Abstract
Chloroplasts arose from a cyanobacterial endosymbiont and multiply by division, reminiscent of their free-living ancestor. However, chloroplasts can not divide by themselves, and the division is performed and controlled by proteins that are encoded by the host nucleus. The continuity of chloroplasts was originally established by synchronization of endosymbiotic cell division with host cell division, as seen in existent algae. In contrast, land plant cells contain multiple chloroplasts, the division of which is not synchronized, even in the same cell. Land plants have evolved cell and chloroplast differentiation systems in which the size and number of chloroplasts (or other types of plastids) change along with their respective cellular function by changes in the division rate. We recently reported that PLASTID DIVISION (PDV) proteins, land-plant specific components of the chloroplast division apparatus, determined the rate of chloroplast division. The level of PDV protein is regulated by the cell differentiation program based on cytokinin, and the increase or decrease of the PDV level gives rise to an increase or decrease in the chloroplast division rate. Thus, the integration of PDV proteins into the chloroplast division machinery enabled land plant cells to change chloroplast size and number in accord with the fate of cell differentiation.
Key words: chloroplast division, cell cycle, cell differentiation, cytokinin, endosymbiosis, evolution
Endosymbiotic Origin of the Chloroplast
Chloroplasts arose from a bacterial endosymbiont related to extant cyanobacteria more than 1 billion years ago.1,2 The ancient alga resulting from this endosymbiotic event evolved into the Glaucophyta, Rhodophyta (red algae) and Viridiplantae (green algae and terrestrial plants), which together are referred to as the Plantae or Archaeplastida (Fig. 1). After the primitive green and red algae were established, chloroplasts then spread into other lineages of eukaryotes through secondary endosymbiotic events in which a red or a green alga was integrated into a previously nonphotosynthetic eukaryote.1,2 The transformation of the cyanobacterium into the chloroplast required several steps. Most of the genes once present in the cyanobacterial endosymbiont have been lost or relocated to the host nucleus, and a protein import system developed, which translocates proteins encoded by the nucleus into the chloroplast. Several transporters spanning the envelope membranes were developed which exchange metabolites between the cytoplasm and the chloroplast. The host and symbiont cell division became synchronized.1 Such synchronization enabled a permanent endosymbiotic relationship in which each daughter host cell inherits an endosymbiont after cytokinesis (Fig. 2). In most algae, which contain one or a few chloroplasts per cell, the size and the number of the chloroplast are held constant by this synchronization of division3 (Fig. 2). In contrast, land plants have evolved cell and chloroplast differentiation systems in which the size and number of chloroplasts change along with their respective cellular functions4 (Fig. 2). The mechanism underlying the regulation of the chloroplast division has been poorly understood. However, recent progress on the understanding of the chloroplast division machinery5–7 (Fig. 2) has allowed us to begin to examine how the host cell regulates proliferation of the chloroplast.
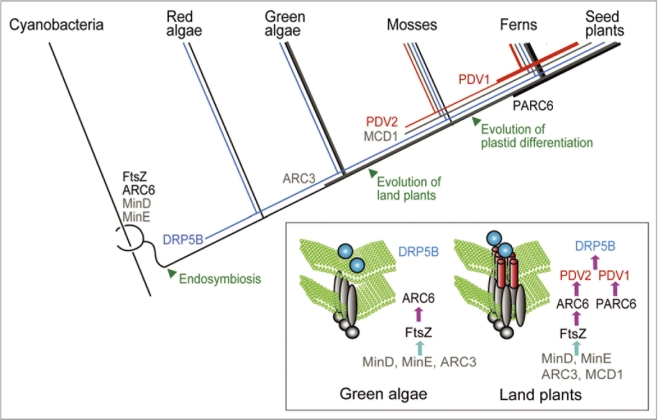
Figure 1.
Evolution of the chloroplast division machinery. The division complex of algae and land plants include FtsZ and ARC 6, both of which are descended from the cell division machinery of cyanobacteria. DR P5B was added after endosymbiosis. PDV2 was acquired by a common ancestor of land plants. PDV1 and PARC 6 emerged by gene duplication of PDV2 and ARC 6, respectively, in a common ancestor of vascular plants (Ferns and seed plants). The site of the FtsZ ring formation is determined by the MinD and MinE proteins which are descended from cyanobacteria, the ARC 3 protein which is specific to green algae and land plants, and land-plant specific protein MCD1. The inset diagram shows the pathway of chloroplast division complex assembly in green algae and land plants.
