Abstract
Plant peroxisomes are essential organelles that house diverse metabolic activities. To understand the full spectrum of peroxisomal functions, we recently performed a proteomic analysis of Arabidopsis leaf peroxisomes followed by in vivo subcellular targeting validation of some novel proteins. Here, we continue with the targeting analysis and demonstrate that the nonapeptide RVx5HF, which is present near the N terminus of the newly identified peroxisomal protein, HIT3 (histidine triad family protein 3), is a functional peroxisome targeting signal type 2 (PTS2). In addition, we have confirmed the peroxisomal localization of UP6 (unknown protein 6) and GLX1 (glyoxalase 1 homolog), two proteins with possible roles in stress responses and glutathione-dependent detoxification, respectively. These data, together with results from our previous analysis of the peroxisomal proteome, reinforce the notion that peroxisomes are involved in various stress responses and suggest glutathione as a major antioxidant in plant peroxisomes.
Key words: peroxisome targeting, PTS2, glyoxalase, defense, arabidopsis
Peroxisomes are single membrane-delimited organelles involved in a suite of metabolic functions in eukaryotic organisms.1 Processes facilitated by plant peroxisomes include photorespiration, fatty acid metabolism, jasmonic acid biosynthesis, metabolism of the proto-auxin indole-3-butyric acid, detoxification, polyamine catabolism, sulfite oxidation, and defense responses to pathogens.2 To identify the complete set of proteins involved in all the biochemical and regulatory pathways in peroxisomes, it is crucial to comprehensively catalog the proteome of these essential organelles. Although the major peroxisomal metabolic pathways (e.g., the glycolate cycle, β-oxidation and the glyoxylate cycle) have been characterized, our knowledge of the molecular factors involved in many minor peroxisomal functions is still limited. To this end, we recently conducted an in-depth analysis of the Arabidopsis leaf peroxisomal proteome, using one-dimensional gel electrophoresis (1-DE) followed by liquid chromatography and tandem mass spectrometry (LC-MS/MS). We identified 150 proteins, including 30 proteins whose association with peroxisomes had only been demonstrated by proteomic data, and 50 proteins whose functions had never been related to peroxisomes. Using in vivo subcellular targeting analysis of yellow fluorescent protein (YFP) fusions for selected proteins, we verified the peroxisomal localization of 12 proteins with predicted peroxisome targeting signals type 1 or 2 (PTS1/2), three proteins that contain PTS-related peptides, and four proteins without apparent PTSs.3
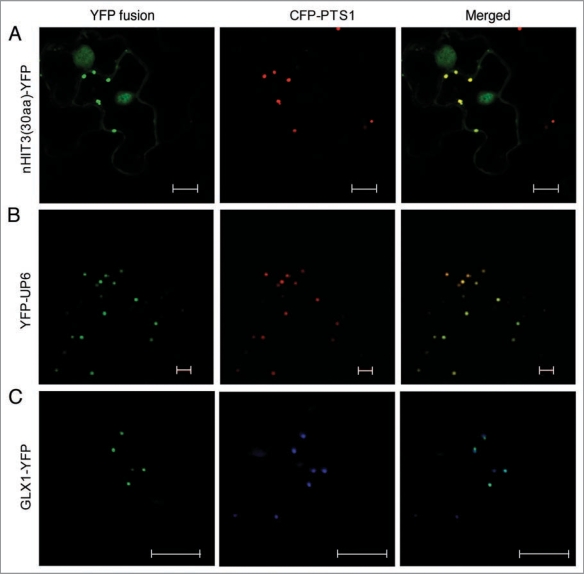
In our previous study,3 we confirmed the peroxisomal localization of HIT3 (histidine triad family protein 3; At3g56490), a member of a new family of plant peroxisomal proteins with putative functions as nucleotide hydrolases and/or transferases.4 Near the N terminus (4th–12th amino acids) of this protein is a nonapeptide, RVx5HF, which is related to the canonical PTS2 sequence, RLx5HL, and is conserved among HIT3 homologs in diverse plant species.3 To test whether RVx5HF is a functional PTS2, we fused the first 30 amino acids of the HIT3 protein to the N terminus of YFP and examined the subcellular localization of the fusion protein. The YFP fusion was transiently coexpressed with the peroxisomal marker protein, CFP-PTS1 (PTS1, composed of ser-lys-leu) in tobacco (Nicotiana tabacum) plants. Two days after inoculation with Agrobacteria containing the constructs, we performed confocal laser scanning microscopic analysis on tobacco epidermal cells. The YFP fusion protein is strongly co-localized with CFP-PTS1, with a weak localization in the nucleus and cytosol (Fig. 1A). We thus conclude that RVx5HF is a functional but “minor”; PTS2 peptide (defined by Reumann5) in plants. Minor PTSs often need adjacent basic and hydrophobic resides, the so-called “enhancing elements”, for efficient peroxisomal targeting.5 However, these types of amino acids are not enriched in sequences around the PTS2 in HIT3, which may explain the weak cytosolic and nuclear localization of the protein.
