Abstract
We have recently reported the identification and characterization of Sad1/UNC84 (SUN) domain proteins in various plant species. In animals and yeasts, SUN domain proteins are localized at the inner nuclear membrane and form a bridge across the nuclear envelope (NE) by interacting with outer nuclear membrane-localized Klarsicht/Anc-1/Syne-1 homology (KASH) domain proteins. This bridge physically connects cytoskeletal elements with chromatin and nucleoskeletal components. These multiprotein complexes are essential for various cellular and nuclear processes. The identification of SUN domain proteins provides the first evidence of putative NE bridging complexes in plants. Here we speculate on the composition and functions of these in regards to our current understanding of plant SUN domain proteins.
Key words: SUN domain protein, LINC complex, plant nuclear envelope, cytoskeleton, KASH domain proteins, Arabidopsis
Introduction
Nucleo-cytoskeletal bridging complexes span the nuclear envelope (NE) of animal and yeast cells and physically connect the cytoskeleton with the nucleoskeleton and chromatin. Also called linker of nucleoskeleton and cytoskeleton (LINC), they consist of two key components; inner nuclear membrane (INM)—localized Sad1/UNC84 (SUN) domain proteins and outer nuclear membrane (ONM)—localized Klarsicht/Anc-1/Syne-1 homology (KASH) domain proteins.1–3 On the nucleoplasmic side, SUN domain proteins associate with chromatin and nucleoskeletal elements such as lamins. In the periplasm, the SUN and KASH domain proteins bind to each other forming a bridge across the NE membranes. On the cytoplasmic face, KASH domain proteins link to various cytoskeletal components including actin, microtubule motors kinesin and dynein, centrosomes and spindle pole body (SPB) components, completing the linkage (Fig. 1).3–5 Such physical connections between nucleo-and cytoskeleton facilitate communication and coordination of cellular and nuclear processes during the various phases of the cell cycle. In interphase, LINC complexes and their individual components are essential for positioning and anchorage of the nucleus, positioning and duplication of centrosomes and anchorage of SPB. During cell division anchorage of telomeres (Fig. 1), formation of the meiotic chromosome bouquet and decondensation of chromatin involves LINC complex components.4–6 Most of these processes are also essential to the plant cell cycle. Our identification of plant SUN domain proteins is a first concrete indication of the existence of putative LINC complexes at the plant NE.7
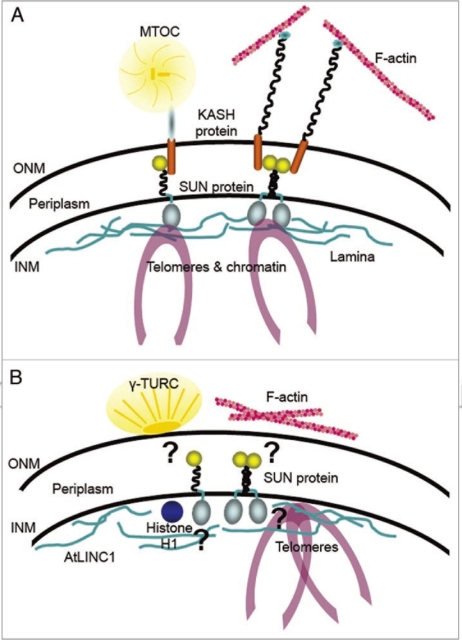
Figure 1.
The LINC complex in animal and yeast systems (A) connects nuclear and cytoskeletal components across the NE. The outer nuclear membrane (ONM ) contains Klarsicht/ANC -1/SYNE -1 homology (KASH) domain proteins, which link actin, microtubule (MT ) associated elements such as dynamin, kinesin and centrosomes to the NE . The inner nuclear membrane (INM ) associated Sad1/UNC −84 (SUN ) domain proteins anchor chromatin, telomeres and lamins to the nuclear face of the NE . SUN and KASH domain proteins interact with each other in the periplasm completing the linkage. In plants (B), cytoskeletal and nuclear components are also associated with the NE—MT and actin elements on the cytoplasmic face, chromatin and filamentous lamin-like elements on the nuclear side. The nature of their NE anchorage remains unknown but the identification of plant SUN domain proteins suggests the presence of putative plant LINC complexes. Their structure and functions, however, may differ to those in animal and yeast systems.
