMed] [Google Scholar]
- 43.Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 44.Naik RR, Kirkpatrick SM, Stone MO. The thermostabilty of an α-helical coiled-coil protein and its potential use in sensor applications. Biosen Bioelect. 2001;16:1051–1057. doi: 10.1016/s0956-5663(01)00226-3. [DOI] [PubMed] [Google Scholar]
- 45.Lupas A. Coiled coils: new structures and functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 46.Eftink MR, Ghiron CA. Fluorescence quenching studies with proteins. Anal Biochem. 1981;114:199–227. doi: 10.1016/0003-2697(81)90474-7. [DOI] [PubMed] [Google Scholar]
- 47.Ghatge MS, Deshpande VV. Evidence for specific interaction of guanidine hydrochloride with carboxy groups of enzymes/proteins. Biochem Biophy Res Comm. 1993;193:979–984. doi: 10.1006/bbrc.1993.1721. [DOI] [PubMed] [Google Scholar]
- 48.Palleros DR, Shi L, Reid KL, Fink AL. Hsp70-protein complexes. Complex stability and conformation of bound substrate protein. J Biol Chem. 1994;269:13107–13114. [PubMed] [Google Scholar]
- 49.Lu Y, Stemler K. Extrinsic phytosystem II carbonic anhydrase in maize mesophyll chloroplasts. Plant Physiol. 2002;128:643–649. doi: 10.1104/pp.010643. [DOI] [PMC free article] [PubMed] [Google Scholar]