Abstract
Mitochondrial atftsh4 protease, whose catalytic site is exposed to the intermembrane space, is one of four inner membrane-bound ftsh proteases in arabidopsis. we found that the loss of atftsh4 altered arabidopsis leaf morphology at the late stage of rosette growth under short-day photoperiod, while such changes were not observed in ftsh4 mutants grown under long days. these morphological changes were correlated with elevated levels of both reactive oxygen species (ros) and carbonylated proteins, which strongly suggested that ageing ftsh4 plants experienced oxidative stress. this view was supported by the accumulation of electron-dense material, presumably containing aggregated oxidized proteins, in mitochondria of ftsh4 plants with the most strongly malformed leaf blades. taken together, our data published in the may issue of plant journal1 suggest a link between the lack of AtFtsH4 protease, oxidative stress and altered leaf morphology at the late rosette stage under short days. Here, we present evidence that the onset of altered leaf morphology in ftsh4 correlates with an increase in the abundance of AtFtsH4 transcript observed in wildtype Arabidopsis growing under the same conditions. We also discuss how the lack of AtFtsH4 may cause oxidative stress towards the end of the vegetative growth in short days.
Key words: plant mitochondria, FtsH proteases, AAA proteases, oxidative stress, oxidized proteins, leaf morphology, photoperiod, Arabidopsis
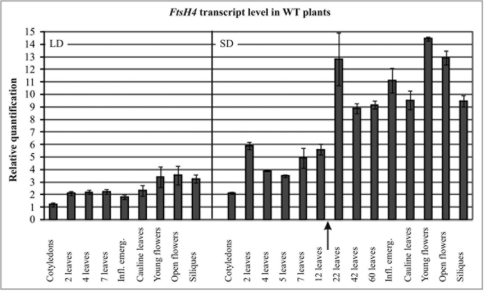
In Arabidopsis, four membrane-bound FtsH proteases, members of the ATP-dependent metalloprotease family, reside in mitochondria.2 One of them is AtFtsH4 with catalytic sites exposed to the intermembrane space.3 In yeast and mammals, FtsH proteases control mitochondrial protein quality, act as processing peptidases or serve as chaperones independently of their proteolytic function.4–7 Using two T-DNA null mutants we investigated the role of AtFtsH4 in plant growth and development.1 We found that the loss of this protease significantly affects morphogenesis of Arabidopsis rosette leaves at the late stage of vegetative growth in shortday conditions (SD, 8 h light per day). In contrast, no morphological consequences of the lack of AtFtsH4 were detected at the early stage of vegetative growth in SD as well as during the entire rosette vegetative growth under long day conditions (LD, 16 h light per day). The most striking features of ageing ftsh4 grown under SD conditions were a distinct asymmetry and irregular serration of leaf blades. The strength of these developmental disturbances increased towards the end of the vegetative growth and was well correlated with elevated levels of reactive oxygen species (ROS) and carbonylated proteins, which strongly indicated that ageing ftsh4 plants suffered from oxidative stress. This view was supported by the presence of tiny accumulations of electron-dense material, presumably aggregates of oxidized proteins, in the mitochondrial matrix of ftsh4 plants with the leaf-blade morphology severely affected. However, no such accumulations were visible in mitochondria of young ftsh4 plants. In this addendum, we present additional data supporting the role of AtFtsH4 in the late phase of Arabidopsis vegetative growth under short-day conditions. The time-course of the AtFtsH4 transcript accumulation was followed in wild-type plants grown under LD and SD conditions using quantitative PCR (Fig. 1). In the LD, AtFtsH4 transcripts remained almost constant in rosette leaves during the entire vegetative growth. Under SD conditions, the abundance of AtFtsH4 transcript was almost the same in leaves appearing at the early stage of rosette development but this level turned out to be substantially higher in leaves of ageing Arabidopsis plants. Moreover, the similar high level was also observed in all generative organs. Interestingly, the increase of the AtFtsH4 transcript level in wild-type plants occurs at exactly the same developmental time, when first, however subtle, malformations in leaf-blade morphology become visible in ftsh4 plants.
Figure 1.
Time-course of AtFtsH4 transcript level detected by quantitative PCR in wild-type plants under long (LD) and short (SD) day conditions. At the early growth stage the transcript level was measured in cotyledons or two-leaved seedlings. Later, between the fourth leaf stage and inflorescence emergence (Inf.emerg.) the youngest leaf was always sampled. In the generative phase, cauline leaves, buds, open flowers and siliques were tested. The arrow indicates the time when subtle leaf malformations become visible in the ftsh4 mutant. Error bars indicate SE.
Taken together, our data suggest a link between the lack of AtFtsH4 protease, oxidative stress and altered leaf morphology under certain developmental and environmental conditions. Our results indicate that the loss of AtFtsH4 leads to the mild deficiency of the mitochondrial oxidative system caused by impaired assembly/stability of respiratory complexes regardless of the growth stage and the day-length.1,8 The impaired respiratory activity may result in an increased production of ROS and oxidized proteins with deleterious effects on mitochondrial function. On the other hand, it is well known that mitochondria have a threshold below which they can accommodate the increased level of ROS and oxidized proteins as by-products of a dysfunctional oxidative system.9 We believe that the defense system devoid of the AtFtsH4 protease is sufficiently efficient only in the short term and in optimal growth conditions (long days, early stage of growth in short days). However, in the SD photoperiod when the vegetative phase is much longer than in LD, plants have to deal with longer dark periods which enhance the role of mitochondria in energy production, which in turn results in increased oxidative parameters in ageing rosettes of the ftsh4 mutant.
