Abstract
The plant cell wall is a complex polysaccharide network and performs important developmental and physiological functions far beyond supplying the physical constrains. Plant cells have the ability to react to cell wall defects as exhibited by changes in gene expression, accumulation of ectopic lignin, stress responses and growth arrest. It is a major challenge to understand how plants sense and respond to wall integrity since very little is known about the signaling involved in the responses. Cellulose synthase-like D (CSLD) proteins mediating the biosynthesis of a wall polysaccharide polymer make up a common subfamily to all plants. Recently, we have reported the functional characterization of CSLD4 in rice. Mutations in OsCSLD4 show morphological alterations and pleiotropic effects on wall compositions and structure. Our study demonstrates that OsCSLD4 play a critical role in cell wall formation and plant growth. Here we show the subtle wall alterations by separating the culm residues into five fractions. Quantitative RT-PCR analysis further revealed that the expression of various genes involved in xylan synthesis and cell cycle regulation was altered in mutant plants, as the responses to OsCSLD4 disruption. Therefore, plants may have fine sensory machinery to react to wall defects and modulate growth for adapting to the changes.
Key words: OsCSLD4, cell wall biosynthesis, plant development, wall integrity, rice
Introduction
The plant cell wall is a complex network of polysaccharides mainly containing cellulose, hemicelluloses and pectins.1 Hemicellulosic polysaccharides, synthesized and modulated by numerous putative glycosyltransferases (GT) or glycosly hydrolases (GH), are macro molecules responsible for the crosslinking of cellulose microfibrils for providing a load-bearing matrix.2,3 Cellulose synthase like (CSL) genes belonging to an important GT superfamily encode proteins presumed to catalyze the biosynthesis of various β-linked glycan backbones.3
Traditional biochemical techniques and identification of relative mutants are general ways for understanding the specific functions of CSLs. Using these approaches, the polysaccharides synthesized by CSLA, CSLC, CSLF and CSLH have been discovered.4–7 However, the enzymatic activities of the other CSLs, such as CSLD, are less understood, even though several csld mutants have been reported in Arabidopsis and rice.8–13
Although the biochemical function of CSLD is still unknown, through identification of mutants in CSLD subfamily, all the CSLD members in Arabidopsis exhibit diverse developmental roles in root hair and pollen tube elongation and plant growth,8–13 indicating that the polysaccharide synthesized by CSLDs is essential for plant growth. Increasing evidence has shown that plants have the ability to sense the wall perturbations and trigger a set of cellular changes, including growth inhibition, lignin ectopic accumulation and wall compositional alterations.1,14 In contrast to yeast, in which a cell-wall-integrity-signaling pathway has been reasonably well characterized,15,16 very little is known about the signaling involved in plants response to wall defects. We recently reported mutation in OsCSLD4 causes pleiotropic effects on polysaccharides synthesis and plant growth.13 Here, we describe the alterations in xylan synthesis and cell growth in oscsld4 being translated into changes in gene expression, to understand how plants monitor the integrity of cell walls and possess roles in plant development.
Mutation in OsCSLD4 Causes Decreased Xylan and Cellulose Content in Mutant Culms
OsCSLD4 is an ortholog of AtCSLD5. The Arabidopsis atcsld5 knock out and T-DNA insertion mutants exhibited reduced growth and altered xylan levels.10 Without chemical analysis of the wall, the direct or indirect alterations in wall composition resulted from the disruption of AtCSLD5 have not been evaluated in detail. To further characterize the board changes of polysaccharides in oscsld4, we sequentially extracted the wall residues from the wild-type and mutant culms and performed sugar compositional analysis in the five fractions. As shown in Table 1, a number of wall compositional changes were observed. Some sugars in oscsld4, such as arabinose, xylose and glucose, were slightly increased in the fractions of pectin digested with endopolygalaturonase (EPG) plus pectin methylesterase (PME) or extracted with EGTA-Na2CO3. Contrastingly, xylose was decreased by ∼32.5% in hemicellulose fractions extracted with 1 N and 4 N KOH, resulting in the significantly decreased total sugars in these fractions. Because most arabinoxylan can be extracted by 1 N KOH, the decreased xylose in oscsld4 may also be a consequence of the reduced arabinoxylan solubility. Another major wall defect is a significant reduction in cellulose content. Our report on its Golgi apparatus localization suggests that OsCSLD4 is unlikely to participate in the synthesis of crystalline cellulose occurring at the plasma membrane.13 Deficiency in the synthesis of polysaccharide matrix might result in aberrant assembly of nascent cellulose microfibrils at the plasma membrane, which could, in turn, impede the further cellulose synthesis.17 The reduced cellulose is thus one of the possible responses to OsCSLD4 disruption.