Figure 1.
Validation of a new PTS2 peptide and the peroxisomal localization of two novel proteins. Confocal (A and B) or epifluorescent (C) microscopic images were obtained from leaf epidermal cells of 4-week-old tobacco (Nicotiana tabacum) plants co-expressing the indicated YFP fusion and the CFP-PTS1 peroxisomal marker proteins. YFP fusions are shown in green, and the CFP-PTS1-labeled peroxisomes are in red (A and B) or blue (C). Scale bars = 10 µm.
Next we tested the subcellular localization of two additional putative novel peroxisomal proteins identified from our previous proteomic analysis of leaf peroxisomes.3 UP6 (unknown protein 6; At1g16730) carries a canonical C-terminal PTS1 tripeptide (SKL>), which is conserved in UP6 homologs in other plant species,3 strongly suggesting the localization of this protein in the peroxisomal matrix. Indeed, when fused to the C terminus of YFP (YFP-UP6), UP6 showed strong peroxisomal targeting in tobacco cells (Fig. 1B). Although its biochemical function cannot be predicted from the sequence, the UP6 gene has a much higher expression level in seeds compared with in any other tissue, and is dramatically upregulated by stresses such as drought, heat and cold, and the “stress hormone” abscisic acid (ABA) (www.genevestigator.com). This interesting expression pattern indicates that UP6 may be involved in plant stress responses.
The second novel protein we examined is a glyoxalase 1 homolog (GLX1; At1g11840), which does not contain a predicted PTS. When attached to the C terminus of YFP, GLX1 remained cytosolic (data not shown). However, when fused to the N-terminal end of YFP, this protein showed co-localization with the peroxisomal marker protein in tobacco leaf epidermal cells (Fig. 1C). Hence, GLX1, together with the previously identified sarcosine oxidase,6 dehydroascorbate reductase (DHAR1),3 dephospho-CoA kinase (COAE),3 and nucleoside diphosphate kinase type 1 (NDPK1),3 represent a group of peroxisomal proteins lacking predicted conventional PTSs or transmembrane domains. These proteins are most likely located in the peroxisome matrix and imported via a yet-to-be-identified pathway(s).