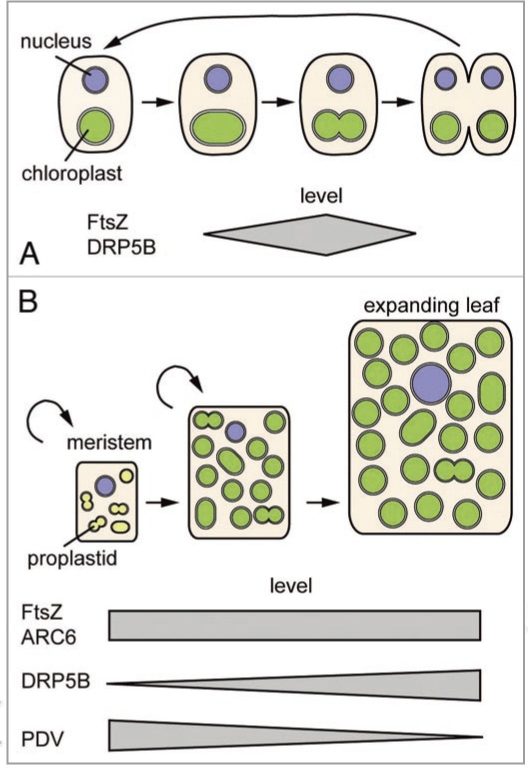
Figure 2.
Regulation of the chloroplast division in algae and land plants. (A) The chloroplast divides once per host cell cycle in algae. Components of the chloroplast division machinery are expressed in a specific phase of the host cell cycle. (B) The rate of the chloroplast division changes during cell differentiation in land plants. In angiosperms, proplastids actively divide in the meristematic cells, while the division rate slows down as leaves and chloroplasts mature. The PDV level, but not that of the other division components, decreases in parallel with the division rate.
The Mechanism of Chloroplast Division
Chloroplast division is performed by the constriction of a protein complex that forms at the division site, spanning both the inside and the outside of the two chloroplast envelope membranes5–7 (Fig. 1). Earlier observations by electron microscopy identified ring structures both the inside and the outside of the chloroplast at the division site.8 The components of these ring structures are still under investigation by recently established biochemical isolation of the structures from a unicellular red alga.9,10 However, recent molecular genetic studies have identified several proteins that localize around the rings, and have clarified that the proteins form a large complex at the division site5–7 (Fig. 2).
Consistent with the endosymbiotic origin of chloroplasts, the division complex includes nucleus-encoded FtsZ, a selfassembling tubulin-like GTPase, and ARC6, a J-domain containing protein, both of which are descended from the cell division machinery of the cyanobacterium5,6 (Fig. 1). In addition, the division complex includes a member of the eukaryotic dynamin family of self-assembling GTPase proteins (DRP5B).5,6 DRP5B is well conserved in algae and land plants and is suggested to have evolved from a dynamin-related protein that is involved in eukaryotic cytokinesis11 (Fig. 1). The chloroplast division machinery in land plants contains additional components. PDV1 and PDV2 proteins contain coiledcoil domains on the cytosolic side and are paralogous to each other.12 PDV2 is unique to land plants and PDV1 is unique to vascular plants13 (Fig. 1).
Chloroplast division in land plants is initiated by stromal FtsZ ring formation at the division site, which is stabilized by the inner envelope spanning protein ARC6.6 The site of the FtsZ ring formation is determined by the MinD and MinE proteins, which are descended from cyanobacteria, and the ARC3 protein, which contains an FtsZlike domain and a membrane occupation and recognition nexus (MORN) motif.5,6 In addition, the land-plant specific inner envelope-spanning protein MCD1 is also involved in the determination of the FtsZ ring formation.14 After the FtsZ ring forms, PDV1 and PDV2 are recruited to the division site through direct interaction between PDV2 and ARC6.6,15 In addition, a recent study showed that the recruitment of PDV1 is mediated by PARC6 (a paralog of ARC6 unique to vascular plants).16 Finally, the dynamin-related protein DRP5B is recruited by PDV1 and PDV2, and the entire division complex is involved in the fission of the chloroplast at the division site6 (Fig. 1).
This composition points out that the chloroplast division machinery is derived from both endosymbiotic (bacterial) and host (eukaryotic) membrane fission machinery, and that several new components have been added during the land plant evolution (F1|Fig. 1).
Division Synchronization of the Host Cell and the Chloroplast in Algae
Many algae (both unicellular and multicellular) have just one or only a few chloroplasts per cell, and it is obvious that a direct and precise relationship must exist between the host cell and the chloroplast division. That is, chloroplasts must divide once per cell cycle before the host cell completes cytokinesis (Fig. 2).
