Abstract
Higher plants respond to fluctuating Fe availability by regulating the expression of genes involved in Fe homeostasis. Transcriptional responses to Fe deficiency in plants are mediated via various interactions between cisacting elements and trans-acting factors. The transcription factor IDEF1 regulates the response to Fe deficiency in Oryza sativa (rice) by recognizing the CATGC sequence within the Fe deficiency-responsive cis-acting element IDE1. We recently presented evidence that IDEF1 mediates two-phase responses to Fe deficiency. During the early stages of Fe deficiency, the majority of known Fe uptake/utilization-related genes are positively regulated by IDEF1. In subsequent stages, IDEF1-mediated regulation of these Fe uptake/utilization-related genes are less obvious. In turn, expression of several Fe deficiency-induced genes encoding late embryogenesis abundant proteins is increasingly regulated by IDEF1 at the subsequent stages. We propose a dual function of IDEF1 for Fe deficiency response, namely, (1) the coordinated transactivation of Fe utilization-related genes via CATGC-containing IDE1-like elements, especially at the early stage, and (2) the transactivation of seed maturation-related genes via RY elements, especially during the subsequent stages of Fe deficiency. IDEF1 appears to have evolved to mediate an interface between Fe deficiency-inducible and seed maturation-related gene expression.
Key words: iron deficiency, transcriptional regulation, early response, IDE1, RY element, late embryogenesis abundant proteins, rice
Iron (Fe) is an essential mineral nutrient for all plants. To acquire enough Fe, while circumventing toxicity of excess Fe, plants tightly control the level of Fe uptake, utilization and storage in response to Fe availability in the environment. In spite of its high abundance in the Earth’s crust, Fe is sparingly soluble in aerobic conditions, especially in calcareous and high pH soils.1 Under conditions of low Fe availability, higher plants adopt two major strategies for Fe acquisition from the rhizosphere: Strategy I, utilized by non-graminaceous plants, and Strategy II by graminaceous plants.2 In both strategies, expression of the genes participating in Fe acquisition is strongly induced in response to Fe deficiency. Molecular components regulating these Fe deficiency responses are now being elucidated. The Strategy I response is mediated by Fe deficiency-inducible basic helix-loop-helix (bHLH) transcription factors (TFs), among which tomato FER and its Arabidopsis thaliana ortholog FIT (formerly FIT1/FRU/AtbHLH29) are the most characterized.3–5 Interactions of these bHLH TFs are thought to play key roles in regulation.6 However, neither the functional cis sequences nor their upstream regulation pathways have been elucidated for these non-graminaceous TFs.
Our recent studies focused on clarification of cis element/trans factor interactions regulating Fe deficiency responses of graminaceous genes. Promoter analysis of the barley Fe deficiency-inducible IDS2 gene led to identification of the Fe deficiency-responsive cis-acting elements IDE1 and IDE2,7 which are functional both in rice (Graminaceae) and tobacco (non-Graminaceae).7,8 We further identified two rice IDE-binding TFs, IDEF1 (ABI3/VP1 family) and IDEF2 (NAC family), which specifically bind to IDE1 and IDE2, respectively.9,10 Both IDEF1 and IDEF2 are constitutively expressed irrespective of Fe status, suggesting their roles in sensing Fe deficiency signals directly or indirectly. Characterization of transgenic rice plants with altered IDEF1 or IDEF2 expression levels revealed physiological functions of these TFs in Fe homeostasis during Fe deficiency.9–11 RNAi-mediated IDEF2 knockdown lines exhibit aberrant Fe distribution between roots and shoots and are defective in inducing a subset of Fe deficiency-induced genes.10