Table 1.
Monosaccharide compositional analysis of the wall fractions derived from culms of mutant and wild-type plants
| Fractions | RHA | FUC | ARA | XYL | MAN | GLC | GAL | Total | |
| EPG | WT | 0.2 | 0.1 | 0.3 | 0.9 | 1.0 | 0.6 | 0.2 | 3.3 |
| oscsld4 | 0.3 | 0.3 | 0.6 | 1.2 | 0.9 | 0.6 | 0.5 | 4.5 | |
| Na2CO3 | WT | 0.1 | 0.4 | 0.2 | 0.4 | 0.1 | 0.3 | 0.2 | 1.6 |
| oscsld4 | 0.1 | 0.2 | 0.4 | 0.6 | 0.1 | 0.7 | 0.3 | 2.4 | |
| 1 N KOH | WT | 0.2 | 0.1 | 6.1 | 34.7 | 0.1 | 2.8 | 1.4 | 45.4 |
| oscsld4 | 0.2 | 0.1 | 5.8 | 23.4 | 0.1 | 5.4 | 2.0 | 37.0 | |
| 4 N KOH | WT | 0.1 | 0.1 | 1.1 | 8.5 | 0.1 | 3.0 | 0.4 | 13.2 |
| oscsld4 | 0.1 | 0.1 | 1.3 | 5.7 | 0.1 | 6.4 | 0.8 | 14.6 | |
| 4 N insol | WT | 294.7 | |||||||
| oscsld4 | 252.6 | ||||||||
| Total | WT | 0.5 | 0.7 | 7.8 | 44.5 | 1.2 | 301.4 | 2.2 | 358.2 |
| oscsld4 | 0.7 | 0.8 | 8.2 | 30.9 | 1.2 | 265.7 | 3.7 | 311.2 |
The alcohol insoluble wall residues (AIR) were extracted from the second internodes of oscsld4 and wild-type culms; The destarched AIR was fractioned sequentially with endopolygalacturonase (EPG) plus pectin methylesterase (PME), 50 mM Na2CO3, 1 N KOH, and 4 N KOH; The fractions were hydrolyzed with sulfuric acid and the alditol acetates were analyzed by GC-MS; The insoluble residue after 4 N KOH extraction was used to determine cellulose content by colorimetric assay; The results are given as means (mg g−1 of AIR) of four independent assays; The variance was not shown but is <15%.
The pleiotropic polysaccharide modifications in oscsld4 are possible results of that plants adapt to the loss of one polysaccharide component by altering the quantity of an alternative, or by modifying the structure of the wall.
The Expression of Genes Involved in Xylan Synthesis and Cell Cycle Regulation Is Altered in oscsld4
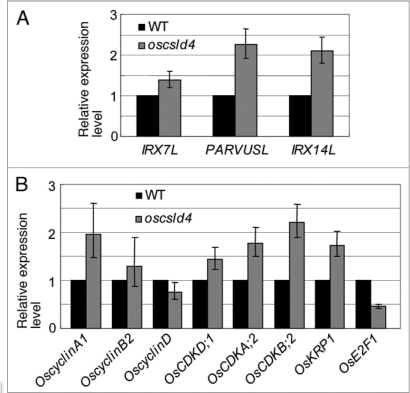
Xylan is a major component of hemicellulose in dicot and monocot walls, which is believed to be synthesized by protein complexes that include CSL proteins along with several GT and GH.18 Although CSL involved in xylan synthesis remains elusive, previous studies have identified five GTs required for its formation.19–21 IRX7, IRX8 and PARVUS are probably responsible for the synthesis of a xylan primer, whereas IRX9 and IRX14 may be involved in xylan backbone production.21 The reduced xylan content promoted us to explore the expression of IRX7, IRX14 and PARVUS homologs in oscsld4 mutant and wild-type plants. Their expression levels were unexpectedly upregulated (Fig. 1A), indicating that the rice plants may perceive the defect in xylan and increase the expression of some genes involved in xylan synthesis as the compensatory reaction.