Another possibility that does not exclude the first one is that AtFtsH4 may play a more specific role at the end of the vegetative growth in addition to the housekeeping function required during the entire Arabidopsis life. The AtFtsH protease, like its yeast or bacterial homologues, is expected to have numerous substrates and therefore may be involved in multiple molecular pathways preventing the oxidative stress. The increased abundance of AtFtsH4 transcripts in ageing wild-type rosettes in the SD photoperiod supports the view that AtFtsH4 may have an important function towards the end of the vegetative growth. It was reported by Johansson et al.10 that a carbonylated protein level in Arabidopsis rosette leaves increases progressively with age and then declines rapidly at the end of the vegetative growth. Thus, it is possible that one of the putative functions of the AtFtsH4 protease at this stage of plant development is to prevent accumulation of carbonylated proteins produced by ROS-generated oxidative stress. It has been suggested that protein oxidation leads to a partial loss of its secondary structure without disturbing the overall folding pattern, resulting in flexible regions that serve as targeting signals for degradation.11 In agreement with this view, degradation of oxidized proteins requires enzymes sensing the local protein unfolding. Unlike other ATP-dependent proteases, FtsH proteases lack a robust unfoldase activity and use the folding state of their substrates as a criterion for degradation.12 Thus, the AtFtsH4 may recognize oxidized proteins and then degrade them or deliver them for degradation mediated by other proteases. In mammals, the Lon protease, an ATP-dependent serine protease, plays a crucial role in the degradation of oxidized proteins.13 Likely, the lack of AtFtsH4 limits the capacity of mitochondrial system controlling the level of oxidized proteins. This defect can be perceptible mostly under conditions favorable for accumulation of oxidized proteins, as in ageing rosettes of ftsh4 mutant in SD, causing mitochondrial dysfunctions and, subsequently, impairing cell and leaf development. We believe that the abnormality in leaf formation could be associated with an accumulation of oxidatively damaged proteins in mitochondria of both meristematic and growing leaf cells, which is progressive with the age of ftsh4 plants. This assumption could explain why the malformation of emerging leaves progressed with every initiated leaf of aging ftsh4 rosettes. Furthermore, the developmental retardation and the inability of 85% of ftsh4 plants to enter flowering phase under SD conditions could result from the impairment of the meristem function due to progressive accumulation of oxidatively damaged proteins. However, the exact relations between observed malformations at the organismal level and molecular and physiological reasons laying at their background require further investigations.
Acknowledgement
This research was supported by a grant from the Ministry of Education and Science, Poland (grant number N N303 3504 33) to H.J.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10303
References
- 1.Gibala M, Kicia M, Sakamoto W, Gola EM, Kubrakiewicz J, Smakowska E, et al. The lack of mitochondria AtFtsH4 protease alters Arabidopsis leaf morphology at the late stage of rosette development under short-day photoperiod. Plant J. 2009;59:685–699. doi: 10.1111/j.1365-313X.2009.03907.x. [DOI] [PubMed] [Google Scholar]
- 2.Janska H. ATP-dependent proteases in plant mitochondria: What do we know about them today? Physiol Plant. 2005;123:399–405. [Google Scholar]
- 3.Urantowka A, Knorpp C, Olczak T, Kolodziejczak M, Janska H. Plant mitochondria contain at least two i-AAA-like complexes. Plant Mol Biol. 2005;59:239–252. doi: 10.1007/s11103-005-8766-3. [DOI] [PubMed] [Google Scholar]
- 4.Koppen M, Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol. 2007;42:221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- 5.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatsuta T, Augustin S, Nolden M, Friedrichs B, Langer T. m-AAA protease-driven membrane dislocation allows intermembrane claveage by rhomboid in mitochondria. EMBO J. 2007;26:325–335. doi: 10.1038/sj.emboj.7601514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rainey RN, Glavin JD, Chen HW, French SW, Teitell MA, Koehler CM. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol Cell Biol. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodziejczak M, Gibala M, Urantowka A, Janska H. The significance of Arabidopsis AAA proteases for activity and assembly/stability of mitochondria OXPHOS complexes. Physiol Plant. 2007;129:135–142. [Google Scholar]
- 9.Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem J. 2003;370:751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansson E, Olsson O, Nystrom T. Progression and specificity of protein oxidation in the life cycle of Arabidopsi thaliana. J Biol Chem. 2004;279:22204–22208. doi: 10.1074/jbc.M402652200. [DOI] [PubMed] [Google Scholar]
- 11.Kurepa J, Smalle JA. To misfold or to lose structure? Detection and degradation of oxidized proteins by the 20S proteasome. Plant Signal Behav. 2008;3:6. doi: 10.4161/psb.3.6.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman C, Prakash S, Lu CZ, Matouschek A, Gross CA. Lack of robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol Cell. 2003;11:659–669. doi: 10.1016/s1097-2765(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 13.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]