Plant SUN Domain Proteins
Using a bioinformatics approach we identified SUN-domain protein homologues in various plant species including dicots Arabidopsis thaliana and Vitis vinifera, monocots Oryza sativa and Zea mays and the moss Physcomitella patens.7 Most plants, apart from the moss, contain two SUN domain proteins and are identifiable as members of the SUN domain protein family by similarities in domain structure to animal and yeast members. Although the plant homologues generally appear to be smaller, they all contain the highly conserved C-terminal SUN domain. Most of them also contain coiled coil domains and a transmembrane domain towards the N-terminus, which anchors the proteins into the INM.7 Closer examination of the Arabidopsis homologues AtSUN1 and AtSUN2 also revealed the presence of a bipartite nuclear localization signal (NLS) at the N-terminus. Similar to animal and yeast SUN domain proteins, the two Arabidopsis homologues localize to the plant NE, in particular the INM, when expressed as fluorescent protein fusions.7 A further similarity to animal and yeast SUN domain proteins is their ability to form multimeric complexes facilitated by their coiled coil domains.2,4 Both AtSUN1 and AtSUN2 were shown to form homomers and heteromers (Fig. 1) and deletion of the coiled coil domains abolished these interactions.7 These complex formations are thought to increase the number and diversity of binding partners to any one complex so that various KASH domain proteins and their associated cytoskeletal components can be connected to multiple nuclear components.2,4
Putative Plant LINC Complexes and their Components
Mobility studies of AtSUN1 and AtSUN2 fluorescent fusion proteins revealed both to be highly immobile at the plant NE, which is evidence for binding of the proteins to either nuclear, membrane-localized or periplasmic components. In support of this we found that deletion of the N-terminus increased the mobility of the two proteins indicating that the N-terminus is involved in binding interactions strong enough to affect protein movement.7 While work is ongoing to identify interaction partners of AtSUN1 and AtSUN2, speculations based on recent research can be made as to the nature of putative plant LINC complex components.
Nakayama et al.8 have shown that the nuclear-localized histone H1 induces the radial organization of MT at the ONM of tobacco BY-2 cells (Fig. 1). This is first evidence that connection of nuclear components to cytoskeletal elements also occurs in plants. MT nucleation at the ONM requires the γ-tubulin ring complex (γ-TURC), two of whose components, AtGCP2 and AtGCP3, contain nuclear targeting domains, which mediate the localization of the complex to the ONM (Fig. 1).9 However, as both proteins are soluble, it has been speculated that an ONM-intrinsic anchor is needed, to which the γ-TURC is linked.9 The identity of this anchor remains unknown, but as LINC complex components anchor MTOC and SPB to the animal and yeast NE (Fig. 1), it could be envisaged that AtSUN1 and AtSUN2 are part of a similar anchoring complex. Further, while lamin sequence homologues are not present in plants, structurally similar plant-specific nuclear proteins are thought to fulfil similar functions.10,11 One of those is Arabidopsis little nuclei 1 (AtLINC1), which localizes to the periphery of the NE (Fig. 1) when expressed as a fluorescent fusion and is involved in maintaining nuclear size and morphology.12 In addition to this, Fiserova et al.13 showed by electron microscopy that a filamentous meshwork similar to the animal lamina underlies the plant INM. Apart from histone H1 and AtLINC1, other putative nuclear interaction partners of AtSUN1 and AtSUN2 could include telomere components. Similar to the anchorage of telomeres to the animal NE, plant telomeres are also anchored in this fashion (Fig. 1) during prophase 1.14 The increased expression of AtSUN2 in proliferating tissue suggests that it may be involved in mitosis/meiosis-specific processes.7 Finally, while cytoskeletal components such as γ-TURC and actin are known to be linked to the plant NE, the proteins facilitating such anchorage are unknown. While KASH domain proteins mediate such anchorage in animal and yeast cells (Fig. 1), it has been reported that plants lack KASH domain sequence homologues.11,15 Unlike the SUN domain, however, the KASH domain is not very well conserved, even between animal and yeast species.4,16 Similar to lamin-like plant proteins, it could be speculated that plants have KASH-like proteins that are structurally similar to animal and yeast KASH domain proteins and may fulfil similar functions. Using bioinformatics we have recently found a few proteins with a similar structure to KASH domain proteins (unpublished observations)—N-terminal domain with coiled coils and a C-terminal transmembrane domain.4 Current investigations on the likelihood of those proteins being candidates for KASH-like proteins are underway.
