Abstract
Serine proteases of the subtilase family are present in Archaea, Bacteria and Eukarya. Many more subtilases are found in plants as compared to other organisms, implying adaptive significance for the expansion of the subtilase gene family in plants. Structural data, however, were hitherto available only for non-plant subtilases. We recently solved the first structure of a plant subtilase, SlSBT3 from tomato (Solanum lycopersicum). SlSBT3 is a multidomain enzyme displaying a subtilisin, a Protease-Associated (PA) and a fibronectin (Fn) III-like domain. Two prominent features set SlSBT3 apart from other structurally elucidated subtilases: (i) activation by PA domain-mediated homo-dimerization and (ii) calcium-independent activity and thermostability. To address the question whether these characteristics are unique features of SlSBT3, or else, general properties of plant subtilases, homology models were calculated for representative proteases from tomato and Arabidopsis using the SlSBT3 structure as template. We found the major structural elements required for the stabilization of the subtilisin domain to be conserved among all enzymes analyzed. PA domain-mediated dimerization as an auto-regulatory mechanism of enzyme activation, on the other hand, appears to be operating in only a subset of the analyzed subtilases
Key words: Arabidopsis thaliana, enzyme structure, homology modeling, proprotein convertase, protease-associated domain, proteolysis, Solanum lycopersicum, SBT3, subtilisin, thermostability
Subtilases constitute the second largest family of serine peptidases, both in terms of number of sequences and characterized enzymes (merops.sanger.ac.uk/).1 The function of subtilases ranges from the non-selective degradation of proteins by e.g., subtilisin Carlsberg in Bacillus licheniformis, to the highly specific maturation of peptide hormones and processing of protein precursors by e.g., kexin in Saccharomyces cerevisiae and proprotein convertases in mammals. In higher plants, subtilases are represented by large gene families comprising 56 members in Arabidopsis thaliana and 63 in rice.2,3 The expansion of the subtilase family in plants as compared to animals has apparently been accompanied by the acquisition of novel physiological roles that are plant-specific. Plant subtilases were shown to be involved in stomata and seed development,4,5 in the maintenance of the shoot apical meristem and the cell wall,6,7 in the processing of peptide growth factors,8,9 and in responses to the biotic and abiotic environment.10,11 To address the question whether the adoption of specific roles in plant physiology is reflected in unique structural or biochemical features that distinguish subtilases in plants from those in other organisms, we recently characterized the subtilase SlSBT3 from tomato12 and solved its structure by X-ray crystallography.13,14
SlSBT3 is an extracellular 79 kDa glycoprotein that exhibits a remarkable level of stability at elevated temperatures and alkaline pH.12 Like most other subtilases, SlSBT3 is synthesized as a pre-pro-protein and targeted for secretion by an N-terminal signal peptide. Maturation of SlSBT3 involves cleavage of its prodomain, which is a prerequisite for passage through the secretory pathway.12 Mature SlSBT3 features a protease-associated (PA) domain as a large insertion between the His and Ser active site residues of the protease domain and a fibronectin (Fn) III-like domain as C-terminal extension.14 This domain architecture (Fig. 1A) is shared with the majority of plant subtilases (e.g., 54 of the 56 subtilases in Arabidopsis). Unlike other structurally elucidated subtilases from bacteria, fungi and animals, SlSBT3 was found to be free of Ca2+ in its native state and independent of Ca2+ with respect to activity and thermostability. The ability of SlSBT3 to form homodimers is also unique and appears to be critical for enzyme activity and stability.14 For this addendum, we calculated homology models for representative subtilases from Arabidopsis and tomato to investigate whether PA domain-mediated homo-dimerization and calcium-independent thermostability are unique features of the SlSBT3 structure, or whether they can serve as a first paradigm for the structural biology of plant subtilases in general.
Figure 1.