The identification of the chloroplast division proteins and the synchronization of the algal cell cycle with the light-dark cycle led to the finding that chloroplast division gene expression is regulated by the host cell division cycle. For example, in the unicellular red alga Cyanidioschyzon merolae, the nucleus-encoded ftsZ and the dynamin-related gene drp5B are expressed in the S-phase, and the respective proteins are degraded at the late M-phase.17–19 In the green alga Chlamydomonas reinhardtii, the nucleus-encoded ftsZ, minD and minE transcripts accumulate during cell division, when the chloroplast divides.20 In addition, in the diatom (a stramenopile) Seminavis robusta, a single cell of which contains two chloroplasts of a red algal secondary endosymbiotic origin, the nucleus-encoded ftsZ transcript accumulates during the S/G2 phase, when the chloroplasts divide.21 Overall, these reports suggest that the cell cycle-based expression of the chloroplast division genes allows the chloroplast to divide in a specific phase of the host cell division cycle.
Regulation of the Chloroplast Division Rate by PDV Proteins in Land Plants
Similar to algal chloroplasts, proplastid division keeps pace with cell division to maintain the plastid number per cell in land plant meristematic cells.3 However, the expression of the chloroplast division genes in A. thaliana is apparently constant during the cell cycle, based on the experimental results of cell cycle synchronization in a cultured cell line.22 In addition, chloroplast division occurs without cell division during leaf cell expansion.3,4 Land plant cells usually contain dozens of chloroplasts per cell and the chloroplasts divide nonsynchroously, even in the same cell (Fig. 2).
For example, in Arabidopsis thaliana, the shoot meristematic cells contain 10 to 20 proplastids.23 During leaf development, chloroplasts divide in accord with cell expansion without cell division, and the number of chloroplasts per mesophyll cell reaches more than 100 in fully expanding leaves.4 During leaf development, the rate of the chloroplast division decreases and as a result, the size of the chloroplasts increases3 (Fig. 2). In the moss Physcomitrella patens, the protonemal cell contains 40–50 chloroplasts and chloroplast division occurs non-synchronously in the same cell.24 When the formation of buds (from which gametophytes arise) is induced by cytokinin, the rate of chloroplast division increases.13 As a result, the apical cell of the bud contains chloroplasts more numerous and smaller than those in protonemal cells. Thus, land plant cells possess mechanisms to control the rate of chloroplast division upon cell differentiation which are not linked to the cell cycle.
We recently reported that the level of PDV proteins, land-plant-specific components of the division apparatus, determine the rate of chloroplast division in land plants.13 When PDV proteins are overexpressed in A. thaliana, the number of chloroplasts increases and the size is reduced compared to the wild type. The PDV level is highest in the apical meristem and young emerging leaves, but decreases during leaf development, during which the chloroplast division rate also decreases. In contrast, the DRP5B level increases, but the FtsZ and ARC6 levels remain constant, during leaf development13 (Fig. 2). Our analyses also showed that cytokinin treatment or CRF2 (CYTOKININ RESPONSE FACTOR2) overexpression increases the PDV levels, but not those of the other division components, in parallel with the increase in the chloroplast division rate.13
Similarly, overexpression of the PDV gene accelerates chloroplast division in the moss P. patens. When bud formation is induced by cytokinin, where chloroplast division is accelerated, transcription of PDV, but not those of other chloroplast division genes, is upregulated.13 Since mosses branched earliest in land plant evolution25 (Fig. 1), the results suggest that the integration of PDV proteins into the division machinery enabled land plant cells to change the chloroplast size and number in accord with the fate of cell differentiation.
Mosses contain chloroplasts throughout the life cycle, as do algae. Therefore, PDV proteins were probably acquired to modulate photosynthetic chloroplast division prior to the evolutionary emergence of the differentiation system based on the proplastids. However, mosses have PDV2 and ARC6, but not PDV1 and PARC6, which are conserved in vascular plants.13,16 Therefore, the modulation of the chloroplast division rate by PDV proteins, and the evolution of the PDV and ARC6 interactions by gene duplication and functional differentiation, appear to have been critical for the evolution of the plastid differentiation system in vascular plants.