Conclusion
The characterization of plant SUN domain proteins is a first step in the identification of putative plant LINC complexes and their involvement in cellular and nuclear processes. However, the lack of sequence homologues of major interaction partners of animal and yeast SUN domain proteins, such as lamins and KASH domain proteins, suggests that putative plant LINC complexes may be differently structured and involved in different, plant specific processes. Identification and description of plant SUN domain protein interaction partners may therefore provide the opportunity to explore nucleo-cytoskeletal signaling in plants in an entirely novel way.
Abbreviations
- NE
nuclear envelope
- INM
inner nuclear membrane
- ONM
outer nuclear membrane
- SUN
Sad1/UNC84
- KASH
klarsicht/anc-1/syne-1 homology
- LINC
linker of nucleoskeleton and cytoskeleton
- MTOC
microtubule organising centre
- NLS
nuclear localisation signal
- SPB
spindle pole body
- γ-TURC
gamma tubulin ring complex
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10458
References
- 1.Crisp M, Qian Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- 3.Worman HJ, Gundersen GG. Here come the SUNs: a nucleocytoskeletal missing link. Trends Cell Biol. 2006;16:67–69. doi: 10.1016/j.tcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Starr DA. A nuclear envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi YH, Haller K, Peleponese JM, Jang KT. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in decondensation of mitotic chromosomes. J Biol Chem. 2007;282:27447–27458. doi: 10.1074/jbc.M703098200. [DOI] [PubMed] [Google Scholar]
- 7.Graumann K, Runions J, Evans DE. Characterisation of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 2010;61:134–144. doi: 10.1111/j.1365-313X.2009.04038.x. [DOI] [PubMed] [Google Scholar]
- 8.Nakayama T, Ishii T, Hotta T, Mizuno K. Radial microtubule organisation by histone H1 on nuclei of cultured tobacco BY-2 cells. J Biol Chem. 2008;283:16632–16640. doi: 10.1074/jbc.M705764200. [DOI] [PubMed] [Google Scholar]
- 9.Seltzer V, Janski N, Canaday J, Herzog E, Erhardt M, Evrard J-L, Schmit A-C. Arabidopsis GCP2 and GCP3 are part of a soluble γ-tubulin complex and have nuclear envelope targeting domains. Plant. 2007;52:322–331. doi: 10.1111/j.1365-313X.2007.03240.x. [DOI] [PubMed] [Google Scholar]
- 10.Brandizzi F, Irons SL, Evans DE. The plant nuclear envelope: new prospects for a poorly understood structure. New Phytol. 2004;163:227–246. doi: 10.1111/j.1469-8137.2004.01118.x. [DOI] [PubMed] [Google Scholar]
- 11.Evans DE, Irons SL, Graumann K, Runions J. The Plant Nuclear Envelope. In: Meier I, editor. Functional Organisation of the Plant Nucleus. Berlin: Springer; 2009. p. 20. [Google Scholar]
- 12.Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–2803. doi: 10.1105/tpc.107.053231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organisation in tobacco BY-2 cells. Plant J. 2009;59:243–255. doi: 10.1111/j.1365-313X.2009.03865.x. [DOI] [PubMed] [Google Scholar]
- 14.Roberts NY, Osman K, Armstrong SJ. Telomere distribution and dynamics in somatic and meiotic nuclei of Arabidopsis thaliana. Cytogenet Genome Res. 2009;124:193–201. doi: 10.1159/000218125. [DOI] [PubMed] [Google Scholar]
- 15.Meier I. Composition of the plant nuclear envelope: theme and variations. J Exp Bot. 2007;58:27–34. doi: 10.1093/jxb/erl009. [DOI] [PubMed] [Google Scholar]
- 16.McGee MD, Stagljar I, Starr DA. KDP-1 is a nuclear envelope KASH proteins required for cell cycle progression. J Cell Sci. 2009;122:2895–2905. doi: 10.1242/jcs.051607. [DOI] [PMC free article] [PubMed] [Google Scholar]