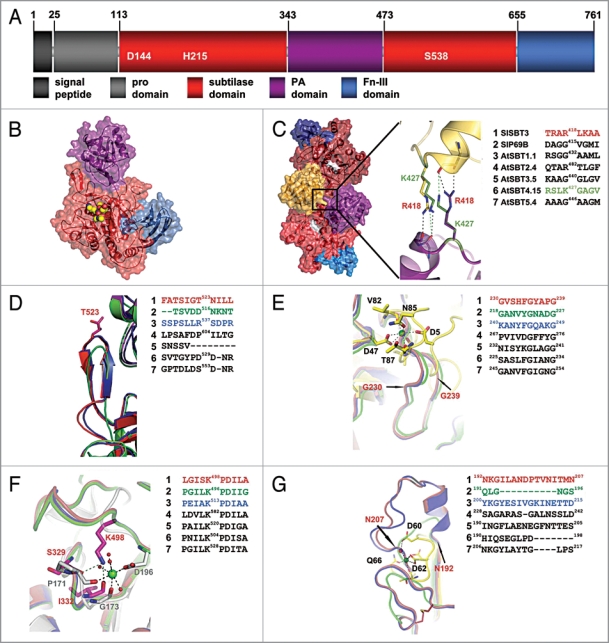
Structural comparison of plant subtilases. (A) Domain architecture of SlSBT3. In addition to the domain borders the three residues constituting the active site are displayed. (B) Structure of the SlSBT3 monomer. Color coding of the domains is like in (A). The bound chloromethylketone (cmk)-inhibitor is shown as ball model in yellow (carbon), red (oxygen) and blue (nitrogen). (C) Functional homodimer of SlSBT3. The region of the direct contact between the two PA domains (gold, purple) is highlighted. In this and all the following panels, a sequence alignment of the relevant regions in SlSBT3 and the modeled subtilases is shown on the right. The sequences highlighted in color were included in the structural alignment on the left. (D) Structure of the partially conserved β-hairpin. (E) Structure of the region corresponding to the conserved calcium-binding site 1 (Ca-1) in thermitase (yellow sticks, PDB code: 1THM). (F) Functional substitution of the conserved Ca-2 (white sticks, PDB code 1S2N) site by a lysine side chain in plant subtilases. (G) Structure of the region corresponding to the less conserved Ca-3 site in thermitase (yellow sticks, PDB code: 1THM). For details, see text.
Structural Conservation among Plant Subtilases
The 56-membered subtilase gene family in Arabidopsis comprises six subfamilies, AtSBT1 to AtSBT6. The AtSBT6 subfamily had to be excluded from our comparison because its two members are only distantly related to SlSBT3 and could not be modeled reliably on basis of the SlSBT3 structure. One representative was selected from each of the other subfamilies including AtSBT1.1 (At1g01900; involved in the maturation of phytosulfokines8), AtSBT2.4 (At1g62340; required for epidermal differentiation15), AtSBT3.5 (At1g32940; function unknown), AtSBT4.15 (At5g03620; function unknown), and AtSBT5.4 (implicated in the clavata signaling pathway for restriction of the stem cell niche in the apical meristem6). Also included was the subtilase P69 from tomato (accession number Y17276) which attracted considerable interest because of it’s role in plant pathogen interactions.10–17 The deduced amino acid sequence were used to build homology models employing MODELLER 9v7.18 Subsequently, the resulting models were compared to the structure of SlSBT3 and to each other by visual inspections with PYMOL (www.pymol.org).