Acknowledgements
We would like to thank Drs. Tananari Ichikawa and Minami Matsui for technical helps and critical comments on the study of PDV proteins. This research has been supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to S.M.).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10461
References
- 1.Rodriguez-Ezpeleta N, Philippe H. Plastid origin: replaying the tape. Curr Biol. 2006;16:53–56. doi: 10.1016/j.cub.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 3.Possingham JV, Lawrence ME. Controls to plastid division. International Review of Cytology. 1983;84:1–56. [Google Scholar]
- 4.Lopez-Juez E, Pyke KA. Plastids unleashed: their development and their integration in plant development. Int J Dev Biol. 2005;49:557–577. doi: 10.1387/ijdb.051997el. [DOI] [PubMed] [Google Scholar]
- 5.Maple J, Moller SG. Plastid division: evolution, mechanism and complexity. Ann Bot (Lond) 2007;99:565–579. doi: 10.1093/aob/mcl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y, Glynn JM, Olson BJ, Schmitz AJ, Osteryoung KW. Plastid division: across time and space. Curr Opin Plant Biol. 2008;11:577–584. doi: 10.1016/j.pbi.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Kuroiwa T, Misumi O, Nishida K, Yagisawa F, Yoshida Y, Fujiwara T, et al. Vesicle, mitochondrial and plastid division machineries with emphasis on dynamin and electron-dense rings. Int Rev Cell Mol Biol. 2008;271:97–152. doi: 10.1016/S1937-6448(08)01203-3. [DOI] [PubMed] [Google Scholar]
- 8.Kuroiwa T, Kuroiwa H, Sakai A, Takahashi H, Toda K, Itoh R. The division apparatus of plastids and mitochondria. Int Rev Cytol. 1998;181:1–41. doi: 10.1016/s0074-7696(08)60415-5. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida Y, Kuroiwa H, Hirooka S, Fujiwara T, Ohnuma M, Yoshida M, et al. The bacterial ZapAlike protein ZED is required for mitochondrial division. Curr Biol. 2009 doi: 10.1016/j.cub.2009.07.035. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida Y, Kuroiwa H, Misumi O, Nishida K, Yagisawa F, Fujiwara T, et al. Isolated chloroplast division machinery can actively constrict after stretching. Science. 2006;313:1435–1438. doi: 10.1126/science.1129689. [DOI] [PubMed] [Google Scholar]
- 11.Miyagishima SY, Kuwayama H, Urushihara H, Nakanishi H. Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc Natl Acad Sci USA. 2008;105:15202–15207. doi: 10.1073/pnas.0802412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyagishima SY, Froehlich JE, Osteryoung KW. PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell. 2006;18:2517–2530. doi: 10.1105/tpc.106.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okazaki K, Kabeya Y, Suzuki K, Mori T, Ichikawa T, Matsui M, et al. The PLASTID DIVISION 1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell. 2009;21:1769–1780. doi: 10.1105/tpc.109.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakanishi H, Suzuki K, Kabeya Y, Miyagishima SY. Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr Biol. 2009;19:151–156. doi: 10.1016/j.cub.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Glynn JM, Froehlich JE, Osteryoung KW. Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell. 2008;20:2460–2470. doi: 10.1105/tpc.108.061440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glynn JM, Yang Y, Vitha S, Schmitz AJ, Hemmes M, Miyagishima SY, et al. PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 2009;59:700–711. doi: 10.1111/j.1365-313X.2009.03905.x. [DOI] [PubMed] [Google Scholar]
- 17.Takahara M, Takahashi H, Matsunaga S, Miyagishima S, Takano H, Sakai A, et al. A putative mitochondrial ftsZ gene is present in the unicellular primitive red alga Cyanidioschyzon merolae. Mol Gen Genet. 2000;264:452–460. doi: 10.1007/s004380000307. [DOI] [PubMed] [Google Scholar]
- 18.Miyagishima SY, Nishida K, Mori T, Matsuzaki M, Higashiyama T, Kuroiwa H, et al. A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell. 2003;15:655–665. doi: 10.1105/tpc.009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara T, Misumi O, Tashiro K, Yoshida Y, Nishida K, Yagisawa F, et al. Periodic gene expression patterns during the highly synchronized cell nucleus and organelle division cycles in the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2009;16:59–72. doi: 10.1093/dnares/dsn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams S, Maple J, Moller SG. Functional conservation of the MIN plastid division homologues of Chlamydomonas reinhardtii. Planta. 2008;227:1199–1211. doi: 10.1007/s00425-008-0692-6. [DOI] [PubMed] [Google Scholar]
- 21.Gillard J, Devos V, Huysman MJ, De Veylder L, D’Hondt S, Martens C, et al. Physiological and transcriptomic evidence for a close coupling between chloroplast ontogeny and cell cycle progression in the pennate diatom Seminavis robusta. Plant Physiol. 2008;148:1394–1411. doi: 10.1104/pp.108.122176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menges M, Hennig L, Gruissem W, Murray JA. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol. 2003;53:423–442. doi: 10.1023/B:PLAN.0000019059.56489.ca. [DOI] [PubMed] [Google Scholar]
- 23.Segui-Simarro JM, Staehelin LA. Mitochondrial reticulation in shoot apical meristem cells of Arabidopsis provides a mechanism for homogenization of mtDNA prior to gamete formation. Plant Signal Behav. 2009;4:168–171. doi: 10.4161/psb.4.3.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strepp R, Scholz S, Kruse S, Speth V, Reski R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA. 1998;95:4368–4373. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenrick P, Crane PR. The origin and early evolution of plants on land. Nature. 1997;389:33–39. [Google Scholar]