Transgenic rice lines with an introduced IDEF1 under the control of the Fe deficiency-inducible IDS2 promoter exhibit a slower decrease in leaf chlorophyll under low Fe availability and enhanced expression of an Fe deficiency-induced bHLH TF gene OsIRO2.9 OsIRO2 binds to the CACGTGG sequence12 and regulates the majority of the genes involved in Strategy II Fe acquisition.13 These findings indicate the presence of a gene regulation cascade involving IDEF1 and OsIRO2.9,13
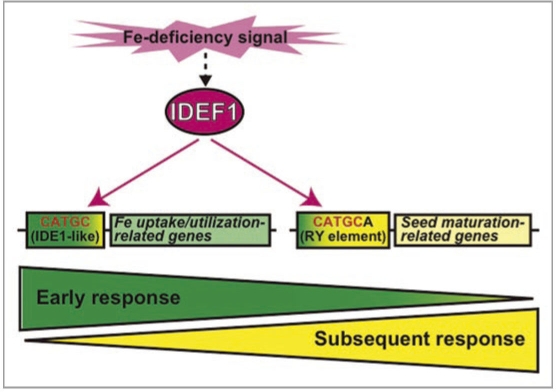
Through time-course expression analysis of transformants with induced or repressed IDEF1 expression, we previously revealed that IDEF1-mediated gene regulation consists of two phases during the progression of Fe deficiency.11 On day 1 of Fe deficiency treatment in hydroponic culture, IDEF1 mediates transactivation of the majority of the Fe deficiency-induced genes currently known to be involved in Fe uptake and/or utilization. On day 2, and thereafter, IDEF1-mediated transactivation of these Fe uptake/utilization-related genes becomes weaker and restricted to only some of the members. In silico analysis of rice promoters revealed that IDEF1 target genes, as estimated by microarray analyses, exhibit a significantly higher presence of IDEF1-binding core sites (CATGC) and IDE1-like sequences in the proximal promoter regions. The highest occurrence of these sequences was found on day 1 of Fe deficiency, confirming the predominance of an early response that is mediated, at least in part, through direct binding of IDEF1 to CATGC sequences (Fig. 1, left).
Figure 1.
A simplified model of dual regulation of Fe deficiency response mediated by IDEF1.
We further investigated the occurrences of the RY element (CATGCA) among the IDEF1 target genes, because the RY element is thought to be the minimal recognition sequence of all the ABI/VP1 family TFs analyzed,14–16 except for the IDEF1 subfamily.9 Interestingly, IDEF1 target gene promoters possess not only CATGC but also RY elements at significantly high levels, even though IDEF1 does not require the sixth base of the RY element (CATGCA) for efficient binding in vitro.9 In general, RY elements confer seed-specific expression of various genes, including those encoding late embryogenesis abundant (LEA) proteins.16 Our expression analyses revealed that a subset of LEA genes including Osem is induced even in vegetative organs (leaves and roots) in response to Fe deficiency in an IDEF1-dependent manner, especially at the subsequent stages (Fig. 1, right). Thus, IDEF1 is thought to possess a dual function for Fe deficiency response: the coordinated transactivation of Fe uptake/utilization-related genes via CATGC-containing IDE1-like elements, and the transactivation of seed maturationrelated genes via RY elements. The former is predominant at the early stage, while the latter becomes dominant at subsequent stages of Fe deficiency.