We first addressed the question whether the ability to form homodimers is likely to be a common feature of plant subtilases. SlSBT3 homo-dimerizes via binding of the PA domain of one protomer to the subtilase domain of the other (Fig. 1C).14 Most of the extensive interface between the PA domain and the subtilisin domain is conserved between SlSBT3 and the modeled subtilases (data not shown). An additional prominent interaction is established in the SlSBT3 homodimer between the two PA domains involving the side chain of Arg418 of one protomer and two mainchain carbonyls of the other (Fig. 1C). This arginine is conserved in AtSBT2.4 and replaced by a functionally similar lysine residue in AtSBT4.15 (Fig. 1C). With all the specific interactions retained, homo-dimerization is likely to occur in AtSBT2.4 and 4.15. In most other plant subtilases, Arg418 is replaced by glycine or another small uncharged residue (Fig. 1C). However, homo-dimerization remains a possibility also for subtilases lacking this stabilizing interaction, since it was shown by site-directed mutagenesis that Arg418 is not absolutely required for dimer formation in SlSBT3.14
SlSBT3 possesses a potentially autoinhibitory β-hairpin which was suggested to obstruct the active site of the monomeric enzyme. Upon homo-dimerization, this β-hairpin is immobilized by intimate binding to the PA domain of the second protomer, its auto-inhibitory activity is relieved, and SlSBT3 is activated.14 While allosteric regulation is a common theme in proteases,19 a regulatory role for the PA domain has not been described in other proteases and the auto-inhibitory β-hairpin is not found in subtilases from bacteria, fungi and animals. We show here by homology modeling that the β-hairpin is present in most of the studied Arabidopsis subtilases (AtSBT1.1, AtSBT2.4, AtSBT4.15, AtSBT5.4; Fig. 1D). SlSBT3’s auto-regulatory mechanism involving PA domain-mediated homo-dimerization and immobilization of the β-hairpin, concomitant with enzyme activation, is thus likely to be widespread among plant subtilases. There are exceptions, however. In AtSBT3.5 the hairpin is missing entirely, and in tomato P69B it is replaced by a loop which is considerably shorter and lacks secondary structure elements (Fig. 1D). The regulation of proteolytic activity, if any, must be accomplished by different means in these two and other related subtilases. A different auto-regulatory mechanism has in fact been identified in SlSBT1 from tomato. This subtilase possesses an amino-terminal auto-inhibitory region of 21 amino acids, which is cleaved off in an auto-catalytic pH-dependent reaction when the enzyme reaches the acidic environment of the cell wall.20
In previously published subtilase structures, there are two highly (Fig. 1E and F) and one less (Fig. 1G) conserved calcium binding sites and the binding of calcium is an important factor contributing to enzyme stability.21 Surprisingly, no calcium ions could be identified in the structure of SlSBT3 despite the fact that the general organization of the calcium binding regions is retained. The apparent calcium independence was corroborated by the finding that thermostability and activity of SlSBT3 are not influenced by the addition of Ca2+ or chelating agents.14 Hence, thermostability in SlSBT3 must be achieved by mechanisms independent from Ca2+ binding.
In the region corresponding to the highly conserved Ca1 site (in for example thermitase), an additional, elongated loop spanning residues 230–239 (Fig. 1E) confers extra stability in SlSBT3. This loop is perfectly conserved among the seven plant subtilases analyzed in this study. The backbone geometry of the second highly conserved Ca2+ binding site (Ca2) in SlSBT3 resembles that of known Ca2+-dependent subtilases. The function of the bound calcium ion, however, is mimicked by the positively charged side chain of Lys498. This region including the stabilizing lysine residue is also highly conserved in all plant subtilases studied (Fig. 1F). The region of the third, less conserved Ca2+ site is stabilized in SlSBT3 by an elongated loop with a short helical segment (Fig. 1G). These additional structural features are also present in AtSBT1.1, AtSBT2.4 and AtSBT3.5 but not in P69B, AtSBT4.15 and AtSBT5.4, which is consistent with the lower degree of conservation of Ca3 compared to Ca1 and Ca2 in non-plant subtilases. We conclude that plant subtilases, like those in other organisms, rely on structural elements for the stabilization of the subtilisin domain. In contrast to subtilases from bacteria, fungi and animals, however, stabilization is independent of calcium binding.