Figure 1.
Real-time PCR analysis of the gene expression levels in oscsld4 and wild-type plants. (A) The expression level of genes involved in xylan synthesis. (B) The expression level of genes of CD K/Cyclin complexes. The expression levels are given by counting that of wild type as 1.
oscsld4 mutant also shows significant reduction in plant growth due to retarded cell cycle progression.13 Because OsCSLD4 is mainly expressed in tissues undergoing rapid growth, we therefore detected the expression of several genes involved in cell cycle regulation. As shown in Figure 1B, two cyclins, three CDKs, and KRP1 (Kip-Related Protein 1), which govern the key G1-to-S or G2-to-M transition points,22 were upregulated in mutant plants. However, E2F1, a transcription factor that is in the down-stream of CDK/Cyclin complexes regulation pathway and activates the expression of S phase genes,22 was downregulated in oscsld4. This result, as well as diverse functions of CSLDs let us reason that the polysaccharide produced by CSLDs may have a signaling role in development. Cell wall is an extremely complex reservoir of chemical and mechanical information,1 which provides a wealth of potential signaling molecules that are capable of regulating plant growth.23,24 For example, galactoglucomannan can inhibit auxin-induced stem elongation in peas and pine stems.25,26 CSLA7 that catalyzes glucomannan backbone formation was then found playing a role in embryogenesis.27,28 Screening suppressors of atcesa6 null mutant that attenuate growth effects led to the identification of a novel plasma-membrane receptor kinase (THE1) as a sensor of wall integrity, making a prominent progress in plant cell-wall-integrity-signaling pathway.14 Besides the growth responses, defective cell walls also trigger local and systemic stress responses. Mutations in the AtCESA3 gene constitutively activate ethylene and jasmonate mediated signaling, thus increase powdery mildew resistance. 29 And the disruption of AtCESA8 causes drought resistance.30
Conclusion
Our study has revealed that rice plants have complex responses to the disruption of OsCSLD4, including the effects on various wall components and the expression of genes related to xylan synthesis and cell cycle regulation. All those probably reflect the ability of plants to perceive and react to the changes in cell wall. Further analysis is required to understand the systemic signaling mechanism that transducts the wall integrity information in specific cells to the machinery that regulates the wall remodeling and plant growth throughout the plant. Arabidopsis and rice each has five CSLD members. All those proteins could make the same glycan product, but contribute to different developmental roles. The wall-integrity-signaling system may be a conceivable way to decipher their diverse biological functions. Our findings here provide essential clues in understanding the co-regulated mechanism in response to the disruption of OsCSLD4.