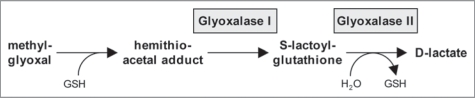
Validation of the peroxisomal localization of GLX1 is significant, because, to the best of our knowledge, glyoxalases have not been shown to associate with peroxisomes in any organism. The glyoxalase system is a ubiquitous detoxification system in both eukaryotes and prokaryotes. The two major enzymes in this system are glyoxalase I (lactoylglutathione lyase) and glyoxalase II (hydroxyacylglutathione hydrolase), which together convert a variety of α-keto aldehydes into hydroxyacids in the presence of glutathione. Although the exact role of this system in the cell has still not been clearly defined, its major function is speculated to be in preventing the accumulation of methylglyoxal, a toxic by-product of glycolysis and metabolism of amino acids and ketone bodies, and its adducts (Fig. 2).7,8 Methylglyoxal has recently been found in higher plants, where it is detoxified mainly via the glyoxalase system.9 Cytosolic- and mitochondrial-targeted isoforms of glyoxalase II have been detected in Arabidopsis,10,11 whereas little information is available on plant glyoxalase I. Our data suggests that the glyoxalate detoxification system is partially compartmentalized in peroxisomes in Arabidopsis. It remains to be investigated whether any of the five Arabidopsis glyoxalase II isoforms, none of which contain a predicted PTS, is also targeted to peroxisomes. To date, a number of glutathione-dependent enzymes have been identified from proteome analyses of Arabidopsis leaf peroxisomes and verified to be peroxisome targeted.3,12 Glutathione is thus emerging as a major antioxidant in plant peroxisomes, protecting peroxisomal and cellular enzymes against a range of peroxides, xenobiotics and possibly heavy metals.13
Figure 2.
A proposed model for the action of glyoxalase I and II in detoxification. See text for a description of the pathway. GSH, reduced glutathione (γ-L-glutamyl-L-cysteinyl-glycine).
Acknowledgements
This work was supported by grants from the National Science Foundation (MCB 0618335) and the U.S. Department of Energy (DE-FG02-91ER20021) to J.H.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10412
References
- 1.Schrader M, Fahimi HD. The peroxisome: still a mysterious organelle. Histochem Cell Biol. 2008;129:421–440. doi: 10.1007/s00418-008-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaur N, Reumann S, Hu J. Peroxisome biogenesis and function. The Arabidopsis Book. Rockville MD,: American Society of Plant Biologists; 2009. doi: 10.1199/tab.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D, et al. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 2009;150:125–143. doi: 10.1104/pp.109.137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner C. Hint, Fhit and Gal T: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry. 2002;41:9003–9014. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reumann S. Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 2004;135:783–800. doi: 10.1104/pp.103.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goyer A, Johnson TL, Olsen LJ, Collakova E, Shachar-Hill Y, Rhodes D, et al. Characterization and metabolic function of a peroxisomal sarcosine and pipecolate oxidase from Arabidopsis. J Biol Chem. 2004;279:16947–16953. doi: 10.1074/jbc.M400071200. [DOI] [PubMed] [Google Scholar]
- 7.Thornalley PJ. The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem J. 1990;269:1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thornalley PJ. Modification of the glyoxalase system in disease processes and prospects for therapeutic strategies. Biochem Soc Trans. 1993;21:531–534. doi: 10.1042/bst0210531. [DOI] [PubMed] [Google Scholar]
- 9.Yadav SK, Singla-Pareek SL, Ray M, Reddy MK, Sopory SK. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem Biophys Res Commun. 2005;337:61–67. doi: 10.1016/j.bbrc.2005.08.263. [DOI] [PubMed] [Google Scholar]
- 10.Maiti MK, Krishnasamy S, Owen HA, Makaroff CA. Molecular characterization of glyoxalase II from Arabidopsis thaliana. Plant Mol Biol. 1997;35:471–481. doi: 10.1023/a:1005891123344. [DOI] [PubMed] [Google Scholar]
- 11.Zang TM, Hollman DA, Crawford PA, Crowder MW, Makaroff CA. Arabidopsis glyoxalase II contains a zinc/iron binuclear metal center that is essential for substrate binding and catalysis. J Biol Chem. 2001;276:4788–4795. doi: 10.1074/jbc.M005090200. [DOI] [PubMed] [Google Scholar]
- 12.Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, et al. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways and defense mechanisms. Plant Cell. 2007;19:3170–3193. doi: 10.1105/tpc.107.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noctor G, Gomez L, Vanacker H, Foyer CH. Interactions between biosynthesis, compartmentation and transport in the control of glutathione homeostasis and signalling. J Exp Bot. 2002;53:1283–12304. doi: 10.1093/jexbot/53.372.1283. [DOI] [PubMed] [Google Scholar]