Acknowledgements
Support by a research grant (SCHA591/2) from the German Research Foundation to A.S. is gratefully acknowledged.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/11069
References
- 1.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucl Acids Res. 2008;36:320–325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rautengarten C, Steinhauser D, Bussis D, Stintzi A, Schaller A, Kopka J, et al. Inferring hypotheses on functional relationships of genes: Analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput Biol. 2005;1:e40. doi: 10.1371/journal.pcbi.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tripathi L, Sowdhamini R. Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics. 2006;7:200. doi: 10.1186/1471-2164-7-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- 5.Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Bussis D, Altmann T. A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 2008;54:466–480. doi: 10.1111/j.1365-313X.2008.03437.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu J-X, Srivastava R, Howell S. Overexpression of an Arabidopsis gene encoding a subtilase (AtSBT5.4) produces a clavata-like phenotype. Planta. 2009;230:687–697. doi: 10.1007/s00425-009-0976-5. [DOI] [PubMed] [Google Scholar]
- 7.Wolf WM, Rausch T, Greiner S. The N-terminal pro region mediates retention of unprocessed type-I PME in the Golgi apparatus. Plant J. 2009;58:361–375. doi: 10.1111/j.1365-313X.2009.03784.x. [DOI] [PubMed] [Google Scholar]
- 8.Srivastava R, Liu JX, Howell SH. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 2008;56:219–227. doi: 10.1111/j.1365-313X.2008.03598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava R, Liu J-X, Guo H, Yin Y, Howell SH. Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 2009;59:930–939. doi: 10.1111/j.1365-313X.2009.03926.x. [DOI] [PubMed] [Google Scholar]
- 10.Tornero P, Conejero V, Vera P. Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J Biol Chem. 1997;272:14412–14419. doi: 10.1074/jbc.272.22.14412. [DOI] [PubMed] [Google Scholar]
- 11.Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cedzich A, Huttenlocher F, Kuhn BM, Pfannstiel J, Gabler L, Stintzi A, et al. The protease-associated (PA) domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3) J Biol Chem. 2009;284:14068–14078. doi: 10.1074/jbc.M900370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose R, Huttenlocher F, Cedzich A, Kaiser M, Schaller A, Ottmann C. Purification, crystallization and preliminary X-ray diffraction analysis of a plant subtilase. Acta Cryst F. 2009;65:522–525. doi: 10.1107/S1744309109013979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ottmann C, Rose R, Huttenlocher F, Cedzich A, Hauske P, Kaiser M, et al. Structural basis for Ca2+-independence and activation by homodimerization of tomato subtilase 3. Proc Natl Acad Sci USA. 2009;106:17223–17228. doi: 10.1073/pnas.0907587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka H, Onouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Machida C, et al. A subtilisin-like serine protease is required for epidermal surface formation in Arabidopsis embryos and juvenile plants. Development. 2001;128:4681–4689. doi: 10.1242/dev.128.23.4681. [DOI] [PubMed] [Google Scholar]
- 16.Tian MY, Huitema E, da Cunha L, Torto-Alalibo T, Kamoun S. A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem. 2004;279:26370–26377. doi: 10.1074/jbc.M400941200. [DOI] [PubMed] [Google Scholar]
- 17.Tian MY, Benedetti B, Kamoun S. A second kazallike protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 2005;138:1785–1793. doi: 10.1104/pp.105.061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eswar M, Marti-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, et al. Current Protocols in Bioinformatics. John Wiley & Sons, Inc; 2006. Comparative protein structure modeling with MODELLER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauske P, Ottmann C, Meltzer M, Ehrmann M, Kaiser M. Allosteric regulation of proteases. ChemBioChem. 2008;9:2920–2928. doi: 10.1002/cbic.200800528. [DOI] [PubMed] [Google Scholar]
- 20.Janzik I, Macheroux P, Amrhein N, Schaller A. LeSBT1, a subtilase from tomato plants. Overexpression in insect cells, purification and characterization. J Biol Chem. 2000;275:5193–5199. doi: 10.1074/jbc.275.7.5193. [DOI] [PubMed] [Google Scholar]
- 21.Alexander PA, Ruan B, Bryan PN. Cation-dependent stability of subtilisin. Biochemistry. 2001;40:10634–10639. doi: 10.1021/bi010797m. [DOI] [PubMed] [Google Scholar]