Because IDEF1 is expressed during both vegetative and reproductive stages,17 this TF might positively regulate seed maturation-related genes in various organs. In addition, the unique DNAbinding property of IDEF1 to efficiently bind to CATGC and IDE1-like elements lead to this TF being capable of regulating the Fe deficiency response. Thus, IDEF1 appears to have evolved to mediate an interface between Fe deficiency-inducible and seed maturation-related gene expression. Curiously, gene homologs of the IDEF1 subfamily have been found only in graminaceous plants, even though IDE1 is also functional in non-graminaceous tobacco plants.7,9
The essentiality of IDEF1 for Fe deficiency response is also confirmed by the phenotype of IDEF1 knockdown lines, which are hypersensitive to Fe deficiency, as observed by an earlier decrease in leaf chlorophyll. 11 Clarification of factors interacting with IDEF1 and/or IDEF2 might provide a significant step toward our understanding of the precise mechanisms of Fe deficiency response in plants, including the nature of the Fe deficiency signal. In addition, the IDEF1-mediated transactivation of seed maturation-related genes suggests possible roles for these genes under low Fe availability, thus providing a basis for exploring new functions for LEA proteins.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10459
References
- 1.Marschner H. Mineral Nutrition of Higher Plants. 2nd edn. London UK: Academic press,; 1995. [Google Scholar]
- 2.Marschner H, Römheld V, Kissel M. Different strategies in higher plants in mobilization and uptake of iron. J Plant Nutr. 1986;9:695–713. [Google Scholar]
- 3.Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA. 2002;99:13938–13943. doi: 10.1073/pnas.212448699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer P, Ling HQ, Guerinot ML. FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol Biochem. 2007;45:260–261. doi: 10.1016/j.plaphy.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Walker EL, Connolly EL. Time to pump iron: irondeficiency-signaling mechanisms of higher plants. Curr Opin Plant Biol. 2008;11:530–535. doi: 10.1016/j.pbi.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, et al. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008;18:385–397. doi: 10.1038/cr.2008.26. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi T, Nakayama Y, Itai RN, Nakanishi H, Yoshihara T, Mori S, et al. Identification of novel cisacting elements, IDE1 and IDE2, of the barley IDS2 gene promoter conferring iron-deficiency-inducible, root-specific expression in heterogeneous tobacco plants. Plant J. 2003;36:780–793. doi: 10.1046/j.1365-313x.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi T, Nakayama Y, Takahashi M, Inoue H, Nakanishi H, Yoshihara T, et al. Construction of artificial promoters highly responsive to iron deficiency. Soil Sci Plant Nutr. 2004;50:1167–1175. [Google Scholar]
- 9.Kobayashi T, Ogo Y, Itai RN, Nakanishi H, Takahashi M, Mori S, et al. The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc Natl Acad Sci USA. 2007;104:19150–19155. doi: 10.1073/pnas.0707010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogo Y, Kobayashi T, Itai RN, Nakanishi H, Kakei Y, Takahashi M, et al. A novel NAC transcription factor IDEF2 that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J Biol Chem. 2008;283:13407–13417. doi: 10.1074/jbc.M708732200. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T, Itai RN, Ogo Y, Kakei Y, Nakanishi H, Takahashi M, et al. The rice transcription factor IDEF1 is essential for the early response to iron deficiency, and induces vegetative expression of late embryogenesis abundant genes. Plant J. 2009. DOI:10._1111/j.1365-313X.2009.04015.x. [DOI] [PubMed]
- 12.Ogo Y, Itai RN, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, et al. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J Exp Bot. 2006;57:2867–2878. doi: 10.1093/jxb/erl054. [DOI] [PubMed] [Google Scholar]
- 13.Ogo Y, Itai RN, Nakanishi H, Kobayashi T, Takahashi M, Mori S, et al. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 2007;51:366–377. doi: 10.1111/j.1365-313X.2007.03149.x. [DOI] [PubMed] [Google Scholar]
- 14.Reidt W, Wohlfarth T, Ellerström M, Czihal A, Tewes A, Ezcurra I, et al. Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 2000;21:401–408. doi: 10.1046/j.1365-313x.2000.00686.x. [DOI] [PubMed] [Google Scholar]
- 15.Mönke G, Altschmied L, Tewes A, Reidt W, Mock HP, Bäumlein H, et al. Seed-specific transcription factor ABI3 and FUS3: molecular interaction with DNA. Planta. 2004;219:158–166. doi: 10.1007/s00425-004-1206-9. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki M, McCarty D. Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol. 2008;11:548–553. doi: 10.1016/j.pbi.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Ogo Y, Aung MS, Kobayashi T, Nozoye T, Nakanishi H, Yamakawa T, et al. Analysis of the spatial expression patterns of rice iron-deficiency-responsive element-binding factors IDEF1 and IDEF2. In Proceedings of XVI International Plant Nutrition Colloquium. 2009:1195. [Google Scholar]