Acknowledgements
This work was supported by the grant from the National Natural Science Foundation of China (90717117) and the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-YW-G-033).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10369
References
- 1.Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, et al. Toward a systems approach to understanding plant cell walls. Science. 2004;306:2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- 2.Henrissat B, Coutinho PM, Davies GJ. A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol Biol. 2001;47:55–72. [PubMed] [Google Scholar]
- 3.Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. Biosynthesis of plant cell wall polysaccharides—a complex process. Curr Opin Plant Biol. 2006;9:621–630. doi: 10.1016/j.pbi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA. 2005;102:2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, et al. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- 6.Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, et al. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci USA. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, et al. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-Dglucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci USA. 2009;106:5996–6001. doi: 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, et al. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 2001;15:79–89. doi: 10.1101/gad.188801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Cnops G, Vanderhaeghen R, De Block S, Van Montagu M, Van Lijsebettens M. AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol. 2001;126:575–586. doi: 10.1104/pp.126.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernal AJ, Jensen JK, Harholt J, Sorensen S, Moller I, Blaukopf C, et al. Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 2007;52:791–802. doi: 10.1111/j.1365-313X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- 11.Bernal AJ, Yoo CM, Mutwil M, Jensen JK, Hou G, Blaukopf C, et al. Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2 and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 2008;148:1238–1253. doi: 10.1104/pp.108.121939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, et al. OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol. 2007;143:1220–1230. doi: 10.1104/pp.106.091546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, et al. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009. doi: 10.1111/j.1365-313X.2009.04022.x. [DOI] [PubMed]
- 14.Hématy K, Sado P, Tuinen AV, Rochange S, Desnos T, Balzergue S, et al. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol. 2007;17:922–931. doi: 10.1016/j.cub.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Heinisch JJ. Baker's yeast as a tool for the development of antifungal kinase inhibitors—targeting protein kinase C and the cell integrity pathway. Biochim Biophys Acta. 2005;1754:171–182. doi: 10.1016/j.bbapap.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 16.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong R, Péna MJ, Zhou GK, Nairn CJ, Wood-Jones A, Richardson EA, et al. Arabidopsis fragile fiber8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell. 2005;17:3390–3408. doi: 10.1105/tpc.105.035501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.York WS, O’Neill MA. Biochemical control of xylan biosynthesis—which end is up? Curr Opin Plant Biol. 2008;11:258–265. doi: 10.1016/j.pbi.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Péna MJ, Zhong R, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, et al. Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. 2007;19:549–563. doi: 10.1105/tpc.106.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, et al. The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell. 2007;19:237–255. doi: 10.1105/tpc.106.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, et al. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 2007;52:1154–1168. doi: 10.1111/j.1365-313X.2007.03307.x. [DOI] [PubMed] [Google Scholar]
- 22.Inzé D, Veylder LD. Cell cycle regulation in plant development. Ann Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 23.McDougall GJ, Fry SC. Purification and analysis of growth-regulating xyloglucan-derived oligosaccharides by high-pressure liquid chromatography. Carbohydr Res. 1991;219:123–132. doi: 10.1016/0008-6215(91)89047-j. [DOI] [PubMed] [Google Scholar]
- 24.Takeda T, Furuta Y, Awano T, Mizuno K, Mitsuishi Y, Hayashi T. Suppression and acceleration of cell elongation by integration of xyloglucans in pea stem segments. Proc Natl Acad Sci USA. 2002;99:9055–9060. doi: 10.1073/pnas.132080299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auxtová O, Lišková D, Kákoniová D, Kubaková M, Kará csonyi S, Bilisics L. Effect of galactoglucomannan-derived oligosaccharides on elongation growth of pea and spruce stem segments stimulated by auxin. Planta. 1995;196:420–424. [Google Scholar]
- 26.Auxtová-Šamajová O, Lišková D, Kákoniová D, Kubacková MM, Karácsonyi S, Bilisics L. Inhibition of auxin stimulated short-term elongation growth of pea stem segments by galactoglucomannan-derived oligosaccharides. J Plant Physiol. 1996;147:611–613. [Google Scholar]
- 27.Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P. AtCslA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 2003;131:547–557. doi: 10.1104/pp.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goubet F, Barton CJ, Mortimer JC, Yu X, Zhang Z, Miles GP, et al. Cell wall glucomannan in Arabidopsis is synthesized by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 2009. doi: 10.1111/j.1365-313X.2009.03977.x. [DOI] [PubMed]
- 29.Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell. 2002;14:1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z, Hong X, Zhang H, Wang Y, Li X, Zhu JK, Gong Z. Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J. 2007;43:273–283. doi: 10.1111/j.1365-313X.2005.02452.x. [DOI] [PubMed] [Google Scholar]